295:
decade and introduced a variety of improvements, including significant expansion of the free energy capabilities, accommodation for modern parallel and array processing hardware platforms (Cray, Star, etc.), restructuring of the code and revision control for greater maintainability, PME Ewald summations, tools for NMR refinement, and many others.
1265:
To use the AMBER force field, it is necessary to have values for the parameters of the force field (e.g. force constants, equilibrium bond lengths and angles, charges). A fairly large number of these parameter sets exist, and are described in detail in the AMBER software user manual. Each parameter
549:
294:
The next iteration of AMBER was started around 1987 by a group of developers in (and associated with) the
Kollman lab, including David Pearlman, David Case, James Caldwell, William Ross, Thomas Cheatham, Stephen DeBolt, David Ferguson, and George Seibel. This team headed development for more than a
37:, Version1: +Paul Weiner. Version 2: + U Chandra Singh; V3. + David Pearlman, James Caldwell, William Ross, Thomas Cheatham, Stephen Debolt, David Ferguson, George Seibel; Later versions: David Case, Tom Cheatham, Ken Merz, Adrian Roitberg, Carlos Simmerling, Ray Luo, Junmei Wang, Ross Walker
968:
1758:
Duan, Yong; Wu, Chun; Chowdhury, Shibasish; Lee, Mathew C.; Xiong, Guoming; Zhang, Wei; Yang, Rong; Cieplak, Piotr; et al. (2003). "A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations".
998:
Third term (summing over torsions): represents the energy for twisting a bond due to bond order (e.g., double bonds) and neighboring bonds or lone pairs of electrons. One bond may have more than one of these terms, such that the total torsional energy is expressed as a
240:
1256:
The form of the electrostatic energy used here assumes that the charges due to the protons and electrons in an atom can be represented by a single point charge (or in the case of parameter sets that employ lone pairs, a small number of point charges.)
341:
generally refers to the functional form used by the family of AMBER force fields. This form includes several parameters; each member of the family of AMBER force fields provides values for these parameters and has its own name.
356:
1604:
Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM Jr, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA (1995). "A Second
Generation Force Field for the Simulation of Proteins, Nucleic Acids, and Organic Molecules".
675:
679:
1319:. Development is conducted by a loose association of mostly academic labs. New versions are released usually in the spring of even numbered years; AMBER 10 was released in April 2008. The software is available under a
1281:
parameters are provided by parameter sets with names starting with "ff" and containing a two digit year number, for instance "ff99". As of 2018 the primary protein model used by the AMBER suit is the ff14SB force
992:
over bonds): represents the energy between covalently bonded atoms. This harmonic (ideal spring) force is a good approximation near the equilibrium bond length, but becomes increasingly poor as atoms separate.
1567:"AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules"
2402:
1251:
1424:
1413:
is a built-in nucleic acid building environment made to aid in the process of manipulating proteins and nucleic acids where an atomic level of description will aid computing.
1166:
1091:
1111:
1186:
1565:
Pearlman, David A.; Case, David A.; Caldwell, James W.; Ross, Wilson S.; Cheatham, Thomas E.; DeBolt, Steve; Ferguson, David; Seibel, George; Kollman, Peter (1995).
1895:
1131:
1044:
1024:
1303:
The AMBER software suite provides a set of programs to apply the AMBER forcefields to simulations of biomolecules. It is written in the programming languages
1842:
544:{\displaystyle V(r^{N})=\sum _{i\in {\text{bonds}}}{k_{b}}_{i}(l_{i}-l_{i}^{0})^{2}+\sum _{i\in {\text{angles}}}{k_{a}}_{i}(\theta _{i}-\theta _{i}^{0})^{2}}
1429:
1288:(GAFF) provides parameters for small organic molecules to facilitate simulations of drugs and small molecule ligands in conjunction with biomolecules.
963:{\displaystyle +\sum _{j=1}^{N-1}\sum _{i=j+1}^{N}f_{ij}{\biggl \{}\epsilon _{ij}{\biggl }+{\frac {q_{i}q_{j}}{4\pi \epsilon _{0}r_{ij}}}{\biggr \}}}
557:
157:
2224:
263:
47:
1347:(SANDER) is the central simulation program and provides facilities for energy minimizing and molecular dynamics with a wide variety of options.
2594:
2367:
291:
Subsequently, U Chandra Singh expanded AMBER as a post-doc in
Kollman's laboratory, adding molecular dynamics and free energy capabilities.
2889:
2699:
2049:
2894:
1323:
agreement, which includes full source, currently priced at US$ 500 for non-commercial and US$ 20,000 for commercial organizations.
2654:
1835:
977:, this equation defines the potential energy of the system; the force is the derivative of this potential relative to position.
2659:
2283:
2184:
995:
Second term (summing over angles): represents the energy due to the geometry of electron orbitals involved in covalent bonding.
285:
1528:"AMBER : Assisted model building with energy refinement. A general program for modeling molecules and their interactions"
2421:
2844:
2512:
2352:
2129:
1642:
Maier, James A; Martinez, Carmenza; Kasavajhala, Koushik; Wickstrom, Lauren; Hauser, Kevin E; Simmerling, Carlos (2015).
2684:
2564:
2194:
1705:
Dickson, Callum J; Madej, Benjamin D; Skjevik, Åge A; Betz, Robin M; Teigen, Knut; Gould, Ian R; Walker, Ross C (2014).
1828:
76:
2609:
1464:
1387:
numerically analyzes simulation results. AMBER includes no visualizing abilities, which is commonly performed with
209:
2884:
2834:
2538:
2204:
1890:
1444:
1401:
to give faster analysis of simulation results. Several actions have been made parallelizable with OpenMP and MPI.
1388:
1191:
2043:
1364:
1308:
323:
105:
2164:
1933:
1851:
337:
267:
1614:
315:
17:
2709:
2599:
2543:
307:
2344:
2265:
1982:
1925:
1479:
278:
that can be used both within the AMBER software suite and with many modern computational platforms.
217:
204:
141:
1619:
2788:
2624:
2218:
2159:
2068:
2037:
1449:
1047:
288:
was written by Paul Weiner as a post-doc in Peter
Kollman's laboratory, and was released in 1981.
186:
2723:
2604:
2497:
2372:
2297:
2003:
1977:
1784:
1439:
1434:
1354:
303:
299:
271:
191:
148:
1136:
1061:
1353:
is a somewhat more feature-limited reimplementation of SANDER by Bob Duke. It was designed for
1096:
2629:
2396:
2139:
2063:
2018:
1957:
1776:
1736:
1673:
1586:
1547:
1459:
311:
125:
1171:
2850:
2527:
2457:
2452:
1943:
1768:
1726:
1718:
1663:
1655:
1624:
1578:
1539:
1527:
198:
120:
1357:, and performs significantly better than SANDER when running on more than 8–16 processors.
258:) is the name of a widely-used molecular dynamics software package originally developed by
2839:
2273:
1972:
1860:
1644:"Ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB"
1046:): represents the non-bonded energy between all atom pairs, which can be decomposed into
2765:
2664:
2134:
1731:
1706:
1668:
1643:
1407:
allows implicit solvent calculations on snap shots from molecular dynamics simulations.
1116:
1051:
1029:
1009:
1000:
239:
1291:
The GLYCAM force fields have been developed by Rob Woods for simulating carbohydrates.
2878:
2533:
2306:
2245:
2232:
1869:
1582:
259:
213:
34:
2793:
2199:
2169:
1885:
1820:
1494:
1320:
1278:
1058:
The form of the van der Waals energy is calculated using the equilibrium distance (
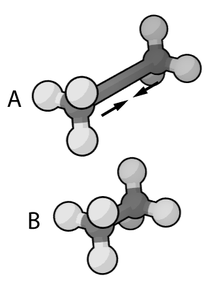
981:
1788:
2619:
2614:
1938:
298:
Currently, AMBER is maintained by an active collaboration between David Case at
275:
1566:
670:{\displaystyle +\sum _{i\in {\text{torsions}}}\sum _{n}{\frac {1}{2}}V_{i}^{n}}
2783:
2739:
2548:
2174:
2149:
1910:
1304:
42:
29:
1659:
1590:
1551:
2860:
2770:
2749:
2744:
2694:
2674:
1880:
1312:
989:
319:
168:
161:
1780:
1740:
1677:
1341:
automates the process of parameterizing small organic molecules using GAFF.
1543:
2823:
2778:
2689:
2679:
2649:
2517:
2442:
2437:
2240:
2179:
2144:
2088:
1998:
1454:
1316:
1628:
1514:
1266:
set has a name, and provides parameters for certain types of molecules.
2477:
2462:
2319:
2250:
2108:
2098:
2083:
2023:
2013:
2008:
1474:
1274:
1270:
113:
1772:
1722:
2855:
2704:
2669:
2644:
2639:
2634:
2589:
2584:
2579:
2507:
2502:
2492:
2472:
2432:
2427:
2416:
2377:
2362:
2334:
2329:
2324:
2278:
2189:
2113:
2078:
1900:
1691:
1489:
1469:
1294:
The primary force field used in the AMBER suit for lipids is lipid14.
244:
1398:
1373:
handles the extra parameters in the polarizable AMOEBA force field.
109:
2818:
2813:
2574:
2569:
2522:
2482:
2467:
2255:
2154:
2103:
1915:
1905:
238:
133:
1253:, as used e.g. in the implementation of the softcore potentials.
2487:
2447:
2382:
2314:
2093:
2073:
1815:
1484:
137:
129:
1824:
1810:
153:
243:
AMBER is used to minimize the bond stretching energy of this
266:. It has also, subsequently, come to designate a family of
2403:
List of quantum chemistry and solid-state physics software
1425:
Comparison of software for molecular mechanics modeling
1803:
1345:
Simulated
Annealing with NMR-Derived Energy Restraints
228:
24:
Assisted Model
Building with Energy Refinement (AMBER)
1194:
1174:
1139:
1119:
1099:
1064:
1032:
1012:
682:
560:
359:
1391:(VMD). Ptraj is now unsupported as of AmberTools 13.
2806:
2758:
2732:
2722:
2557:
2409:
2395:
2343:
2305:
2296:
2264:
2231:
2217:
2122:
2056:
2036:
1991:
1965:
1956:
1924:
1868:
1859:
1168:. The energy is sometimes reformulated in terms of
223:
197:
185:
177:
167:
147:
119:
101:
75:
53:
41:
28:
1245:
1180:
1160:
1125:
1105:
1085:
1038:
1018:
962:
669:
543:
1335:prepares input files for the simulation programs.
955:
888:
775:
755:
350:The functional form of the AMBER force field is
252:Assisted Model Building with Energy Refinement
82:Amber23, AmberTools23 / April 21, 2023
1836:
8:
23:
1526:Weiner, Paul K.; Kollman, Peter A. (1981).
1246:{\displaystyle r_{ij}^{0}=2^{1/6}(\sigma )}
2729:
2406:
2302:
2228:
2053:
1962:
1865:
1843:
1829:
1821:
1816:Amber on the German HPC-C5 Cluster-Systems
1711:Journal of Chemical Theory and Computation
1648:Journal of Chemical Theory and Computation
22:
1730:
1667:
1618:
1430:Comparison of force field implementations
1224:
1220:
1207:
1199:
1193:
1173:
1152:
1144:
1138:
1133:ensures that the equilibrium distance is
1118:
1098:
1077:
1069:
1063:
1031:
1011:
954:
953:
941:
931:
913:
903:
896:
887:
886:
880:
865:
855:
847:
841:
824:
809:
799:
791:
785:
774:
773:
764:
754:
753:
744:
734:
717:
701:
690:
681:
655:
642:
611:
606:
592:
586:
575:
568:
559:
535:
525:
520:
507:
494:
487:
482:
474:
467:
454:
444:
439:
426:
413:
406:
401:
393:
386:
370:
358:
310:, Ken Merz at Michigan State University,
1397:is a rewritten version of ptraj made in
2225:List of protein-ligand docking software
1507:
264:University of California, San Francisco
48:University of California, San Francisco
1707:"Lipid14: The Amber Lipid Force Field"
7:
1054:(second term of summation) energies.
1006:Fourth term (double summation over
2050:List of molecular graphics systems
1761:Journal of Computational Chemistry
1532:Journal of Computational Chemistry
1363:runs simulations on machines with
14:
980:The meanings of right hand side
2284:Molecular Operating Environment
2185:Molecular Operating Environment
1571:Computer Physics Communications
1877:Avalon Cheminformatics Toolkit
1311:, with support for most major
1240:
1234:
1050:(first term of summation) and
664:
661:
632:
617:
532:
500:
451:
419:
376:
363:
1:
1583:10.1016/0010-4655(95)00041-d
281:The original version of the
2890:Molecular dynamics software
1515:Amber 2023 Reference Manual
59:; 43 years ago
2911:
1811:AMBER mailing list archive
1465:BOSS (molecular mechanics)
1161:{\displaystyle r_{ij}^{0}}
1086:{\displaystyle r_{ij}^{0}}
15:
1891:Chemistry Development Kit
1445:Molecular design software
1389:Visual Molecular Dynamics
1365:graphics processing units
1286:General AMBER force field
1106:{\displaystyle \epsilon }
97:
71:
2895:Force fields (chemistry)
1692:"The Amber Force Fields"
1660:10.1021/acs.jctc.5b00255
1381:calculates normal modes.
324:University of Pittsburgh
1852:Computational chemistry
1181:{\displaystyle \sigma }
84:; 16 months ago
1315:operating systems and
1247:
1182:
1162:
1127:
1107:
1087:
1040:
1020:
971:
964:
739:
712:
671:
545:
316:Stony Brook University
302:, Tom Cheatham at the
248:
18:Amber (disambiguation)
1544:10.1002/jcc.540020311
1248:
1183:
1163:
1128:
1108:
1088:
1041:
1021:
965:
713:
686:
672:
553:
546:
322:, and Junmei Wang at
308:University of Florida
306:, Adrian Roitberg at
242:
1480:Reverse Pharmacology
1192:
1172:
1137:
1117:
1097:
1062:
1030:
1010:
680:
558:
357:
16:For other uses, see
2789:JME Molecule Editor
2038:Molecular modelling
1629:10.1021/ja00124a002
1450:Molecular mechanics
1212:
1157:
1082:
860:
804:
616:
530:
449:
25:
2724:Skeletal structure
2298:Molecular dynamics
2004:Chemical WorkBench
1440:Molecular geometry
1435:Molecular dynamics
1355:parallel computing
1243:
1195:
1178:
1158:
1140:
1123:
1103:
1093:) and well depth (
1083:
1065:
1036:
1016:
960:
843:
787:
667:
602:
591:
581:
541:
516:
480:
435:
399:
304:University of Utah
300:Rutgers University
272:molecular dynamics
249:
192:Molecular dynamics
30:Original author(s)
2872:
2871:
2802:
2801:
2718:
2717:
2397:Quantum chemistry
2391:
2390:
2292:
2291:
2219:Molecular docking
2213:
2212:
2140:Atomistix ToolKit
2064:Ascalaph Designer
2032:
2031:
1958:Chemical kinetics
1952:
1951:
1773:10.1002/jcc.10349
1767:(16): 1999–2012.
1723:10.1021/ct4010307
1613:(19): 5179–5197.
1460:Ascalaph Designer
1126:{\displaystyle 2}
1113:). The factor of
1039:{\displaystyle j}
1019:{\displaystyle i}
973:Despite the term
951:
874:
818:
600:
582:
578:
564:
477:
463:
396:
382:
312:Carlos Simmerling
237:
236:
2902:
2885:Fortran software
2851:Materials Studio
2730:
2528:Quantum ESPRESSO
2407:
2303:
2229:
2054:
1963:
1944:Discovery Studio
1866:
1845:
1838:
1831:
1822:
1807:
1806:
1804:Official website
1792:
1745:
1744:
1734:
1702:
1696:
1695:
1688:
1682:
1681:
1671:
1654:(8): 3696–3713.
1639:
1633:
1632:
1622:
1607:J. Am. Chem. Soc
1601:
1595:
1594:
1562:
1556:
1555:
1523:
1517:
1512:
1252:
1250:
1249:
1244:
1233:
1232:
1228:
1211:
1206:
1187:
1185:
1184:
1179:
1167:
1165:
1164:
1159:
1156:
1151:
1132:
1130:
1129:
1124:
1112:
1110:
1109:
1104:
1092:
1090:
1089:
1084:
1081:
1076:
1045:
1043:
1042:
1037:
1025:
1023:
1022:
1017:
969:
967:
966:
961:
959:
958:
952:
950:
949:
948:
936:
935:
919:
918:
917:
908:
907:
897:
892:
891:
885:
884:
879:
875:
873:
872:
859:
854:
842:
829:
828:
823:
819:
817:
816:
803:
798:
786:
779:
778:
772:
771:
759:
758:
752:
751:
738:
733:
711:
700:
676:
674:
673:
668:
660:
659:
647:
646:
615:
610:
601:
593:
590:
580:
579:
576:
550:
548:
547:
542:
540:
539:
529:
524:
512:
511:
499:
498:
493:
492:
491:
479:
478:
475:
459:
458:
448:
443:
431:
430:
418:
417:
412:
411:
410:
398:
397:
394:
375:
374:
286:software package
262:'s group at the
233:
230:
121:Operating system
92:
90:
85:
67:
65:
60:
26:
2910:
2909:
2905:
2904:
2903:
2901:
2900:
2899:
2875:
2874:
2873:
2868:
2840:CrystalExplorer
2798:
2754:
2714:
2553:
2405:
2387:
2339:
2288:
2260:
2227:
2209:
2118:
2052:
2042:
2040:
2028:
1987:
1948:
1920:
1861:Cheminformatics
1855:
1849:
1802:
1801:
1798:
1757:
1754:
1752:Related reading
1749:
1748:
1704:
1703:
1699:
1690:
1689:
1685:
1641:
1640:
1636:
1620:10.1.1.323.4450
1603:
1602:
1598:
1564:
1563:
1559:
1525:
1524:
1520:
1513:
1509:
1504:
1499:
1420:
1329:
1301:
1263:
1216:
1190:
1189:
1170:
1169:
1135:
1134:
1115:
1114:
1095:
1094:
1060:
1059:
1028:
1027:
1008:
1007:
937:
927:
920:
909:
899:
898:
861:
837:
836:
805:
781:
780:
760:
740:
678:
677:
651:
638:
556:
555:
531:
503:
483:
481:
450:
422:
402:
400:
366:
355:
354:
348:
346:Functional form
332:
227:
207:
93:
88:
86:
83:
63:
61:
58:
54:Initial release
21:
12:
11:
5:
2908:
2906:
2898:
2897:
2892:
2887:
2877:
2876:
2870:
2869:
2867:
2866:
2863:
2858:
2853:
2848:
2842:
2837:
2832:
2829:
2826:
2821:
2816:
2810:
2808:
2804:
2803:
2800:
2799:
2797:
2796:
2791:
2786:
2781:
2776:
2773:
2768:
2766:ACD/ChemSketch
2762:
2760:
2756:
2755:
2753:
2752:
2747:
2742:
2736:
2734:
2727:
2720:
2719:
2716:
2715:
2713:
2712:
2707:
2702:
2697:
2692:
2687:
2682:
2677:
2672:
2667:
2662:
2657:
2652:
2647:
2642:
2637:
2632:
2627:
2622:
2617:
2612:
2607:
2602:
2597:
2592:
2587:
2582:
2577:
2572:
2567:
2561:
2559:
2555:
2554:
2552:
2551:
2546:
2541:
2536:
2531:
2525:
2520:
2515:
2510:
2505:
2500:
2495:
2490:
2485:
2480:
2475:
2470:
2465:
2460:
2455:
2450:
2445:
2440:
2435:
2430:
2425:
2419:
2413:
2411:
2401:
2399:
2393:
2392:
2389:
2388:
2386:
2385:
2380:
2375:
2370:
2365:
2360:
2355:
2349:
2347:
2341:
2340:
2338:
2337:
2332:
2327:
2322:
2317:
2311:
2309:
2300:
2294:
2293:
2290:
2289:
2287:
2286:
2281:
2276:
2270:
2268:
2262:
2261:
2259:
2258:
2253:
2248:
2243:
2237:
2235:
2223:
2221:
2215:
2214:
2211:
2210:
2208:
2207:
2202:
2197:
2192:
2187:
2182:
2177:
2172:
2167:
2162:
2157:
2152:
2147:
2142:
2137:
2135:ACD/ChemSketch
2132:
2126:
2124:
2120:
2119:
2117:
2116:
2111:
2106:
2101:
2096:
2091:
2086:
2081:
2076:
2071:
2066:
2060:
2058:
2048:
2046:
2034:
2033:
2030:
2029:
2027:
2026:
2021:
2016:
2011:
2006:
2001:
1995:
1993:
1989:
1988:
1986:
1985:
1980:
1975:
1969:
1967:
1960:
1954:
1953:
1950:
1949:
1947:
1946:
1941:
1936:
1930:
1928:
1922:
1921:
1919:
1918:
1913:
1908:
1903:
1898:
1893:
1888:
1883:
1878:
1874:
1872:
1863:
1857:
1856:
1850:
1848:
1847:
1840:
1833:
1825:
1819:
1818:
1813:
1808:
1797:
1796:External links
1794:
1753:
1750:
1747:
1746:
1717:(2): 865–879.
1697:
1683:
1634:
1596:
1557:
1538:(3): 287–303.
1518:
1506:
1505:
1503:
1500:
1498:
1497:
1492:
1487:
1482:
1477:
1472:
1467:
1462:
1457:
1452:
1447:
1442:
1437:
1432:
1427:
1421:
1419:
1416:
1415:
1414:
1408:
1402:
1392:
1382:
1376:
1375:
1374:
1368:
1348:
1342:
1336:
1328:
1325:
1300:
1297:
1296:
1295:
1292:
1289:
1283:
1262:
1261:Parameter sets
1259:
1242:
1239:
1236:
1231:
1227:
1223:
1219:
1215:
1210:
1205:
1202:
1198:
1177:
1155:
1150:
1147:
1143:
1122:
1102:
1080:
1075:
1072:
1068:
1056:
1055:
1035:
1015:
1004:
1001:Fourier series
996:
993:
957:
947:
944:
940:
934:
930:
926:
923:
916:
912:
906:
902:
895:
890:
883:
878:
871:
868:
864:
858:
853:
850:
846:
840:
835:
832:
827:
822:
815:
812:
808:
802:
797:
794:
790:
784:
777:
770:
767:
763:
757:
750:
747:
743:
737:
732:
729:
726:
723:
720:
716:
710:
707:
704:
699:
696:
693:
689:
685:
666:
663:
658:
654:
650:
645:
641:
637:
634:
631:
628:
625:
622:
619:
614:
609:
605:
599:
596:
589:
585:
574:
571:
567:
563:
552:
551:
538:
534:
528:
523:
519:
515:
510:
506:
502:
497:
490:
486:
473:
470:
466:
462:
457:
453:
447:
442:
438:
434:
429:
425:
421:
416:
409:
405:
392:
389:
385:
381:
378:
373:
369:
365:
362:
347:
344:
331:
328:
235:
234:
225:
221:
220:
201:
195:
194:
189:
183:
182:
179:
175:
174:
171:
165:
164:
151:
145:
144:
123:
117:
116:
103:
99:
98:
95:
94:
81:
79:
77:Stable release
73:
72:
69:
68:
55:
51:
50:
45:
39:
38:
32:
13:
10:
9:
6:
4:
3:
2:
2907:
2896:
2893:
2891:
2888:
2886:
2883:
2882:
2880:
2864:
2862:
2859:
2857:
2854:
2852:
2849:
2847:(ICM-Browser)
2846:
2843:
2841:
2838:
2836:
2833:
2830:
2827:
2825:
2822:
2820:
2817:
2815:
2812:
2811:
2809:
2805:
2795:
2792:
2790:
2787:
2785:
2782:
2780:
2777:
2774:
2772:
2769:
2767:
2764:
2763:
2761:
2757:
2751:
2748:
2746:
2743:
2741:
2738:
2737:
2735:
2733:Free software
2731:
2728:
2725:
2721:
2711:
2708:
2706:
2703:
2701:
2698:
2696:
2693:
2691:
2688:
2686:
2683:
2681:
2678:
2676:
2673:
2671:
2668:
2666:
2663:
2661:
2658:
2656:
2653:
2651:
2648:
2646:
2643:
2641:
2638:
2636:
2633:
2631:
2628:
2626:
2623:
2621:
2618:
2616:
2613:
2611:
2608:
2606:
2603:
2601:
2598:
2596:
2593:
2591:
2588:
2586:
2583:
2581:
2578:
2576:
2573:
2571:
2568:
2566:
2563:
2562:
2560:
2556:
2550:
2547:
2545:
2542:
2540:
2537:
2535:
2532:
2529:
2526:
2524:
2521:
2519:
2516:
2514:
2511:
2509:
2506:
2504:
2501:
2499:
2496:
2494:
2491:
2489:
2486:
2484:
2481:
2479:
2476:
2474:
2471:
2469:
2466:
2464:
2461:
2459:
2456:
2454:
2451:
2449:
2446:
2444:
2441:
2439:
2436:
2434:
2431:
2429:
2426:
2423:
2420:
2418:
2415:
2414:
2412:
2410:Free software
2408:
2404:
2400:
2398:
2394:
2384:
2381:
2379:
2376:
2374:
2371:
2369:
2366:
2364:
2361:
2359:
2356:
2354:
2351:
2350:
2348:
2346:
2342:
2336:
2333:
2331:
2328:
2326:
2323:
2321:
2318:
2316:
2313:
2312:
2310:
2308:
2307:Free software
2304:
2301:
2299:
2295:
2285:
2282:
2280:
2277:
2275:
2272:
2271:
2269:
2267:
2263:
2257:
2254:
2252:
2249:
2247:
2246:AutoDock Vina
2244:
2242:
2239:
2238:
2236:
2234:
2233:Free software
2230:
2226:
2222:
2220:
2216:
2206:
2203:
2201:
2198:
2196:
2193:
2191:
2188:
2186:
2183:
2181:
2178:
2176:
2173:
2171:
2168:
2166:
2163:
2161:
2158:
2156:
2153:
2151:
2148:
2146:
2143:
2141:
2138:
2136:
2133:
2131:
2128:
2127:
2125:
2121:
2115:
2112:
2110:
2107:
2105:
2102:
2100:
2097:
2095:
2092:
2090:
2087:
2085:
2082:
2080:
2077:
2075:
2072:
2070:
2067:
2065:
2062:
2061:
2059:
2057:Free software
2055:
2051:
2047:
2045:
2044:visualization
2039:
2035:
2025:
2022:
2020:
2017:
2015:
2012:
2010:
2007:
2005:
2002:
2000:
1997:
1996:
1994:
1990:
1984:
1981:
1979:
1976:
1974:
1971:
1970:
1968:
1966:Free software
1964:
1961:
1959:
1955:
1945:
1942:
1940:
1937:
1935:
1932:
1931:
1929:
1927:
1923:
1917:
1914:
1912:
1909:
1907:
1904:
1902:
1899:
1897:
1894:
1892:
1889:
1887:
1884:
1882:
1879:
1876:
1875:
1873:
1871:
1870:Free software
1867:
1864:
1862:
1858:
1853:
1846:
1841:
1839:
1834:
1832:
1827:
1826:
1823:
1817:
1814:
1812:
1809:
1805:
1800:
1799:
1795:
1793:
1790:
1786:
1782:
1778:
1774:
1770:
1766:
1762:
1751:
1742:
1738:
1733:
1728:
1724:
1720:
1716:
1712:
1708:
1701:
1698:
1693:
1687:
1684:
1679:
1675:
1670:
1665:
1661:
1657:
1653:
1649:
1645:
1638:
1635:
1630:
1626:
1621:
1616:
1612:
1608:
1600:
1597:
1592:
1588:
1584:
1580:
1577:(1–3): 1–41.
1576:
1572:
1568:
1561:
1558:
1553:
1549:
1545:
1541:
1537:
1533:
1529:
1522:
1519:
1516:
1511:
1508:
1501:
1496:
1493:
1491:
1488:
1486:
1483:
1481:
1478:
1476:
1473:
1471:
1468:
1466:
1463:
1461:
1458:
1456:
1453:
1451:
1448:
1446:
1443:
1441:
1438:
1436:
1433:
1431:
1428:
1426:
1423:
1422:
1417:
1412:
1409:
1406:
1403:
1400:
1396:
1393:
1390:
1386:
1383:
1380:
1377:
1372:
1369:
1366:
1362:
1359:
1358:
1356:
1352:
1349:
1346:
1343:
1340:
1337:
1334:
1331:
1330:
1326:
1324:
1322:
1318:
1314:
1310:
1306:
1298:
1293:
1290:
1287:
1284:
1280:
1276:
1272:
1269:
1268:
1267:
1260:
1258:
1254:
1237:
1229:
1225:
1221:
1217:
1213:
1208:
1203:
1200:
1196:
1175:
1153:
1148:
1145:
1141:
1120:
1100:
1078:
1073:
1070:
1066:
1053:
1052:electrostatic
1049:
1048:van der Waals
1033:
1013:
1005:
1002:
997:
994:
991:
987:
986:
985:
983:
978:
976:
970:
945:
942:
938:
932:
928:
924:
921:
914:
910:
904:
900:
893:
881:
876:
869:
866:
862:
856:
851:
848:
844:
838:
833:
830:
825:
820:
813:
810:
806:
800:
795:
792:
788:
782:
768:
765:
761:
748:
745:
741:
735:
730:
727:
724:
721:
718:
714:
708:
705:
702:
697:
694:
691:
687:
683:
656:
652:
648:
643:
639:
635:
629:
626:
623:
620:
612:
607:
603:
597:
594:
587:
583:
572:
569:
565:
561:
536:
526:
521:
517:
513:
508:
504:
495:
488:
484:
471:
468:
464:
460:
455:
445:
440:
436:
432:
427:
423:
414:
407:
403:
390:
387:
383:
379:
371:
367:
360:
353:
352:
351:
345:
343:
340:
339:
329:
327:
325:
321:
318:, Ray Luo at
317:
313:
309:
305:
301:
296:
292:
289:
287:
284:
279:
277:
273:
269:
265:
261:
260:Peter Kollman
257:
253:
246:
241:
232:
226:
222:
219:
215:
214:public domain
211:
206:
202:
200:
196:
193:
190:
188:
184:
180:
176:
172:
170:
166:
163:
159:
155:
152:
150:
146:
143:
139:
135:
131:
127:
124:
122:
118:
115:
111:
107:
104:
100:
96:
80:
78:
74:
70:
56:
52:
49:
46:
44:
40:
36:
35:Peter Kollman
33:
31:
27:
19:
2794:MarvinSketch
2357:
2200:UCSF Chimera
2170:MarvinSketch
1886:Blue Obelisk
1764:
1760:
1755:
1714:
1710:
1700:
1686:
1651:
1647:
1637:
1610:
1606:
1599:
1574:
1570:
1560:
1535:
1531:
1521:
1510:
1495:Folding@home
1410:
1404:
1394:
1384:
1378:
1371:pmemd.amoeba
1370:
1360:
1350:
1344:
1338:
1332:
1321:site license
1302:
1285:
1279:nucleic acid
1264:
1255:
1057:
988:First term (
979:
974:
972:
554:
349:
335:
333:
297:
293:
290:
282:
280:
276:biomolecules
268:force fields
255:
251:
250:
208:AmberTools:
178:Available in
43:Developer(s)
2771:BIOVIA Draw
2759:Proprietary
2620:GAMESS (US)
2615:GAMESS (UK)
2558:Proprietary
2345:Proprietary
2266:Proprietary
2123:Proprietary
1992:Proprietary
1939:Chemicalize
1926:Proprietary
1339:Antechamber
975:force field
338:force field
330:Force field
218:open-source
205:Proprietary
158:Nvidia GPUs
2879:Categories
2784:ChemWindow
2775:ChemDoodle
2740:JChemPaint
2549:YAMBO code
2503:OpenMolcas
2175:MarvinView
2150:ChemWindow
1911:Open Babel
1502:References
1361:pmemd.cuda
1305:Fortran 90
102:Written in
89:2023-04-21
2861:OpenChrom
2750:XDrawChem
2745:Molsketch
2695:TURBOMOLE
2675:Quantemol
1881:Bioclipse
1615:CiteSeerX
1591:0010-4655
1552:0192-8651
1317:compilers
1313:Unix-like
1238:σ
1176:σ
1101:ϵ
929:ϵ
925:π
831:−
762:ϵ
715:∑
706:−
688:∑
653:γ
649:−
640:ω
630:
584:∑
573:∈
566:∑
518:θ
514:−
505:θ
472:∈
465:∑
433:−
391:∈
384:∑
334:The term
320:UC Irvine
247:molecule.
162:Blue Gene
2824:EXC code
2779:ChemDraw
2690:TeraChem
2680:Scigress
2650:OpenAtom
2625:Gaussian
2518:PyQuante
2443:CONQUEST
2438:COLUMBUS
2241:AutoDock
2180:MODELLER
2160:Gaussian
2145:ChemDraw
2089:Ghemical
2069:Avogadro
1999:Autochem
1854:software
1781:14531054
1741:24803855
1678:26574453
1455:MDynaMix
1418:See also
1327:Programs
1299:Software
1188:, where
577:torsions
216:, other
149:Platform
2865:SASHIMI
2835:Mercury
2726:drawing
2685:Spartan
2610:Firefly
2605:CRYSTAL
2530:(PWscf)
2498:Octopus
2478:MADNESS
2463:DP code
2424:(CFOUR)
2373:Desmond
2353:Abalone
2320:GROMACS
2251:FlexAID
2195:Spartan
2165:Maestro
2130:Abalone
2109:QuteMol
2099:Molekel
2084:Gabedit
2024:Khimera
2014:COSILAB
2009:CHEMKIN
1978:Cantera
1732:3985482
1669:4821407
1475:GROMACS
1405:MM-PBSA
1395:cpptraj
1367:(GPUs).
1275:protein
1271:Peptide
990:summing
229:ambermd
224:Website
203:Amber:
199:License
181:English
126:Windows
114:Fortran
87: (
62: (
2856:Molden
2807:Others
2705:WIEN2k
2670:Q-Chem
2645:ONETEP
2640:MOLPRO
2635:MOLCAS
2630:Jaguar
2600:CRUNCH
2590:CASTEP
2585:CASINO
2580:CADPAC
2544:VB2000
2539:SIESTA
2508:PARSEC
2493:NWChem
2473:FreeON
2453:Dalton
2433:BigDFT
2428:AIMAll
2417:ABINIT
2378:GROMOS
2363:CHARMM
2335:PLUMED
2330:OpenMM
2325:LAMMPS
2279:LeDock
2190:SAMSON
2114:RasMol
2079:Biskit
2019:DelPhi
1934:Canvas
1901:JOELib
1789:283317
1787:
1779:
1739:
1729:
1676:
1666:
1617:
1589:
1550:
1490:Yasara
1470:CHARMM
1282:field.
1277:, and
476:angles
336:AMBER
245:ethane
173:Varies
2819:Eulim
2814:Aqion
2660:PLATO
2575:DMol3
2570:AMPAC
2523:PySCF
2483:MOPAC
2468:FLEUR
2458:DIRAC
2358:AMBER
2274:Glide
2256:rDock
2155:EzMol
2104:PyMOL
1916:RDKit
1906:OELib
1785:S2CID
1385:ptraj
1379:nmode
1351:pmemd
984:are:
982:terms
395:bonds
283:AMBER
256:AMBER
134:Linux
2831:GSim
2828:GenX
2710:XMVB
2700:VASP
2655:ORCA
2595:CPMD
2488:MPQC
2448:CP2K
2422:ACES
2383:NAMD
2368:CPMD
2315:CP2K
2094:Jmol
2074:BALL
2041:and
1973:APBS
1896:ECCE
1777:PMID
1737:PMID
1674:PMID
1587:ISSN
1548:ISSN
1485:OPLS
1333:LEaP
1307:and
1026:and
270:for
231:.org
187:Type
169:Size
138:Unix
130:OS X
64:1981
57:1981
2845:ICM
2665:PQS
2565:ADF
2534:RMG
2513:PSI
2205:VMD
1983:KPP
1769:doi
1756:1.
1727:PMC
1719:doi
1664:PMC
1656:doi
1625:doi
1611:117
1579:doi
1540:doi
1411:NAB
1399:C++
627:cos
314:at
274:of
210:GPL
154:x86
142:CNK
110:C++
2881::
1783:.
1775:.
1765:24
1763:.
1735:.
1725:.
1715:10
1713:.
1709:.
1672:.
1662:.
1652:11
1650:.
1646:.
1623:.
1609:.
1585:.
1575:91
1573:.
1569:.
1546:.
1534:.
1530:.
1273:,
826:12
326:.
212:,
160:,
156:,
140:,
136:,
132:,
128:,
112:,
108:,
1844:e
1837:t
1830:v
1791:.
1771::
1743:.
1721::
1694:.
1680:.
1658::
1631:.
1627::
1593:.
1581::
1554:.
1542::
1536:2
1309:C
1241:)
1235:(
1230:6
1226:/
1222:1
1218:2
1214:=
1209:0
1204:j
1201:i
1197:r
1154:0
1149:j
1146:i
1142:r
1121:2
1079:0
1074:j
1071:i
1067:r
1034:j
1014:i
1003:.
956:}
946:j
943:i
939:r
933:0
922:4
915:j
911:q
905:i
901:q
894:+
889:]
882:6
877:)
870:j
867:i
863:r
857:0
852:j
849:i
845:r
839:(
834:2
821:)
814:j
811:i
807:r
801:0
796:j
793:i
789:r
783:(
776:[
769:j
766:i
756:{
749:j
746:i
742:f
736:N
731:1
728:+
725:j
722:=
719:i
709:1
703:N
698:1
695:=
692:j
684:+
665:]
662:)
657:i
644:i
636:n
633:(
624:+
621:1
618:[
613:n
608:i
604:V
598:2
595:1
588:n
570:i
562:+
537:2
533:)
527:0
522:i
509:i
501:(
496:i
489:a
485:k
469:i
461:+
456:2
452:)
446:0
441:i
437:l
428:i
424:l
420:(
415:i
408:b
404:k
388:i
380:=
377:)
372:N
368:r
364:(
361:V
254:(
106:C
91:)
66:)
20:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.