302:
279:
176:
201:
2148:
553:
308:
207:
29:
2934:
1157:
ClpP protease is a major contributor for mitochondrial protein quality control system and removing damaged or misfolded proteins in mitochondrial matrix. Defects in mitochondrial Clp proteases have been associated with the progression of neurodegenerative diseases while upregulation of ClpP proteases
1064:
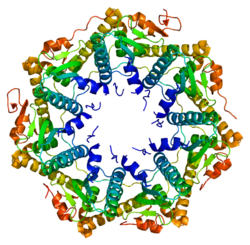
and chloroplasts of eukaryotic cells. The ClpP monomer is folded into three subdomains: the "handle", the globular "head", and the N-terminal region. By itself, ClpP can assemble into a tetradecamer complex (14-members) and form a closed proteolytic chamber. A fully assembled Clp protease complex has
1169:
Intracellular proteases have a role in bacterial virulence. Deletion of ClpP causes growth inhibition or loss of virulence in many bacterial species which makes them a good target for developing new antimicrobial agents. Currently there are no approved antimicrobial agents that target bacterial
1161:
Recessive CLPP mutations were recently observed in the human
Perrault variant associating with ovarian failure and sensorineural hearing loss, in parallel with growth retardation. The clinical phenotype was accompanied by the accumulation of ClpP associating partner chaperon ClpX, mtRNA, and
1124:
ClpP1 is expressed constitutively throughout growth whereas ClpP2 expression is induced 10-fold in stationary phase. Quorum-sensing transcription factor LasR activates expression of ClpP2 in stationary phase. ClpP1 and ClpP2 have differential cleavage specificities which contributes to total
1106:. These ClpX chaperons recognize, unfold and transfer protein substrates to proteolytic core formed by ClpP tetradecamer. The proteolytic sites of ClpP subunits contain hydrophobic grooves which recruit substrate and host the catalytic triad Asp-His-Ser. In several bacteria, such as
55:
1148:
The protein encoded by this gene belongs to the peptidase family S14 and hydrolyzes proteins into small peptides in the presence of ATP and magnesium. The protein is transported into mitochondrial matrix and is associated with the inner mitochondrial membrane.
1116:
are digested by Clp proteases. Proteases target damaged or misfolded proteins, transcription factors and signaling proteins in bacteria to coordinate complex cell responses and thus they have robust importance for the physiology and virulence of bacteria.
1069:
or ClpY). ClpXP is presented in almost all bacteria while ClpA is found in the Gram-negative bacteria, ClpC in Gram-Positive bacteria and cyanobacteria. ClpAP, ClpXP and ClpYQ coexist in E. Coli while only ClpXP complex in present in humans.
1165:
ClpP has been shown to be over-expressed in the tumour cells of a subset of cancer patients. This can be exploited by therapeutic agents, including by the hyperactivation of ClpP to cause selective cancer cell lethality.
1101:
In bacteria, it was shown that ClpP is able to cleave full-length proteins without being associated with ClpA but the degradation is at a much slower rate. Fully functional Clp protease requires the participation of
1065:
a barrel-shaped structure in which two stacked ring of proteolytic subunits (ClpP or ClpQ) are either sandwiched between two rings or single-caped by one ring of ATPase-active chaperon subunits (ClpA, ClpC, ClpE,
2005:
de
Sagarra MR, Mayo I, Marco S, Rodríguez-Vilariño S, Oliva J, Carrascosa JL, Casta ñ JG (October 1999). "Mitochondrial localization and oligomeric structure of HClpP, the human homologue of E. coli ClpP".
2147:
2588:
2448:
315:
214:
2188:
1093:, which in complex with ClpX or ClpA form functional proteases. PaClpP2 is not able to form an active peptidase on its own but it needs PaClpP1 to be active.
1162:
inflammatory factors. The disease pathological cause probably involves deficient clearance of mitochondrial components and inflammatory tissue destruction.
1742:"Loss of mitochondrial peptidase Clpp leads to infertility, hearing loss plus growth retardation via accumulation of CLPX, mtDNA and inflammatory factors"
771:
752:
137:
2964:
2037:"A human homologue of Escherichia coli ClpP caseinolytic protease: recombinant expression, intracellular processing and subcellular localization"
1412:"A human homologue of Escherichia coli ClpP caseinolytic protease: recombinant expression, intracellular processing and subcellular localization"
2954:
2127:
1716:
1285:
1267:
301:
2181:
2087:
978:
278:
2653:
1077:, which has two distinct ClpP isoforms ClpP1 and ClpP2. These isoforms have differences in assembly and functional characteristics.
971:
1455:
Hamon MP, Bulteau AL, Friguet B (September 2015). "Mitochondrial proteases and protein quality control in ageing and longevity".
1355:
1650:
Hall BM, Breidenstein EB, de la Fuente-Núñez C, Reffuveille F, Mawla GD, Hancock RE, Baker TA (February 2017). O'Toole G (ed.).
1254:
1233:
2174:
200:
175:
2809:
1250:
52:
2093:
1229:
2408:
117:
2166:
1603:"The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system"
314:
213:
2924:
2794:
2910:
2897:
2884:
2871:
2858:
2845:
2832:
2323:
2308:
2208:
2120:
307:
206:
2804:
2758:
2701:
2205:
816:
125:
1699:
Luce, K; Weil, AC; Osiewacz, HD (2010). "Mitochondrial
Protein Quality Control Systems in Aging and Disease".
2706:
2598:
2563:
2558:
797:
2156:: Crystallography and mutagenesis point to an essential role for the N-terminus of human mitochondrial ClpP
1939:"Crystallography and mutagenesis point to an essential role for the N-terminus of human mitochondrial ClpP"
1904:"Human mitochondrial ClpP is a stable heptamer that assembles into a tetradecamer in the presence of ClpX"
1205:
1113:
1652:"Two Isoforms of Clp Peptidase in Pseudomonas aeruginosa Control Distinct Aspects of Cellular Physiology"
2727:
2646:
2553:
2403:
189:
2799:
1740:
Gispert S, Parganlija D, Klinkenberg M, Dröse S, Wittig I, Mittelbronn M, et al. (December 2013).
1305:"Human ClpP protease: cDNA sequence, tissue-specific expression and chromosomal assignment of the gene"
2543:
2113:
1356:"Entrez Gene: CLPP ClpP caseinolytic peptidase, ATP-dependent, proteolytic subunit homolog (E. coli)"
104:
2763:
2443:
2433:
2250:
2245:
2099:
1190:
2969:
2959:
2696:
1879:
1583:
1480:
1334:
149:
954:
933:
907:
886:
1174:. Deletion of PaClpP2 or mutation of PaClpP2 active site notably decrease biofilm thickness of
2425:
2062:
2023:
1993:
1958:
1925:
1871:
1863:
1822:
1773:
1722:
1712:
1681:
1632:
1575:
1534:
1472:
1437:
1392:
1326:
1195:
1045:
97:
45:
1503:
Mawla GD, Hall BM, Cárcamo-Oyarce G, Grant RA, Zhang JJ, Kardon JR, et al. (June 2021).
2742:
2737:
2711:
2639:
2217:
2052:
2044:
2015:
1983:
1972:"Functional proteolytic complexes of the human mitochondrial ATP-dependent protease, hClpXP"
1950:
1915:
1853:
1812:
1804:
1763:
1753:
1704:
1671:
1663:
1622:
1614:
1565:
1524:
1516:
1464:
1427:
1419:
1382:
1316:
1108:
394:
325:
269:
224:
2789:
2773:
2686:
2458:
2453:
2225:
2201:
2035:
Corydon TJ, Bross P, Holst HU, Neve S, Kristiansen K, Gregersen N, Bolund L (April 1998).
1554:"The structure of ClpP at 2.3 A resolution suggests a model for ATP-dependent proteolysis"
1410:
Corydon TJ, Bross P, Holst HU, Neve S, Kristiansen K, Gregersen N, Bolund L (April 1998).
552:
369:
145:
1170:
proteases. PaClpP2 is required for proper biofilm development in opportunistic pathogen
2938:
2827:
2768:
2081:
2057:
2036:
1817:
1792:
1768:
1741:
1676:
1651:
1529:
1504:
1432:
1411:
1627:
1602:
1570:
1553:
1387:
1370:
1044:. This protein is an essential component to form the protein complex of Clp protease (
686:
681:
676:
671:
655:
650:
645:
629:
624:
619:
614:
609:
604:
599:
594:
28:
2948:
2732:
2691:
2603:
2476:
2465:
2197:
1791:
Ishizawa J, Zarabi SF, Davis RE, Halgas O, Nii T, Jitkova Y, et al. (May 2019).
1484:
1321:
1304:
581:
1883:
1587:
1338:
129:
2681:
2499:
2393:
2235:
1103:
1061:
387:
166:
1970:
Kang SG, Ortega J, Singh SK, Wang N, Huang NN, Steven AC, Maurizi MR (June 2002).
1793:"Mitochondrial ClpP-Mediated Proteolysis Induces Selective Cancer Cell Lethality"
1703:. Advances in Experimental Medicine and Biology. Vol. 694. pp. 108–25.
1505:"ClpP1P2 peptidase activity promotes biofilm formation in Pseudomonas aeruginosa"
1178:. This founding has relevance in development of new antimicrobial agents against
153:
108:, PRLTS3, DFNB81, caseinolytic mitochondrial matrix peptidase proteolytic subunit
2905:
2840:
2676:
2618:
2613:
2262:
1808:
1708:
1290:
National Center for
Biotechnology Information, U.S. National Library of Medicine
1272:
National Center for
Biotechnology Information, U.S. National Library of Medicine
2933:
470:
2580:
2522:
2303:
2298:
2278:
1954:
1468:
286:
183:
133:
1867:
2879:
2853:
2608:
2594:
2413:
2293:
2283:
716:
530:
408:
353:
340:
252:
239:
141:
2027:
2019:
1997:
1988:
1971:
1962:
1929:
1920:
1903:
1875:
1826:
1777:
1726:
1685:
1538:
1476:
2066:
1636:
1618:
1579:
1441:
1396:
1330:
1018:
1013:
687:
protein quality control for misfolded or incompletely synthesized proteins
2494:
2489:
2438:
2288:
2273:
2240:
1758:
1200:
1057:
1002:
861:
842:
1902:
Kang SG, Dimitrova MN, Ortega J, Ginsburg A, Maurizi MR (October 2005).
1858:
1841:
1667:
1303:
Bross P, Andresen BS, Knudsen I, Kruse TA, Gregersen N (December 1995).
2548:
2538:
2484:
2398:
2230:
828:
783:
2048:
1520:
1423:
74:
2892:
2662:
2575:
2517:
1937:
Kang SG, Maurizi MR, Thompson M, Mueser T, Ahvazi B (December 2004).
1056:
Enzyme ClpP is a highly conserved serine protease present throughout
1034:
986:
738:
1938:
1371:"A multiple-component, ATP-dependent protease from Escherichia coli"
2866:
2584:
2570:
2509:
2383:
2378:
2373:
2368:
2363:
2358:
701:
697:
2353:
2348:
2343:
2338:
2333:
2328:
2318:
2313:
1112:, proteins tagged with the SsrA peptide (ANDENYALAA) encoded by
1066:
1041:
121:
2635:
2170:
2109:
561:
2105:
1141:
produces cleavage products that enhance biofilm formation in
1369:
Katayama-Fujimura Y, Gottesman S, Maurizi MR (April 1987).
2631:
1842:"Bacterial proteases, untapped antimicrobial drug targets"
672:
proteolysis involved in cellular protein catabolic process
377:
2922:
1031:
ATP-dependent Clp protease proteolytic subunit (ClpP)
542:
2818:
2782:
2751:
2720:
2669:
2531:
2508:
2474:
2424:
2261:
2216:
1601:Gottesman S, Roche E, Zhou Y, Sauer RT (May 1998).
1158:has been implicated in preventing premature aging.
947:
926:
900:
879:
1552:Wang J, Hartling JA, Flanagan JM (November 1997).
1246:
1244:
1242:
1225:
1223:
1221:
1081:produces two forms of the ClpP peptidase, PaClpP1
1840:Culp, Elizabeth; Wright, Gerard D. (April 2017).
324:
223:
16:Protein-coding gene in the species Homo sapiens
1251:GRCm38: Ensembl release 89: ENSMUSG00000002660
2647:
2182:
2121:
8:
1701:Protein Metabolism and Homeostasis in Aging
1230:GRCh38: Ensembl release 89: ENSG00000125656
2654:
2640:
2632:
2189:
2175:
2167:
2128:
2114:
2106:
1133:. Peptidase and protease action of PaClpP1
712:
577:
365:
264:
161:
63:
2056:
1987:
1919:
1857:
1816:
1767:
1757:
1675:
1626:
1569:
1528:
1431:
1386:
1320:
1350:
1348:
1073:Some bacteria have multiple ClpPs, like
2929:
2143:
1217:
2202:serine proteases/serine endopeptidases
18:
1498:
1496:
1494:
329:
290:
285:
228:
187:
182:
7:
1976:The Journal of Biological Chemistry
1908:The Journal of Biological Chemistry
1375:The Journal of Biological Chemistry
944:
923:
897:
876:
852:
833:
807:
788:
762:
743:
605:serine-type endopeptidase activity
547:
465:
403:
382:
14:
1037:that in humans is encoded by the
2932:
2146:
625:ATP-dependent peptidase activity
551:
313:
306:
300:
277:
212:
205:
199:
174:
27:
2965:Post-translational modification
2043:. 331 ( Pt 1) (Pt 1): 309–316.
600:serine-type peptidase activity
562:More reference expression data
531:More reference expression data
1:
2414:Urinary plasminogen activator
1943:Journal of Structural Biology
1571:10.1016/s0092-8674(00)80431-6
1388:10.1016/S0021-9258(18)61217-7
1125:peptidase activity of PaClpP1
298:
197:
2955:Genes on human chromosome 19
2409:Tissue plasminogen activator
2008:Journal of Molecular Biology
1418:. 331 ( Pt 1) (1): 309–316.
1322:10.1016/0014-5793(95)01353-9
1809:10.1016/j.ccell.2019.03.014
1709:10.1007/978-1-4419-7002-2_9
677:protein homooligomerization
2986:
2098:gene details page in the
1846:The Journal of Antibiotics
427:mucosa of transverse colon
2810:Michaelis–Menten kinetics
2500:Proteinase 3/Myeloblastin
2141:
1955:10.1016/j.jsb.2004.07.004
1469:10.1016/j.arr.2014.12.010
1286:"Mouse PubMed Reference:"
1268:"Human PubMed Reference:"
1017:
1012:
1008:
1001:
985:
966:
951:
930:
919:
904:
883:
872:
859:
855:
840:
836:
827:
814:
810:
795:
791:
782:
769:
765:
750:
746:
737:
722:
715:
711:
695:
656:endopeptidase Clp complex
620:identical protein binding
580:
576:
559:
550:
541:
528:
477:
468:
415:
406:
376:
368:
364:
347:
334:
297:
276:
267:
263:
246:
233:
196:
173:
164:
160:
115:
112:
102:
95:
90:
71:
66:
49:
44:
39:
35:
26:
21:
2702:Diffusion-limited enzyme
1746:Human Molecular Genetics
2041:The Biochemical Journal
1656:Journal of Bacteriology
1607:Genes & Development
1457:Ageing Research Reviews
1416:The Biochemical Journal
481:interventricular septum
2554:Proprotein convertases
2020:10.1006/jmbi.1999.3121
1989:10.1074/jbc.M201642200
1921:10.1074/jbc.M507240200
1509:Molecular Microbiology
1206:Transfer-messenger RNA
1060:and also found in the
979:Chr 17: 57.3 – 57.3 Mb
972:Chr 19: 6.36 – 6.37 Mb
2795:Eadie–Hofstee diagram
2728:Allosteric regulation
2404:Plasminogen activator
2082:MEROPS entry for ClpP
1619:10.1101/gad.12.9.1338
1153:Clinical significance
331:17 D|17 29.61 cM
292:Chromosome 17 (mouse)
190:Chromosome 19 (human)
2805:Lineweaver–Burk plot
2544:Prolyl endopeptidase
2092:genome location and
646:mitochondrial matrix
489:facial motor nucleus
443:right adrenal cortex
423:gastrocnemius muscle
67:List of PDB id codes
40:Available structures
2100:UCSC Genome Browser
1982:(23): 21095–21102.
1914:(42): 35424–35432.
1859:10.1038/ja.2016.138
1668:10.1128/JB.00568-16
1191:CLP protease family
451:left adrenal cortex
447:right lobe of liver
435:right adrenal gland
2764:Enzyme superfamily
2697:Enzyme promiscuity
1759:10.1093/hmg/ddt338
817:ENSMUSG00000002660
665:Biological process
639:Cellular component
615:hydrolase activity
595:peptidase activity
588:Molecular function
439:left adrenal gland
2920:
2919:
2629:
2628:
2426:Complement system
2218:Digestive enzymes
2164:
2163:
2049:10.1042/bj3310309
1803:(5): 721–737.e9.
1752:(24): 4871–4887.
1718:978-1-4419-7001-5
1521:10.1111/mmi.14649
1424:10.1042/bj3310309
1381:(10): 4477–4485.
1196:Endopeptidase Clp
1140:
1136:
1132:
1128:
1092:
1088:
1084:
1046:Endopeptidase Clp
1028:
1027:
1024:
1023:
997:
996:
962:
961:
941:
940:
915:
914:
894:
893:
868:
867:
849:
848:
823:
822:
804:
803:
778:
777:
759:
758:
707:
706:
572:
571:
568:
567:
537:
536:
524:
523:
462:
461:
360:
359:
259:
258:
86:
85:
82:
81:
50:Ortholog search:
2977:
2937:
2936:
2928:
2800:Hanes–Woolf plot
2743:Enzyme activator
2738:Enzyme inhibitor
2712:Enzyme catalysis
2656:
2649:
2642:
2633:
2191:
2184:
2177:
2168:
2150:
2130:
2123:
2116:
2107:
2070:
2060:
2031:
2001:
1991:
1966:
1933:
1923:
1888:
1887:
1861:
1837:
1831:
1830:
1820:
1788:
1782:
1781:
1771:
1761:
1737:
1731:
1730:
1696:
1690:
1689:
1679:
1647:
1641:
1640:
1630:
1613:(9): 1338–1347.
1598:
1592:
1591:
1573:
1549:
1543:
1542:
1532:
1515:(6): 1094–1109.
1500:
1489:
1488:
1452:
1446:
1445:
1435:
1407:
1401:
1400:
1390:
1366:
1360:
1359:
1352:
1343:
1342:
1324:
1300:
1294:
1293:
1282:
1276:
1275:
1264:
1258:
1248:
1237:
1227:
1138:
1134:
1130:
1126:
1090:
1086:
1082:
1010:
1009:
981:
974:
957:
945:
936:
924:
920:RefSeq (protein)
910:
898:
889:
877:
853:
834:
808:
789:
763:
744:
713:
578:
564:
555:
548:
533:
473:
471:Top expressed in
466:
411:
409:Top expressed in
404:
383:
366:
356:
343:
332:
317:
310:
304:
293:
281:
265:
255:
242:
231:
216:
209:
203:
192:
178:
162:
156:
154:CLPP - orthologs
107:
100:
77:
64:
58:
37:
36:
31:
19:
2985:
2984:
2980:
2979:
2978:
2976:
2975:
2974:
2945:
2944:
2943:
2931:
2923:
2921:
2916:
2828:Oxidoreductases
2814:
2790:Enzyme kinetics
2778:
2774:List of enzymes
2747:
2716:
2687:Catalytic triad
2665:
2660:
2630:
2625:
2527:
2504:
2470:
2420:
2257:
2226:Enteropeptidase
2212:
2195:
2165:
2160:
2157:
2151:
2137:
2134:
2078:
2073:
2034:
2004:
1969:
1936:
1901:
1897:
1895:Further reading
1892:
1891:
1839:
1838:
1834:
1790:
1789:
1785:
1739:
1738:
1734:
1719:
1698:
1697:
1693:
1649:
1648:
1644:
1600:
1599:
1595:
1551:
1550:
1546:
1502:
1501:
1492:
1463:(Pt A): 56–66.
1454:
1453:
1449:
1409:
1408:
1404:
1368:
1367:
1363:
1354:
1353:
1346:
1302:
1301:
1297:
1284:
1283:
1279:
1266:
1265:
1261:
1249:
1240:
1228:
1219:
1214:
1187:
1155:
1099:
1054:
1019:View/Edit Mouse
1014:View/Edit Human
977:
970:
967:Location (UCSC)
953:
932:
906:
885:
798:ENSG00000125656
691:
660:
634:
610:protein binding
560:
529:
520:
515:
511:
509:right ventricle
507:
503:
499:
495:
491:
487:
483:
469:
458:
455:body of stomach
453:
449:
445:
441:
437:
433:
429:
425:
421:
419:muscle of thigh
407:
351:
338:
330:
320:
319:
318:
311:
291:
268:Gene location (
250:
237:
229:
219:
218:
217:
210:
188:
165:Gene location (
116:
103:
96:
73:
51:
17:
12:
11:
5:
2983:
2981:
2973:
2972:
2967:
2962:
2957:
2947:
2946:
2942:
2941:
2918:
2917:
2915:
2914:
2901:
2888:
2875:
2862:
2849:
2836:
2822:
2820:
2816:
2815:
2813:
2812:
2807:
2802:
2797:
2792:
2786:
2784:
2780:
2779:
2777:
2776:
2771:
2766:
2761:
2755:
2753:
2752:Classification
2749:
2748:
2746:
2745:
2740:
2735:
2730:
2724:
2722:
2718:
2717:
2715:
2714:
2709:
2704:
2699:
2694:
2689:
2684:
2679:
2673:
2671:
2667:
2666:
2661:
2659:
2658:
2651:
2644:
2636:
2627:
2626:
2624:
2623:
2622:
2621:
2616:
2606:
2601:
2592:
2578:
2573:
2568:
2567:
2566:
2561:
2551:
2546:
2541:
2535:
2533:
2529:
2528:
2526:
2525:
2520:
2514:
2512:
2506:
2505:
2503:
2502:
2497:
2492:
2487:
2481:
2479:
2472:
2471:
2469:
2468:
2463:
2462:
2461:
2456:
2446:
2441:
2436:
2430:
2428:
2422:
2421:
2419:
2418:
2417:
2416:
2411:
2401:
2389:
2388:
2387:
2386:
2381:
2376:
2371:
2366:
2361:
2356:
2351:
2346:
2341:
2336:
2331:
2326:
2321:
2316:
2311:
2301:
2296:
2291:
2286:
2281:
2276:
2267:
2265:
2259:
2258:
2256:
2255:
2254:
2253:
2248:
2238:
2233:
2228:
2222:
2220:
2214:
2213:
2198:Endopeptidases
2196:
2194:
2193:
2186:
2179:
2171:
2162:
2161:
2159:
2158:
2152:
2145:
2142:
2139:
2138:
2135:
2133:
2132:
2125:
2118:
2110:
2104:
2103:
2084:
2077:
2076:External links
2074:
2072:
2071:
2032:
2014:(4): 819–825.
2002:
1967:
1949:(3): 338–352.
1934:
1898:
1896:
1893:
1890:
1889:
1852:(4): 366–377.
1832:
1783:
1732:
1717:
1691:
1642:
1593:
1564:(4): 447–456.
1544:
1490:
1447:
1402:
1361:
1344:
1315:(2): 249–252.
1295:
1277:
1259:
1238:
1216:
1215:
1213:
1210:
1209:
1208:
1203:
1198:
1193:
1186:
1183:
1180:P. aeruginosa.
1154:
1151:
1122:P. aeruginosa,
1098:
1095:
1053:
1050:
1026:
1025:
1022:
1021:
1016:
1006:
1005:
999:
998:
995:
994:
992:
990:
983:
982:
975:
968:
964:
963:
960:
959:
949:
948:
942:
939:
938:
928:
927:
921:
917:
916:
913:
912:
902:
901:
895:
892:
891:
881:
880:
874:
870:
869:
866:
865:
857:
856:
850:
847:
846:
838:
837:
831:
825:
824:
821:
820:
812:
811:
805:
802:
801:
793:
792:
786:
780:
779:
776:
775:
767:
766:
760:
757:
756:
748:
747:
741:
735:
734:
729:
724:
720:
719:
709:
708:
705:
704:
693:
692:
690:
689:
684:
679:
674:
668:
666:
662:
661:
659:
658:
653:
648:
642:
640:
636:
635:
633:
632:
630:ATPase binding
627:
622:
617:
612:
607:
602:
597:
591:
589:
585:
584:
574:
573:
570:
569:
566:
565:
557:
556:
545:
539:
538:
535:
534:
526:
525:
522:
521:
519:
518:
514:
510:
506:
502:
498:
494:
490:
486:
482:
478:
475:
474:
463:
460:
459:
457:
456:
452:
448:
444:
440:
436:
432:
428:
424:
420:
416:
413:
412:
400:
399:
391:
380:
374:
373:
370:RNA expression
362:
361:
358:
357:
349:
345:
344:
336:
333:
328:
322:
321:
312:
305:
299:
295:
294:
289:
283:
282:
274:
273:
261:
260:
257:
256:
248:
244:
243:
235:
232:
227:
221:
220:
211:
204:
198:
194:
193:
186:
180:
179:
171:
170:
158:
157:
114:
110:
109:
101:
93:
92:
88:
87:
84:
83:
80:
79:
69:
68:
60:
59:
48:
42:
41:
33:
32:
24:
23:
15:
13:
10:
9:
6:
4:
3:
2:
2982:
2971:
2968:
2966:
2963:
2961:
2958:
2956:
2953:
2952:
2950:
2940:
2935:
2930:
2926:
2912:
2908:
2907:
2902:
2899:
2895:
2894:
2889:
2886:
2882:
2881:
2876:
2873:
2869:
2868:
2863:
2860:
2856:
2855:
2850:
2847:
2843:
2842:
2837:
2834:
2830:
2829:
2824:
2823:
2821:
2817:
2811:
2808:
2806:
2803:
2801:
2798:
2796:
2793:
2791:
2788:
2787:
2785:
2781:
2775:
2772:
2770:
2769:Enzyme family
2767:
2765:
2762:
2760:
2757:
2756:
2754:
2750:
2744:
2741:
2739:
2736:
2734:
2733:Cooperativity
2731:
2729:
2726:
2725:
2723:
2719:
2713:
2710:
2708:
2705:
2703:
2700:
2698:
2695:
2693:
2692:Oxyanion hole
2690:
2688:
2685:
2683:
2680:
2678:
2675:
2674:
2672:
2668:
2664:
2657:
2652:
2650:
2645:
2643:
2638:
2637:
2634:
2620:
2617:
2615:
2612:
2611:
2610:
2607:
2605:
2604:Streptokinase
2602:
2600:
2596:
2593:
2590:
2586:
2582:
2579:
2577:
2574:
2572:
2569:
2565:
2562:
2560:
2557:
2556:
2555:
2552:
2550:
2547:
2545:
2542:
2540:
2537:
2536:
2534:
2530:
2524:
2521:
2519:
2516:
2515:
2513:
2511:
2507:
2501:
2498:
2496:
2493:
2491:
2488:
2486:
2483:
2482:
2480:
2478:
2477:immune system
2473:
2467:
2466:C3-convertase
2464:
2460:
2457:
2455:
2452:
2451:
2450:
2447:
2445:
2442:
2440:
2437:
2435:
2432:
2431:
2429:
2427:
2423:
2415:
2412:
2410:
2407:
2406:
2405:
2402:
2400:
2397:
2395:
2391:
2390:
2385:
2382:
2380:
2377:
2375:
2372:
2370:
2367:
2365:
2362:
2360:
2357:
2355:
2352:
2350:
2347:
2345:
2342:
2340:
2337:
2335:
2332:
2330:
2327:
2325:
2322:
2320:
2317:
2315:
2312:
2310:
2307:
2306:
2305:
2302:
2300:
2297:
2295:
2292:
2290:
2287:
2285:
2282:
2280:
2277:
2275:
2272:
2269:
2268:
2266:
2264:
2260:
2252:
2249:
2247:
2244:
2243:
2242:
2239:
2237:
2234:
2232:
2229:
2227:
2224:
2223:
2221:
2219:
2215:
2210:
2207:
2203:
2199:
2192:
2187:
2185:
2180:
2178:
2173:
2172:
2169:
2155:
2149:
2144:
2140:
2131:
2126:
2124:
2119:
2117:
2112:
2111:
2108:
2101:
2097:
2096:
2091:
2090:
2085:
2083:
2080:
2079:
2075:
2068:
2064:
2059:
2054:
2050:
2046:
2042:
2038:
2033:
2029:
2025:
2021:
2017:
2013:
2009:
2003:
1999:
1995:
1990:
1985:
1981:
1977:
1973:
1968:
1964:
1960:
1956:
1952:
1948:
1944:
1940:
1935:
1931:
1927:
1922:
1917:
1913:
1909:
1905:
1900:
1899:
1894:
1885:
1881:
1877:
1873:
1869:
1865:
1860:
1855:
1851:
1847:
1843:
1836:
1833:
1828:
1824:
1819:
1814:
1810:
1806:
1802:
1798:
1794:
1787:
1784:
1779:
1775:
1770:
1765:
1760:
1755:
1751:
1747:
1743:
1736:
1733:
1728:
1724:
1720:
1714:
1710:
1706:
1702:
1695:
1692:
1687:
1683:
1678:
1673:
1669:
1665:
1661:
1657:
1653:
1646:
1643:
1638:
1634:
1629:
1624:
1620:
1616:
1612:
1608:
1604:
1597:
1594:
1589:
1585:
1581:
1577:
1572:
1567:
1563:
1559:
1555:
1548:
1545:
1540:
1536:
1531:
1526:
1522:
1518:
1514:
1510:
1506:
1499:
1497:
1495:
1491:
1486:
1482:
1478:
1474:
1470:
1466:
1462:
1458:
1451:
1448:
1443:
1439:
1434:
1429:
1425:
1421:
1417:
1413:
1406:
1403:
1398:
1394:
1389:
1384:
1380:
1376:
1372:
1365:
1362:
1357:
1351:
1349:
1345:
1340:
1336:
1332:
1328:
1323:
1318:
1314:
1310:
1306:
1299:
1296:
1291:
1287:
1281:
1278:
1273:
1269:
1263:
1260:
1256:
1252:
1247:
1245:
1243:
1239:
1235:
1231:
1226:
1224:
1222:
1218:
1211:
1207:
1204:
1202:
1199:
1197:
1194:
1192:
1189:
1188:
1184:
1182:
1181:
1177:
1176:P. aeruginosa
1173:
1172:P. aeruginosa
1167:
1163:
1159:
1152:
1150:
1146:
1144:
1143:P. aeruginosa
1123:
1118:
1115:
1111:
1110:
1105:
1096:
1094:
1080:
1079:P. aeruginosa
1076:
1075:P. aeruginosa
1071:
1068:
1063:
1059:
1051:
1049:
1047:
1043:
1040:
1036:
1032:
1020:
1015:
1011:
1007:
1004:
1000:
993:
991:
988:
984:
980:
976:
973:
969:
965:
958:
956:
950:
946:
943:
937:
935:
929:
925:
922:
918:
911:
909:
903:
899:
896:
890:
888:
882:
878:
875:
873:RefSeq (mRNA)
871:
864:
863:
858:
854:
851:
845:
844:
839:
835:
832:
830:
826:
819:
818:
813:
809:
806:
800:
799:
794:
790:
787:
785:
781:
774:
773:
768:
764:
761:
755:
754:
749:
745:
742:
740:
736:
733:
730:
728:
725:
721:
718:
714:
710:
703:
699:
694:
688:
685:
683:
680:
678:
675:
673:
670:
669:
667:
664:
663:
657:
654:
652:
651:mitochondrion
649:
647:
644:
643:
641:
638:
637:
631:
628:
626:
623:
621:
618:
616:
613:
611:
608:
606:
603:
601:
598:
596:
593:
592:
590:
587:
586:
583:
582:Gene ontology
579:
575:
563:
558:
554:
549:
546:
544:
540:
532:
527:
516:
513:adrenal gland
512:
508:
504:
500:
496:
492:
488:
484:
480:
479:
476:
472:
467:
464:
454:
450:
446:
442:
438:
434:
431:apex of heart
430:
426:
422:
418:
417:
414:
410:
405:
402:
401:
398:
396:
392:
390:
389:
385:
384:
381:
379:
375:
371:
367:
363:
355:
350:
346:
342:
337:
327:
323:
316:
309:
303:
296:
288:
284:
280:
275:
271:
266:
262:
254:
249:
245:
241:
236:
226:
222:
215:
208:
202:
195:
191:
185:
181:
177:
172:
168:
163:
159:
155:
151:
147:
143:
139:
135:
131:
127:
123:
119:
111:
106:
99:
94:
89:
78:
76:
70:
65:
62:
61:
57:
54:
47:
43:
38:
34:
30:
25:
20:
2906:Translocases
2903:
2890:
2877:
2864:
2851:
2841:Transferases
2838:
2825:
2682:Binding site
2394:fibrinolysis
2392:
2270:
2236:Chymotrypsin
2153:
2094:
2088:
2040:
2011:
2007:
1979:
1975:
1946:
1942:
1911:
1907:
1849:
1845:
1835:
1800:
1796:
1786:
1749:
1745:
1735:
1700:
1694:
1659:
1655:
1645:
1610:
1606:
1596:
1561:
1557:
1547:
1512:
1508:
1460:
1456:
1450:
1415:
1405:
1378:
1374:
1364:
1312:
1309:FEBS Letters
1308:
1298:
1289:
1280:
1271:
1262:
1179:
1175:
1171:
1168:
1164:
1160:
1156:
1147:
1142:
1121:
1119:
1107:
1100:
1078:
1074:
1072:
1062:mitochondria
1055:
1038:
1030:
1029:
952:
931:
905:
884:
860:
841:
815:
796:
770:
751:
731:
726:
517:otic placode
485:right kidney
393:
386:
113:External IDs
72:
2677:Active site
2299:Factor XIIa
2279:Factor VIIa
2263:Coagulation
2136:PDB gallery
1797:Cancer Cell
1104:AAA+ ATPase
1085:and PaClpP1
682:proteolysis
352:57,303,188
339:57,297,305
91:Identifiers
2949:Categories
2880:Isomerases
2854:Hydrolases
2721:Regulation
2581:Subtilisin
2523:Batroxobin
2304:Kallikrein
2294:Factor XIa
2284:Factor IXa
2251:Pancreatic
2246:Neutrophil
1257:, May 2017
1236:, May 2017
1212:References
505:blastocyst
397:(ortholog)
251:6,370,242
238:6,361,531
134:HomoloGene
2970:EC 3.4.21
2960:Proteases
2759:EC number
2609:Cathepsin
2595:Sedolisin
2571:Prostasin
2289:Factor Xa
1868:1881-1469
1485:205667759
1052:Structure
955:NP_059089
934:NP_006003
908:NM_017393
887:NM_006012
717:Orthologs
142:GeneCards
2783:Kinetics
2707:Cofactor
2670:Activity
2510:Venombin
2495:Tryptase
2490:Granzyme
2444:Factor I
2439:Factor D
2434:Factor B
2274:Thrombin
2271:factors:
2241:Elastase
2028:10525407
1998:11923310
1963:15522782
1930:16115876
1884:31720089
1876:27899793
1827:31056398
1778:23851121
1727:20886760
1686:27849175
1588:14136820
1539:33231899
1477:25578288
1339:22019074
1253:–
1232:–
1201:Protease
1185:See also
1097:Function
1058:bacteria
1003:Wikidata
696:Sources:
2939:Biology
2893:Ligases
2663:Enzymes
2549:Pronase
2539:Acrosin
2485:Chymase
2399:Plasmin
2231:Trypsin
2067:9512494
2058:1219353
1818:6620028
1769:7108587
1677:5237113
1637:9573050
1580:9390554
1530:8141546
1442:9512494
1433:1219353
1397:3549708
1331:8543061
1255:Ensembl
1234:Ensembl
1109:E. coli
829:UniProt
784:Ensembl
723:Species
702:QuickGO
501:condyle
372:pattern
230:19p13.3
130:1858213
98:Aliases
2925:Portal
2867:Lyases
2576:Reelin
2518:Ancrod
2475:Other
2209:3.4.21
2086:Human
2065:
2055:
2026:
1996:
1961:
1928:
1882:
1874:
1866:
1825:
1815:
1776:
1766:
1725:
1715:
1684:
1674:
1635:
1628:316764
1625:
1586:
1578:
1537:
1527:
1483:
1475:
1440:
1430:
1395:
1337:
1329:
1035:enzyme
1033:is an
989:search
987:PubMed
862:O88696
843:Q16740
739:Entrez
543:BioGPS
497:morula
493:morula
122:601119
2819:Types
2585:Furin
2532:Other
2459:MASP2
2454:MASP1
2384:KLK15
2379:KLK14
2374:KLK13
2369:KLK12
2364:KLK11
2359:KLK10
1880:S2CID
1662:(3).
1584:S2CID
1481:S2CID
1335:S2CID
1114:tmRNA
772:53895
732:Mouse
727:Human
698:Amigo
395:Mouse
388:Human
335:Start
270:Mouse
234:Start
167:Human
2911:list
2904:EC7
2898:list
2891:EC6
2885:list
2878:EC5
2872:list
2865:EC4
2859:list
2852:EC3
2846:list
2839:EC2
2833:list
2826:EC1
2599:TPP1
2449:MASP
2354:KLK9
2349:KLK8
2344:KLK7
2339:KLK6
2334:KLK5
2329:KLK4
2324:KLK3
2319:KLK2
2314:KLK1
2154:1tg6
2095:CLPP
2089:CLPP
2063:PMID
2024:PMID
1994:PMID
1959:PMID
1926:PMID
1872:PMID
1864:ISSN
1823:PMID
1774:PMID
1723:PMID
1713:ISBN
1682:PMID
1633:PMID
1576:PMID
1558:Cell
1535:PMID
1473:PMID
1438:PMID
1393:PMID
1327:PMID
1067:ClpX
1042:gene
1039:CLPP
753:8192
378:Bgee
326:Band
287:Chr.
225:Band
184:Chr.
146:CLPP
138:4385
118:OMIM
105:CLPP
75:1TG6
56:RCSB
53:PDBe
22:CLPP
2589:S1P
2309:PSA
2053:PMC
2045:doi
2016:doi
2012:292
1984:doi
1980:277
1951:doi
1947:148
1916:doi
1912:280
1854:doi
1813:PMC
1805:doi
1764:PMC
1754:doi
1705:doi
1672:PMC
1664:doi
1660:199
1623:PMC
1615:doi
1566:doi
1525:PMC
1517:doi
1513:115
1465:doi
1428:PMC
1420:doi
1383:doi
1379:262
1317:doi
1313:377
1120:In
1048:).
348:End
247:End
150:OMA
126:MGI
46:PDB
2951::
2206:EC
2200::
2061:.
2051:.
2039:.
2022:.
2010:.
1992:.
1978:.
1974:.
1957:.
1945:.
1941:.
1924:.
1910:.
1906:.
1878:.
1870:.
1862:.
1850:70
1848:.
1844:.
1821:.
1811:.
1801:35
1799:.
1795:.
1772:.
1762:.
1750:22
1748:.
1744:.
1721:.
1711:.
1680:.
1670:.
1658:.
1654:.
1631:.
1621:.
1611:12
1609:.
1605:.
1582:.
1574:.
1562:91
1560:.
1556:.
1533:.
1523:.
1511:.
1507:.
1493:^
1479:.
1471:.
1461:23
1459:.
1436:.
1426:.
1414:.
1391:.
1377:.
1373:.
1347:^
1333:.
1325:.
1311:.
1307:.
1288:.
1270:.
1241:^
1220:^
1145:.
1137:P2
1129:P2
1089:P2
1083:14
700:/
354:bp
341:bp
253:bp
240:bp
148:;
144::
140:;
136::
132:;
128::
124:;
120::
2927::
2913:)
2909:(
2900:)
2896:(
2887:)
2883:(
2874:)
2870:(
2861:)
2857:(
2848:)
2844:(
2835:)
2831:(
2655:e
2648:t
2641:v
2619:G
2614:A
2597:/
2591:4
2587:/
2583:/
2564:2
2559:1
2396::
2211:)
2204:(
2190:e
2183:t
2176:v
2129:e
2122:t
2115:v
2102:.
2069:.
2047::
2030:.
2018::
2000:.
1986::
1965:.
1953::
1932:.
1918::
1886:.
1856::
1829:.
1807::
1780:.
1756::
1729:.
1707::
1688:.
1666::
1639:.
1617::
1590:.
1568::
1541:.
1519::
1487:.
1467::
1444:.
1422::
1399:.
1385::
1358:.
1341:.
1319::
1292:.
1274:.
1139:7
1135:7
1131:7
1127:7
1091:7
1087:7
272:)
169:)
152::
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.