255:
232:
129:
154:
506:
261:
160:
3215:
3235:
1129:
in the liver. The aminophospholipid translocases transport phosphatidylserine and phosphatidylethanolamine from one side of a bilayer to another. Mutations in this gene may result in progressive familial intrahepatic cholestasis type 1 and in benign recurrent intrahepatic cholestasis. Exactly how
1422:
Eppens EF, van Mil SW, de Vree JM, Mok KS, Juijn JA, Oude
Elferink RP, et al. (October 2001). "FIC1, the protein affected in two forms of hereditary cholestasis, is localized in the cholangiocyte and the canalicular membrane of the hepatocyte".
1120:
cation transport ATPase family and specifically belongs to the subfamily of aminophospholipid-transporting ATPases. This protein is highly expressed in the small intestine, stomach, pancreas, and prostate and is also found in
1268:
Carlton VE, Knisely AS, Freimer NB (Oct 1995). "Mapping of a locus for progressive familial intrahepatic cholestasis (Byler disease) to 18q21-q22, the benign recurrent intrahepatic cholestasis region".
2042:
1225:
Bull LN, van Eijk MJ, Pawlikowska L, DeYoung JA, Juijn JA, Liao M, et al. (Mar 1998). "A gene encoding a P-type ATPase mutated in two forms of hereditary cholestasis".
268:
167:
1498:
1701:
Halleck MS, Lawler JF JR, Blackshaw S, Gao L, Nagarajan P, Hacker C, et al. (2001). "Differential expression of putative transbilayer amphipath transporters".
1101:
1834:
Harris MJ, Arias IM (2003). "FIC1, a P-type ATPase linked to cholestatic liver disease, has homologues (ATP8B2 and ATP8B3) expressed throughout the body".
819:
90:
800:
2735:
3314:
2035:
2265:
3280:
3299:
1207:
2234:
1189:
254:
2002:
1506:
Harris MJ, Le
Couteur DG, Arias IM (2006). "Progressive familial intrahepatic cholestasis: genetic disorders of biliary transporters".
2028:
1027:
1034:
231:
2934:
1543:
Clayton RJ, Iber FL, Ruebner BH, McKusick VA (1969). "Byler disease. Fatal familial intrahepatic cholestasis in an Amish kindred".
1307:
3309:
2825:
1176:
1155:
1105:
2820:
153:
128:
3090:
2008:
1172:
2842:
3205:
1151:
2890:
2207:
70:
2812:
267:
166:
1660:"Recurrent familial intrahepatic cholestasis in the Faeroe Islands. Phenotypic heterogeneity but genetic homogeneity"
3273:
1865:"Progressive familial intrahepatic cholestasis, type 1, is associated with decreased farnesoid X receptor activity"
260:
159:
3075:
3191:
3178:
3165:
3152:
3139:
3126:
3113:
2871:
2412:
2310:
2135:
2131:
2108:
2081:
2068:
2055:
1574:"Genome screening by searching for shared segments: mapping a gene for benign recurrent intrahepatic cholestasis"
3085:
3039:
2982:
2426:
2121:
2086:
2059:
1995:
864:
78:
1900:"The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC)"
2987:
2796:
2759:
2098:
1943:"Altered hepatobiliary gene expressions in PFIC1: ATP8B1 gene defect is associated with CFTR downregulation"
1321:
Klomp L.W., Vargas J.C., van Mil S.W., Pawlikowska L, Strautnieks SS, Van Eijk MJ, et al. (July 2004).
845:
2526:
2166:
3266:
3008:
2927:
2656:
2564:
1492:
142:
3080:
1322:
1794:
57:
3044:
2861:
2014:
1991:
1010:
963:
942:
938:
1625:"Multiple members of a third subfamily of P-type ATPases identified by genomic sequences and ESTs"
2977:
2876:
2442:
2020:
1972:
1769:
1726:
1689:
1611:
1531:
1355:
1250:
102:
1783:"Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences"
989:
934:
1572:
Houwen RH, Baharloo S, Blankenship K, Raeymaekers P, Juyn J, Sandkuijl LA, et al. (1995).
505:
3304:
2116:
1964:
1929:
1886:
1851:
1822:
1781:
Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, et al. (2003).
1761:
1718:
1681:
1646:
1603:
1560:
1523:
1480:
1440:
1404:
1347:
1286:
1242:
50:
3250:
3023:
3018:
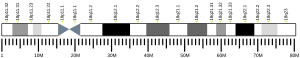
2992:
2920:
2506:
2452:
2434:
2300:
2290:
2244:
1954:
1919:
1911:
1876:
1843:
1812:
1802:
1751:
1710:
1671:
1636:
1593:
1585:
1552:
1515:
1472:
1463:
Knisely AS (2000). "Progressive familial intrahepatic cholestasis: a personal perspective".
1432:
1394:
1386:
1337:
1278:
1234:
347:
278:
222:
177:
98:
2319:
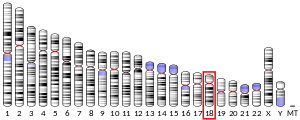
3070:
3054:
2967:
2853:
2742:
2556:
2256:
2076:
322:
1898:
Gerhard DS, Wagner L, Feingold EA, Shenmen CM, Grouse LH, Schuler G, et al. (2004).
1714:
1556:
1941:
Demeilliers C, Jacquemin E, Barbu V, Mergey M, Paye F, Fouassier L, et al. (2006).
1798:
3219:
3108:
3049:
2176:
1863:
Chen F, Ananthanarayanan M, Emre S, Neimark E, Bull LN, Knisely A, et al. (2004).
1399:
1374:
1122:
1924:
1899:
1847:
1817:
1782:
1738:
Klomp LW, Bull LN, Knisely AS, Vanderdoelen M, Juijn J, Berger R, et al. (2001).
1436:
734:
729:
724:
719:
714:
709:
704:
699:
694:
689:
684:
679:
674:
669:
653:
648:
643:
638:
633:
628:
623:
618:
613:
608:
592:
587:
582:
577:
572:
567:
562:
557:
552:
547:
3293:
3246:
3013:
2972:
2720:
2330:
1519:
534:
1976:
1773:
1693:
1535:
1359:
82:
2962:
2705:
2678:
2607:
2600:
2583:
2578:
2552:
1615:
1254:
340:
119:
1730:
1881:
1864:
106:
3186:
3121:
2957:
2801:
1323:"Characterization of mutations in ATP8B1 associated with hereditary cholestasis"
1212:
National Center for
Biotechnology Information, U.S. National Library of Medicine
1194:
National Center for
Biotechnology Information, U.S. National Library of Medicine
1126:
3214:
1740:"A missense mutation in FIC1 is associated with greenland familial cholestasis"
1623:
Halleck MS, Pradhan D, Blackman C, Berkes C, Williamson P, Schlegel RA (1998).
423:
2786:
2475:
239:
136:
86:
3160:
3134:
2492:
2051:
1756:
1739:
764:
483:
361:
306:
293:
205:
192:
94:
61:, ATPIC, BRIC, FIC1, ICP1, PFIC, PFIC1, ATPase phospholipid transporting 8B1
1968:
1933:
1890:
1855:
1826:
1807:
1765:
1722:
1527:
1484:
1444:
1408:
1351:
1282:
1685:
1676:
1659:
1650:
1607:
1564:
1476:
1290:
1246:
1074:
1069:
1117:
1058:
909:
890:
2897:
2866:
2644:
2639:
2398:
2388:
2363:
2358:
2093:
1915:
1641:
1624:
1589:
1390:
1238:
876:
831:
1959:
1942:
1598:
1342:
17:
3173:
2943:
2774:
2416:
2393:
2383:
2353:
2348:
2343:
2338:
2295:
2285:
2280:
2275:
2270:
2239:
2227:
2222:
2217:
2212:
2200:
2195:
2190:
2139:
1090:
1042:
786:
3234:
3147:
2830:
2769:
2752:
2747:
2595:
2573:
2539:
2534:
2519:
2497:
2485:
2480:
2470:
2465:
2460:
2185:
2161:
749:
745:
3242:
2835:
2791:
2779:
2764:
2730:
2725:
2710:
2693:
2688:
2683:
2671:
2666:
2661:
2651:
2627:
2622:
2617:
2612:
2514:
1573:
1130:
mutations result in these diseases is not currently understood.
1097:
74:
2916:
2024:
514:
2885:
2881:
1375:"The molecular genetics of familial intrahepatic cholestasis"
1658:
Tygstrup N, Steig BA, Juijn JA, Bull LN, Houwen RH (1999).
2912:
1308:"Entrez Gene: ATP8B1 ATPase, Class I, type 8B, member 1"
330:
3254:
573:
ATPase-coupled intramembrane lipid transporter activity
3203:
495:
3099:
3063:
3032:
3001:
2950:
2851:
2810:
2550:
2424:
2411:
2376:
2328:
2309:
2255:
2175:
2154:
2147:
2130:
2107:
2067:
1003:
982:
956:
927:
730:
negative regulation of transcription, DNA-templated
1168:
1166:
1164:
1147:
1145:
1143:
277:
176:
27:Protein-coding gene in the species Homo sapiens
1173:GRCm38: Ensembl release 89: ENSMUSG00000039529
3274:
2928:
2036:
1497:: CS1 maint: DOI inactive as of April 2024 (
1102:progressive familial intrahepatic cholestasis
8:
1087:Probable phospholipid-transporting ATPase IC
1152:GRCh38: Ensembl release 89: ENSG00000081923
3281:
3267:
2935:
2921:
2913:
2421:
2151:
2144:
2043:
2029:
2021:
760:
530:
450:transitional epithelium of urinary bladder
318:
217:
114:
1994:at the U.S. National Library of Medicine
1958:
1923:
1880:
1816:
1806:
1755:
1675:
1640:
1597:
1398:
1341:
1106:benign recurrent intrahepatic cholestasis
1302:
1300:
3210:
1139:
1490:
31:
634:integral component of plasma membrane
282:
243:
238:
181:
140:
135:
7:
3231:
3229:
1715:10.1152/physiolgenomics.1999.1.3.139
1557:10.1001/archpedi.1969.02100030114014
710:inner ear receptor cell development
593:aminophospholipid flippase activity
3253:. You can help Knowledge (XXG) by
1116:This gene encodes a member of the
1100:. This protein is associated with
1000:
979:
953:
924:
900:
881:
855:
836:
810:
791:
685:xenobiotic transmembrane transport
670:regulation of microvillus assembly
500:
418:
356:
335:
25:
1373:Jansen PL, Müller M (July 2000).
1125:and the canalicular membranes of
1093:that in humans is encoded by the
725:vestibulocochlear nerve formation
720:bile acid and bile salt transport
3233:
3213:
1520:10.1111/j.1440-1746.2005.03743.x
504:
266:
259:
253:
230:
165:
158:
152:
127:
3315:Human chromosome 18 gene stubs
609:integral component of membrane
515:More reference expression data
484:More reference expression data
1:
1848:10.1016/S1388-1981(03)00107-0
1437:10.1016/S0168-8278(01)00158-1
434:epithelium of small intestine
251:
150:
3300:Genes on human chromosome 18
1882:10.1053/j.gastro.2003.12.013
1787:Proc. Natl. Acad. Sci. U.S.A
2813:Protein-synthesizing GTPase
735:aminophospholipid transport
705:ion transmembrane transport
690:bile acid metabolic process
404:superior vestibular nucleus
3331:
3228:
2013:gene details page in the
715:phospholipid translocation
558:lipid transporter activity
400:superior surface of tongue
3091:Michaelis–Menten kinetics
2872:Guanylate-binding protein
2056:acid anhydride hydrolases
1508:J. Gastroenterol. Hepatol
1208:"Mouse PubMed Reference:"
1190:"Human PubMed Reference:"
1073:
1068:
1064:
1057:
1041:
1022:
1007:
986:
975:
960:
931:
920:
907:
903:
888:
884:
875:
862:
858:
843:
839:
830:
817:
813:
798:
794:
785:
770:
763:
759:
743:
533:
529:
512:
503:
494:
481:
430:
421:
368:
359:
329:
321:
317:
300:
287:
250:
229:
220:
216:
199:
186:
149:
126:
117:
113:
68:
65:
55:
48:
43:
39:
34:
2983:Diffusion-limited enzyme
2427:Heterotrimeric G protein
2122:Phosphoadenylylsulfatase
1996:Medical Subject Headings
1035:Chr 18: 64.66 – 64.79 Mb
2099:Thiamine-triphosphatase
1757:10.1053/jhep.2000.20520
1479:(inactive 2024-04-12).
1028:Chr 18: 57.65 – 57.8 Mb
3310:Transmembrane proteins
1836:Biochim. Biophys. Acta
1808:10.1073/pnas.242603899
695:phospholipid transport
649:apical plasma membrane
388:ventral tegmental area
3076:Eadie–Hofstee diagram
3009:Allosteric regulation
2854:Polymerization motors
2565:Rho family of GTPases
1992:ATP8B1+protein,+human
1677:10.1002/hep.510290214
1477:10.1007/s100240050016
654:endoplasmic reticulum
644:brush border membrane
588:magnesium ion binding
446:epithelium of stomach
245:Chromosome 18 (mouse)
143:Chromosome 18 (human)
3086:Lineweaver–Burk plot
2007:genome location and
1465:Pediatr. Dev. Pathol
1283:10.1093/hmg/4.6.1049
2862:dynamin superfamily
2015:UCSC Genome Browser
1799:2002PNAS...9916899M
553:cardiolipin binding
470:islet of Langerhans
408:subthalamic nucleus
392:trigeminal ganglion
3241:This article on a
3045:Enzyme superfamily
2978:Enzyme promiscuity
1916:10.1101/gr.2596504
1642:10.1101/gr.8.4.354
1590:10.1038/ng1294-380
1391:10.1136/gut.47.1.1
1239:10.1038/ng0398-219
1104:type 1 as well as
865:ENSMUSG00000039529
680:Golgi organization
663:Biological process
602:Cellular component
578:hydrolase activity
548:nucleotide binding
541:Molecular function
396:buccal mucosa cell
107:ATP8B1 - orthologs
3262:
3261:
3201:
3200:
2910:
2909:
2906:
2905:
2407:
2406:
2372:
2371:
2117:Adenylylsulfatase
1960:10.1002/hep.21160
1793:(26): 16899–903.
1703:Physiol. Genomics
1545:Am. J. Dis. Child
1343:10.1002/hep.20285
1084:
1083:
1080:
1079:
1053:
1052:
1018:
1017:
997:
996:
971:
970:
950:
949:
916:
915:
897:
896:
871:
870:
852:
851:
826:
825:
807:
806:
755:
754:
563:metal ion binding
525:
524:
521:
520:
490:
489:
477:
476:
415:
414:
313:
312:
212:
211:
16:(Redirected from
3322:
3283:
3276:
3269:
3237:
3230:
3218:
3217:
3209:
3081:Hanes–Woolf plot
3024:Enzyme activator
3019:Enzyme inhibitor
2993:Enzyme catalysis
2937:
2930:
2923:
2914:
2422:
2152:
2145:
2045:
2038:
2031:
2022:
1980:
1962:
1937:
1927:
1894:
1884:
1869:Gastroenterology
1859:
1830:
1820:
1810:
1777:
1759:
1734:
1697:
1679:
1654:
1644:
1619:
1601:
1568:
1539:
1502:
1496:
1488:
1449:
1448:
1419:
1413:
1412:
1402:
1370:
1364:
1363:
1345:
1327:
1318:
1312:
1311:
1304:
1295:
1294:
1265:
1259:
1258:
1222:
1216:
1215:
1204:
1198:
1197:
1186:
1180:
1170:
1159:
1149:
1066:
1065:
1037:
1030:
1013:
1001:
992:
980:
976:RefSeq (protein)
966:
954:
945:
925:
901:
882:
856:
837:
811:
792:
761:
531:
517:
508:
501:
486:
466:lobe of prostate
426:
424:Top expressed in
419:
364:
362:Top expressed in
357:
336:
319:
309:
296:
285:
270:
263:
257:
246:
234:
218:
208:
195:
184:
169:
162:
156:
145:
131:
115:
109:
60:
53:
32:
21:
3330:
3329:
3325:
3324:
3323:
3321:
3320:
3319:
3290:
3289:
3288:
3287:
3226:
3224:
3212:
3204:
3202:
3197:
3109:Oxidoreductases
3095:
3071:Enzyme kinetics
3059:
3055:List of enzymes
3028:
2997:
2968:Catalytic triad
2946:
2941:
2911:
2902:
2847:
2806:
2557:Ras superfamily
2546:
2530:
2510:
2456:
2446:
2438:
2403:
2368:
2324:
2305:
2251:
2208:Plasma membrane
2171:
2126:
2103:
2077:Pyrophosphatase
2063:
2049:
1988:
1983:
1940:
1910:(10B): 2121–7.
1897:
1862:
1833:
1780:
1737:
1700:
1657:
1622:
1571:
1542:
1505:
1489:
1462:
1458:
1456:Further reading
1453:
1452:
1421:
1420:
1416:
1372:
1371:
1367:
1325:
1320:
1319:
1315:
1306:
1305:
1298:
1267:
1266:
1262:
1224:
1223:
1219:
1206:
1205:
1201:
1188:
1187:
1183:
1171:
1162:
1150:
1141:
1136:
1114:
1075:View/Edit Mouse
1070:View/Edit Human
1033:
1026:
1023:Location (UCSC)
1009:
988:
962:
941:
937:
933:
846:ENSG00000081923
739:
675:lipid transport
658:
629:plasma membrane
619:cell projection
614:Golgi apparatus
597:
568:protein binding
513:
482:
473:
468:
464:
460:
456:
452:
448:
444:
440:
436:
422:
411:
406:
402:
398:
394:
390:
386:
382:
378:
374:
360:
304:
291:
283:
273:
272:
271:
264:
244:
221:Gene location (
203:
190:
182:
172:
171:
170:
163:
141:
118:Gene location (
69:
56:
49:
28:
23:
22:
15:
12:
11:
5:
3328:
3326:
3318:
3317:
3312:
3307:
3302:
3292:
3291:
3286:
3285:
3278:
3271:
3263:
3260:
3259:
3238:
3223:
3222:
3199:
3198:
3196:
3195:
3182:
3169:
3156:
3143:
3130:
3117:
3103:
3101:
3097:
3096:
3094:
3093:
3088:
3083:
3078:
3073:
3067:
3065:
3061:
3060:
3058:
3057:
3052:
3047:
3042:
3036:
3034:
3033:Classification
3030:
3029:
3027:
3026:
3021:
3016:
3011:
3005:
3003:
2999:
2998:
2996:
2995:
2990:
2985:
2980:
2975:
2970:
2965:
2960:
2954:
2952:
2948:
2947:
2942:
2940:
2939:
2932:
2925:
2917:
2908:
2907:
2904:
2903:
2901:
2900:
2895:
2894:
2893:
2888:
2879:
2874:
2869:
2858:
2856:
2849:
2848:
2846:
2845:
2840:
2839:
2838:
2833:
2828:
2817:
2815:
2808:
2807:
2805:
2804:
2799:
2794:
2789:
2784:
2783:
2782:
2777:
2772:
2767:
2757:
2756:
2755:
2750:
2740:
2739:
2738:
2733:
2728:
2716:
2715:
2714:
2713:
2708:
2698:
2697:
2696:
2691:
2686:
2676:
2675:
2674:
2669:
2664:
2654:
2649:
2648:
2647:
2642:
2632:
2631:
2630:
2625:
2620:
2615:
2605:
2604:
2603:
2598:
2588:
2587:
2586:
2581:
2576:
2561:
2559:
2548:
2547:
2545:
2544:
2543:
2542:
2537:
2528:
2524:
2523:
2522:
2517:
2508:
2504:
2503:
2502:
2501:
2500:
2490:
2489:
2488:
2483:
2473:
2468:
2463:
2454:
2450:
2449:
2448:
2444:
2436:
2431:
2429:
2419:
2409:
2408:
2405:
2404:
2402:
2401:
2396:
2391:
2386:
2380:
2378:
2374:
2373:
2370:
2369:
2367:
2366:
2361:
2356:
2351:
2346:
2341:
2335:
2333:
2326:
2325:
2323:
2322:
2316:
2314:
2307:
2306:
2304:
2303:
2298:
2293:
2288:
2283:
2278:
2273:
2268:
2262:
2260:
2253:
2252:
2250:
2249:
2248:
2247:
2242:
2232:
2231:
2230:
2225:
2220:
2215:
2205:
2204:
2203:
2198:
2193:
2182:
2180:
2173:
2172:
2170:
2169:
2164:
2158:
2156:
2155:Cu++ (3.6.3.4)
2149:
2142:
2128:
2127:
2125:
2124:
2119:
2113:
2111:
2105:
2104:
2102:
2101:
2096:
2091:
2090:
2089:
2084:
2073:
2071:
2065:
2064:
2050:
2048:
2047:
2040:
2033:
2025:
2019:
2018:
1999:
1987:
1986:External links
1984:
1982:
1981:
1953:(5): 1125–34.
1938:
1895:
1860:
1831:
1778:
1750:(6): 1337–41.
1735:
1698:
1655:
1620:
1569:
1540:
1503:
1459:
1457:
1454:
1451:
1450:
1414:
1365:
1313:
1296:
1277:(6): 1049–53.
1260:
1217:
1199:
1181:
1160:
1138:
1137:
1135:
1132:
1123:cholangiocytes
1113:
1110:
1082:
1081:
1078:
1077:
1072:
1062:
1061:
1055:
1054:
1051:
1050:
1048:
1046:
1039:
1038:
1031:
1024:
1020:
1019:
1016:
1015:
1005:
1004:
998:
995:
994:
984:
983:
977:
973:
972:
969:
968:
958:
957:
951:
948:
947:
929:
928:
922:
918:
917:
914:
913:
905:
904:
898:
895:
894:
886:
885:
879:
873:
872:
869:
868:
860:
859:
853:
850:
849:
841:
840:
834:
828:
827:
824:
823:
815:
814:
808:
805:
804:
796:
795:
789:
783:
782:
777:
772:
768:
767:
757:
756:
753:
752:
741:
740:
738:
737:
732:
727:
722:
717:
712:
707:
702:
697:
692:
687:
682:
677:
672:
666:
664:
660:
659:
657:
656:
651:
646:
641:
636:
631:
626:
621:
616:
611:
605:
603:
599:
598:
596:
595:
590:
585:
580:
575:
570:
565:
560:
555:
550:
544:
542:
538:
537:
527:
526:
523:
522:
519:
518:
510:
509:
498:
492:
491:
488:
487:
479:
478:
475:
474:
472:
471:
467:
463:
459:
455:
451:
447:
443:
439:
435:
431:
428:
427:
416:
413:
412:
410:
409:
405:
401:
397:
393:
389:
385:
381:
377:
373:
369:
366:
365:
353:
352:
344:
333:
327:
326:
323:RNA expression
315:
314:
311:
310:
302:
298:
297:
289:
286:
281:
275:
274:
265:
258:
252:
248:
247:
242:
236:
235:
227:
226:
214:
213:
210:
209:
201:
197:
196:
188:
185:
180:
174:
173:
164:
157:
151:
147:
146:
139:
133:
132:
124:
123:
111:
110:
67:
63:
62:
54:
46:
45:
41:
40:
37:
36:
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
3327:
3316:
3313:
3311:
3308:
3306:
3303:
3301:
3298:
3297:
3295:
3284:
3279:
3277:
3272:
3270:
3265:
3264:
3258:
3256:
3252:
3248:
3247:chromosome 18
3244:
3239:
3236:
3232:
3227:
3221:
3216:
3211:
3207:
3193:
3189:
3188:
3183:
3180:
3176:
3175:
3170:
3167:
3163:
3162:
3157:
3154:
3150:
3149:
3144:
3141:
3137:
3136:
3131:
3128:
3124:
3123:
3118:
3115:
3111:
3110:
3105:
3104:
3102:
3098:
3092:
3089:
3087:
3084:
3082:
3079:
3077:
3074:
3072:
3069:
3068:
3066:
3062:
3056:
3053:
3051:
3050:Enzyme family
3048:
3046:
3043:
3041:
3038:
3037:
3035:
3031:
3025:
3022:
3020:
3017:
3015:
3014:Cooperativity
3012:
3010:
3007:
3006:
3004:
3000:
2994:
2991:
2989:
2986:
2984:
2981:
2979:
2976:
2974:
2973:Oxyanion hole
2971:
2969:
2966:
2964:
2961:
2959:
2956:
2955:
2953:
2949:
2945:
2938:
2933:
2931:
2926:
2924:
2919:
2918:
2915:
2899:
2896:
2892:
2889:
2887:
2883:
2880:
2878:
2875:
2873:
2870:
2868:
2865:
2864:
2863:
2860:
2859:
2857:
2855:
2850:
2844:
2841:
2837:
2834:
2832:
2829:
2827:
2824:
2823:
2822:
2819:
2818:
2816:
2814:
2809:
2803:
2800:
2798:
2795:
2793:
2790:
2788:
2785:
2781:
2778:
2776:
2773:
2771:
2768:
2766:
2763:
2762:
2761:
2758:
2754:
2751:
2749:
2746:
2745:
2744:
2741:
2737:
2734:
2732:
2729:
2727:
2724:
2723:
2722:
2718:
2717:
2712:
2709:
2707:
2704:
2703:
2702:
2699:
2695:
2692:
2690:
2687:
2685:
2682:
2681:
2680:
2677:
2673:
2670:
2668:
2665:
2663:
2660:
2659:
2658:
2655:
2653:
2650:
2646:
2643:
2641:
2638:
2637:
2636:
2633:
2629:
2626:
2624:
2621:
2619:
2616:
2614:
2611:
2610:
2609:
2606:
2602:
2599:
2597:
2594:
2593:
2592:
2589:
2585:
2582:
2580:
2577:
2575:
2572:
2571:
2570:
2566:
2563:
2562:
2560:
2558:
2554:
2549:
2541:
2538:
2536:
2533:
2532:
2531:
2525:
2521:
2518:
2516:
2513:
2512:
2511:
2505:
2499:
2496:
2495:
2494:
2491:
2487:
2484:
2482:
2479:
2478:
2477:
2474:
2472:
2469:
2467:
2464:
2462:
2459:
2458:
2457:
2451:
2447:
2441:
2440:
2439:
2433:
2432:
2430:
2428:
2423:
2420:
2418:
2414:
2410:
2400:
2397:
2395:
2392:
2390:
2387:
2385:
2382:
2381:
2379:
2375:
2365:
2362:
2360:
2357:
2355:
2352:
2350:
2347:
2345:
2342:
2340:
2337:
2336:
2334:
2332:
2331:P-type ATPase
2327:
2321:
2318:
2317:
2315:
2312:
2308:
2302:
2299:
2297:
2294:
2292:
2289:
2287:
2284:
2282:
2279:
2277:
2274:
2272:
2269:
2267:
2264:
2263:
2261:
2258:
2254:
2246:
2243:
2241:
2238:
2237:
2236:
2233:
2229:
2226:
2224:
2221:
2219:
2216:
2214:
2211:
2210:
2209:
2206:
2202:
2199:
2197:
2194:
2192:
2189:
2188:
2187:
2184:
2183:
2181:
2178:
2174:
2168:
2165:
2163:
2160:
2159:
2157:
2153:
2150:
2146:
2143:
2141:
2137:
2133:
2129:
2123:
2120:
2118:
2115:
2114:
2112:
2110:
2106:
2100:
2097:
2095:
2092:
2088:
2085:
2083:
2080:
2079:
2078:
2075:
2074:
2072:
2070:
2066:
2061:
2057:
2053:
2046:
2041:
2039:
2034:
2032:
2027:
2026:
2023:
2016:
2012:
2011:
2006:
2005:
2000:
1997:
1993:
1990:
1989:
1985:
1978:
1974:
1970:
1966:
1961:
1956:
1952:
1948:
1944:
1939:
1935:
1931:
1926:
1921:
1917:
1913:
1909:
1905:
1901:
1896:
1892:
1888:
1883:
1878:
1875:(3): 756–64.
1874:
1870:
1866:
1861:
1857:
1853:
1849:
1845:
1842:(2): 127–31.
1841:
1837:
1832:
1828:
1824:
1819:
1814:
1809:
1804:
1800:
1796:
1792:
1788:
1784:
1779:
1775:
1771:
1767:
1763:
1758:
1753:
1749:
1745:
1741:
1736:
1732:
1728:
1724:
1720:
1716:
1712:
1709:(3): 139–50.
1708:
1704:
1699:
1695:
1691:
1687:
1683:
1678:
1673:
1669:
1665:
1661:
1656:
1652:
1648:
1643:
1638:
1635:(4): 354–61.
1634:
1630:
1626:
1621:
1617:
1613:
1609:
1605:
1600:
1595:
1591:
1587:
1583:
1579:
1575:
1570:
1566:
1562:
1558:
1554:
1551:(1): 112–24.
1550:
1546:
1541:
1537:
1533:
1529:
1525:
1521:
1517:
1514:(6): 807–17.
1513:
1509:
1504:
1500:
1494:
1486:
1482:
1478:
1474:
1471:(2): 113–25.
1470:
1466:
1461:
1460:
1455:
1446:
1442:
1438:
1434:
1431:(4): 436–43.
1430:
1426:
1418:
1415:
1410:
1406:
1401:
1396:
1392:
1388:
1384:
1380:
1376:
1369:
1366:
1361:
1357:
1353:
1349:
1344:
1339:
1335:
1331:
1324:
1317:
1314:
1309:
1303:
1301:
1297:
1292:
1288:
1284:
1280:
1276:
1272:
1271:Hum Mol Genet
1264:
1261:
1256:
1252:
1248:
1244:
1240:
1236:
1233:(3): 219–24.
1232:
1228:
1221:
1218:
1213:
1209:
1203:
1200:
1195:
1191:
1185:
1182:
1178:
1174:
1169:
1167:
1165:
1161:
1157:
1153:
1148:
1146:
1144:
1140:
1133:
1131:
1128:
1124:
1119:
1111:
1109:
1107:
1103:
1099:
1096:
1092:
1088:
1076:
1071:
1067:
1063:
1060:
1056:
1049:
1047:
1044:
1040:
1036:
1032:
1029:
1025:
1021:
1014:
1012:
1006:
1002:
999:
993:
991:
985:
981:
978:
974:
967:
965:
959:
955:
952:
946:
944:
940:
936:
930:
926:
923:
921:RefSeq (mRNA)
919:
912:
911:
906:
902:
899:
893:
892:
887:
883:
880:
878:
874:
867:
866:
861:
857:
854:
848:
847:
842:
838:
835:
833:
829:
822:
821:
816:
812:
809:
803:
802:
797:
793:
790:
788:
784:
781:
778:
776:
773:
769:
766:
762:
758:
751:
747:
742:
736:
733:
731:
728:
726:
723:
721:
718:
716:
713:
711:
708:
706:
703:
701:
698:
696:
693:
691:
688:
686:
683:
681:
678:
676:
673:
671:
668:
667:
665:
662:
661:
655:
652:
650:
647:
645:
642:
640:
637:
635:
632:
630:
627:
625:
622:
620:
617:
615:
612:
610:
607:
606:
604:
601:
600:
594:
591:
589:
586:
584:
581:
579:
576:
574:
571:
569:
566:
564:
561:
559:
556:
554:
551:
549:
546:
545:
543:
540:
539:
536:
535:Gene ontology
532:
528:
516:
511:
507:
502:
499:
497:
493:
485:
480:
469:
465:
461:
457:
453:
449:
445:
441:
437:
433:
432:
429:
425:
420:
417:
407:
403:
399:
395:
391:
387:
383:
380:renal medulla
379:
375:
371:
370:
367:
363:
358:
355:
354:
351:
349:
345:
343:
342:
338:
337:
334:
332:
328:
324:
320:
316:
308:
303:
299:
295:
290:
284:18|18 E1
280:
276:
269:
262:
256:
249:
241:
237:
233:
228:
224:
219:
215:
207:
202:
198:
194:
189:
179:
175:
168:
161:
155:
148:
144:
138:
134:
130:
125:
121:
116:
112:
108:
104:
100:
96:
92:
88:
84:
80:
76:
72:
64:
59:
52:
47:
42:
38:
33:
30:
19:
3255:expanding it
3240:
3225:
3187:Translocases
3184:
3171:
3158:
3145:
3132:
3122:Transferases
3119:
3106:
2963:Binding site
2700:
2634:
2590:
2568:
2553:Small GTPase
2167:Wilson/ATP7B
2162:Menkes/ATP7A
2009:
2003:
1950:
1946:
1907:
1903:
1872:
1868:
1839:
1835:
1790:
1786:
1747:
1743:
1706:
1702:
1670:(2): 506–8.
1667:
1663:
1632:
1628:
1584:(4): 380–6.
1581:
1577:
1548:
1544:
1511:
1507:
1493:cite journal
1468:
1464:
1428:
1424:
1417:
1382:
1378:
1368:
1336:(1): 27–38.
1333:
1329:
1316:
1274:
1270:
1263:
1230:
1226:
1220:
1211:
1202:
1193:
1184:
1115:
1094:
1086:
1085:
1011:NP_001001488
1008:
987:
964:NM_001001488
961:
943:NM_001374386
939:NM_001374385
932:
908:
889:
863:
844:
818:
799:
779:
774:
639:stereocilium
346:
339:
66:External IDs
29:
2958:Active site
2877:Mitofusin-1
2852:3.6.5.5-6:
2821:Prokaryotic
1127:hepatocytes
583:ATP binding
305:64,794,338
292:64,662,038
204:57,803,315
191:57,646,426
44:Identifiers
3294:Categories
3161:Isomerases
3135:Hydrolases
3002:Regulation
2843:Eukaryotic
2476:Transducin
2313:(3.6.3.10)
2052:Hydrolases
1947:Hepatology
1904:Genome Res
1744:Hepatology
1664:Hepatology
1629:Genome Res
1599:1765/55192
1578:Nat. Genet
1425:J. Hepatol
1385:(1): 1–5.
1330:Hepatology
1179:, May 2017
1158:, May 2017
1134:References
442:left colon
350:(ortholog)
87:HomoloGene
3245:on human
3040:EC number
2811:3.6.5.3:
2551:3.6.5.2:
2493:Gustducin
2425:3.6.5.1:
2259:(3.6.3.9)
2179:(3.6.3.8)
2082:Inorganic
1227:Nat Genet
990:NP_005594
935:NM_005603
765:Orthologs
95:GeneCards
3305:EC 7.6.2
3064:Kinetics
2988:Cofactor
2951:Activity
2087:Thiamine
1977:19185288
1969:16628629
1934:15489334
1891:14988830
1856:12880872
1827:12477932
1774:31234172
1766:11093741
1723:11015572
1694:37536841
1536:21480442
1528:15946126
1485:10679031
1445:11682026
1409:10861251
1360:45979358
1352:15239083
1175:–
1154:–
1112:Function
1059:Wikidata
744:Sources:
624:membrane
183:18q21.31
3220:Biology
3174:Ligases
2944:Enzymes
2898:Tubulin
2867:Dynamin
2719:other:
2399:Katanin
2389:Kinesin
2364:ATP13A3
2359:ATP13A2
2094:Apyrase
1795:Bibcode
1686:9918928
1651:9548971
1616:8131209
1608:7894490
1565:5762004
1400:1727973
1291:7655458
1255:9897047
1247:9500542
1177:Ensembl
1156:Ensembl
877:UniProt
832:Ensembl
771:Species
750:QuickGO
700:hearing
454:jejunum
438:decidua
384:pylorus
325:pattern
83:1859665
51:Aliases
3206:Portal
3148:Lyases
2775:ARL13B
2635:RhoBTB
2529:α12/13
2417:GTPase
2394:Myosin
2384:Dynein
2354:ATP12A
2349:ATP11B
2344:ATP10A
2339:ATP8B1
2329:Other
2301:ATP1B4
2296:ATP1B3
2291:ATP1B2
2286:ATP1B1
2281:ATP1A4
2276:ATP1A3
2271:ATP1A2
2266:ATP1A1
2257:Na+/K+
2245:ATP2C2
2240:ATP2C1
2228:ATP2B4
2223:ATP2B3
2218:ATP2B2
2213:ATP2B1
2201:ATP2A3
2196:ATP2A2
2191:ATP2A1
2140:ATPase
2010:ATP8B1
2004:ATP8B1
2001:Human
1998:(MeSH)
1975:
1967:
1932:
1925:528928
1922:
1889:
1854:
1825:
1818:139241
1815:
1772:
1764:
1731:762447
1729:
1721:
1692:
1684:
1649:
1614:
1606:
1563:
1534:
1526:
1483:
1443:
1407:
1397:
1358:
1350:
1289:
1253:
1245:
1118:P-type
1095:ATP8B1
1091:enzyme
1089:is an
1045:search
1043:PubMed
910:Q148W0
891:O43520
787:Entrez
496:BioGPS
458:cervix
376:nipple
372:cardia
99:ATP8B1
75:602397
58:ATP8B1
35:ATP8B1
18:ATP8B1
3249:is a
3100:Types
2831:EF-Tu
2770:SAR1B
2753:RAB27
2748:RAB23
2701:RhoDF
2591:RhoUV
2574:CDC42
2569:Cdc42
2555:>
2540:GNA13
2535:GNA12
2520:GNA11
2509:αq/11
2498:GNAT3
2486:GNAT2
2481:GNAT1
2471:GNAI3
2466:GNAI2
2461:GNAI1
2413:3.6.5
2377:3.6.4
2320:ATP4A
2311:H+/K+
2186:SERCA
2148:3.6.3
2132:3.6.3
2109:3.6.2
2069:3.6.1
1973:S2CID
1770:S2CID
1727:S2CID
1690:S2CID
1612:S2CID
1532:S2CID
1356:S2CID
1326:(PDF)
1251:S2CID
820:54670
780:Mouse
775:Human
746:Amigo
462:ileum
348:Mouse
341:Human
288:Start
223:Mouse
187:Start
120:Human
91:21151
3251:stub
3243:gene
3192:list
3185:EC7
3179:list
3172:EC6
3166:list
3159:EC5
3153:list
3146:EC4
3140:list
3133:EC3
3127:list
3120:EC2
3114:list
3107:EC1
2891:OPA1
2884:and
2836:EF-G
2826:IF-2
2792:Rheb
2780:ARL6
2765:ARF6
2736:NRAS
2731:KRAS
2726:HRAS
2711:RhoD
2706:RhoF
2652:RhoH
2628:RhoG
2613:Rac1
2601:RhoV
2596:RhoU
2579:TC10
2515:GNAQ
2235:SPCA
2062:3.6)
1965:PMID
1930:PMID
1887:PMID
1852:PMID
1840:1633
1823:PMID
1762:PMID
1719:PMID
1682:PMID
1647:PMID
1604:PMID
1561:PMID
1524:PMID
1499:link
1481:PMID
1441:PMID
1405:PMID
1348:PMID
1287:PMID
1243:PMID
1098:gene
801:5205
331:Bgee
279:Band
240:Chr.
178:Band
137:Chr.
71:OMIM
2886:MX2
2882:MX1
2802:RGK
2797:Rap
2787:Ran
2760:Arf
2743:Rab
2721:Ras
2679:Rnd
2657:Rho
2608:Rac
2584:TCL
2445:olf
2177:Ca+
1955:doi
1920:PMC
1912:doi
1877:doi
1873:126
1844:doi
1813:PMC
1803:doi
1752:doi
1711:doi
1672:doi
1637:doi
1594:hdl
1586:doi
1553:doi
1549:117
1516:doi
1473:doi
1433:doi
1395:PMC
1387:doi
1379:Gut
1338:doi
1279:doi
1235:doi
301:End
200:End
103:OMA
79:MGI
3296::
2567::
2455:αi
2437:αs
2415::
2138::
2060:EC
2054::
1971:.
1963:.
1951:43
1949:.
1945:.
1928:.
1918:.
1908:14
1906:.
1902:.
1885:.
1871:.
1867:.
1850:.
1838:.
1821:.
1811:.
1801:.
1791:99
1789:.
1785:.
1768:.
1760:.
1748:32
1746:.
1742:.
1725:.
1717:.
1705:.
1688:.
1680:.
1668:29
1666:.
1662:.
1645:.
1631:.
1627:.
1610:.
1602:.
1592:.
1580:.
1576:.
1559:.
1547:.
1530:.
1522:.
1512:20
1510:.
1495:}}
1491:{{
1467:.
1439:.
1429:35
1427:.
1403:.
1393:.
1383:47
1381:.
1377:.
1354:.
1346:.
1334:40
1332:.
1328:.
1299:^
1285:.
1273:.
1249:.
1241:.
1231:18
1229:.
1210:.
1192:.
1163:^
1142:^
1108:.
748:/
307:bp
294:bp
206:bp
193:bp
101:;
97::
93:;
89::
85:;
81::
77:;
73::
3282:e
3275:t
3268:v
3257:.
3208::
3194:)
3190:(
3181:)
3177:(
3168:)
3164:(
3155:)
3151:(
3142:)
3138:(
3129:)
3125:(
3116:)
3112:(
2936:e
2929:t
2922:v
2694:3
2689:2
2684:1
2672:C
2667:B
2662:A
2645:2
2640:1
2623:3
2618:2
2527:G
2507:G
2453:G
2443:G
2435:G
2136:4
2134:-
2058:(
2044:e
2037:t
2030:v
2017:.
1979:.
1957::
1936:.
1914::
1893:.
1879::
1858:.
1846::
1829:.
1805::
1797::
1776:.
1754::
1733:.
1713::
1707:1
1696:.
1674::
1653:.
1639::
1633:8
1618:.
1596::
1588::
1582:8
1567:.
1555::
1538:.
1518::
1501:)
1487:.
1475::
1469:3
1447:.
1435::
1411:.
1389::
1362:.
1340::
1310:.
1293:.
1281::
1275:4
1257:.
1237::
1214:.
1196:.
225:)
122:)
105::
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.