720:
743:
778:) can fold and unfold in a "non-cooperative manner." Binding of the substrates causes preference for 'closed' conformations amongst those that are sampled by ADK. These 'closed' conformations are hypothesized to help with removal of water from the active site to avoid wasteful hydrolysis of ATP in addition to helping optimize alignment of substrates for phosphoryl-transfer. Furthermore, it has been shown that the apoenzyme will still sample the 'closed' conformations of the ATP
29:
635:
are known to be related to the ADK family, i.e. yeast uridine monophosphokinase and slime mold UMP-CMP kinase. Some residues are conserved across these isoforms, indicating how essential they are for catalysis. One of the most conserved areas includes an Arg residue, whose modification inactivates the enzyme, together with an Asp that resides in the catalytic cleft of the enzyme and participates in a salt bridge.
4077:
801:
the cellular level. As energy levels change under different metabolic stresses adenylate kinase is then able to generate AMP; which itself acts as a signaling molecule in further signaling cascades. This generated AMP can, for example, stimulate various AMP-dependent receptors such as those involved in glycolytic pathways, K-ATP channels, and 5' AMP-activated protein kinase (
542:
215:
698:
Arg119, which lies in the adenosine binding region of the ADK, and acts to sandwich the adenine in the active site. It has been suggested that the promiscuity of these enzymes in accepting other NTP's is due to this relatively inconsequential interactions of the base in the ATP binding pocket. A network of positive,
739:, aggregate, and perform mechanical work. Large conformational changes in proteins play an important role in cellular signaling. Adenylate Kinase is a signal transducing protein; thus, the balance between conformations regulates protein activity. ADK has a locally unfolded state that becomes depopulated upon binding.
692:
Phosphoryl transfer only occurs on closing of the 'open lid'. This causes an exclusion of water molecules that brings the substrates in proximity to each other, lowering the energy barrier for the nucleophilic attack by the α-phosphoryl of AMP on the γ-phosphoryl group of ATP resulting in formation
615:
of cells; and ADK7 is found in skeletal muscle whereas ADK8 is not. Not only do the locations of the various isoforms within the cell vary, but the binding of substrate to the enzyme and kinetics of the phosphoryl transfer are different as well. ADK1, the most abundant cytosolic ADK isozyme, has a
750:
A 2007 study by
Whitford et al. shows the conformations of ADK when binding with ATP or AMP. The study shows that there are three relevant conformations or structures of ADK—CORE, Open, and Closed. In ADK, there are two small domains called the LID and NMP. ATP binds in the pocket formed by the LID
634:
Some of these isoforms prefer other NTP's entirely. There is a mitochondrial GTP:AMP phosphotransferase, also specific for the phosphorylation of AMP, that can only use GTP or ITP as the phosphoryl donor. ADK has also been identified in different bacterial species and in yeast. Two further enzymes
800:
The ability for a cell to dynamically measure energetic levels provides it with a method to monitor metabolic processes. By continually monitoring and altering the levels of ATP and the other adenyl phosphates (ADP and AMP levels) adenylate kinase is an important regulator of energy expenditure at
697:
with inhibitor Ap5A, the Arg88 residue binds the Ap5A at the α-phosphate group. It has been shown that the mutation R88G results in 99% loss of catalytic activity of this enzyme, suggesting that this residue is intimately involved in the phosphoryl transfer. Another highly conserved residue is
1831:
Lagresle-Peyrou C, Six EM, Picard C, Rieux-Laucat F, Michel V, Ditadi A, Demerens-de
Chappedelaine C, Morillon E, Valensi F, Simon-Stoos KL, Mullikin JC, Noroski LM, Besse C, Wulffraat NM, Ferster A, Abecasis MM, Calvo F, Petit C, Candotti F, Abel L, Fischer A, Cavazzana-Calvo M (January 2009).
813:
Adenylate kinase is present in mitochondrial and myofibrillar compartments in the cell, and it makes two high-energy phosphoryls (β and γ) of ATP available to be transferred between adenine nucleotide molecules. In essence, adenylate kinase shuttles ATP to sites of high energy consumption and
805:). Common factors that influence adenine nucleotide levels, and therefore ADK activity are exercise, stress, changes in hormone levels, and diet. It facilitates decoding of cellular information by catalyzing nucleotide exchange in the intimate “sensing zone” of metabolic sensors.
883:. It is also characterized by an impaired lymphoid maturation and early differentiation arrest in the myeloid lineage. AK2 deficiency results in absent or a large decrease in the expression of proteins. AK2 is specifically expressed in the stria vascularis of the
751:
and CORE domains. AMP binds in the pocket formed by the NMP and CORE domains. The
Whitford study also reported findings that show that localized regions of a protein unfold during conformational transitions. This mechanism reduces the
854:
Knock out of AK1 disrupts the synchrony between inorganic phosphate and turnover at ATP-consuming sites and ATP synthesis sites. This reduces the energetic signal communication in the post-ischemic heart and precipitates inadequate
814:
removes the AMP generated over the course of those reactions. These sequential phosphotransfer relays ultimately result in propagation of the phosphoryl groups along collections of ADK molecules. This process can be thought of as a
786:
domains in the absence of substrates. When comparing the rate of opening of the enzyme (which allows for product release) and the rate of closing that accompanies substrate binding, closing was found to be the slower process.
3604:
818:
of ADK molecules that results in changes in local intracellular metabolic flux without apparent global changes in metabolite concentrations. This process is extremely important for overall homeostasis of the cell.
706:) stabilize the buildup of negative charge on phosphoryl group during the transfer. Two distal aspartate residues bind to the arginine network, causing the enzyme to fold and reduces its flexibility. A magnesium
197:
that catalyzes the interconversion of the various adenosine phosphates (ATP, ADP, and AMP). By constantly monitoring phosphate nucleotide levels inside the cell, ADK plays an important role in cellular energy
574:
The equilibrium constant varies with condition, but it is close to 1. Thus, ΔG for this reaction is close to zero. In muscle from a variety of species of vertebrates and invertebrates, the concentration of
1202:
Henzler-Wildman KA, Thai V, Lei M, Ott M, Wolf-Watz M, Fenn T, Pozharski E, Wilson MA, Petsko GA, Karplus M, Hübner CG, Kern D (December 2007). "Intrinsic motions along an enzymatic reaction trajectory".
710:
is also required, essential for increasing the electrophilicity of the phosphate on AMP, though this magnesium ion is only held in the active pocket by electrostatic interactions and dissociates easily.
3614:
3609:
1075:"The characterization of human adenylate kinases 7 and 8 demonstrates differences in kinetic parameters and structural organization among the family of adenylate kinase isoenzymes"
719:
3729:
3707:
631:
in the protein. Each isoform also has different preference for NTP's. Some will only use ATP, whereas others will accept GTP, UTP, and CTP as the phosphoryl carrier.
2029:
3525:
611:
identified. While some of these are ubiquitous throughout the body, some are localized into specific tissues. For example, ADK7 and ADK8 are both only found in the
326:
140:
505:
1397:"Rational modulation of conformational fluctuations in adenylate kinase reveals a local unfolding mechanism for allostery and functional adaptation in proteins"
524:
3635:
2510:
3530:
3697:
2505:
2596:
2178:
895:
AK1 genetic ablation decreases tolerance to metabolic stress. AK1 deficiency induces fiber-type specific variation in groups of transcripts in
2264:
3336:
1506:
Rundqvist L, Adén J, Sparrman T, Wallgren M, Olsson U, Wolf-Watz M (March 2009). "Noncooperative folding of subdomains in adenylate kinase".
274:
88:
2073:
2022:
1934:"Deficiency of a plastidial adenylate kinase in Arabidopsis results in elevated photosynthetic amino acid biosynthesis and enhanced growth"
988:"The contents of adenine nucleotides, phosphagens and some glycolytic intermediates in resting muscles from vertebrates and invertebrates"
3470:
2364:
2299:
545:
PDB image 3HPQ showing the ADK enzyme skeleton in cartoon and the key residues as sticks and labeled according to their placement in
3796:
3040:
2834:
2309:
2134:
627:
of ADK7 and 8, indicating a much weaker binding of ADK1 to AMP. Sub-cellular localization of the ADK enzymes is done by including a
935:
3181:
2635:
2433:
2068:
1164:
Cooper AJ, Friedberg EC (May 1992). "A putative second adenylate kinase-encoding gene from the yeast
Saccharomyces cerevisiae".
517:
2802:
2630:
2294:
2015:
2625:
2411:
2369:
460:
3952:
1074:
617:
484:
346:
160:
4097:
3754:
3749:
3309:
3127:
843:
4067:
3619:
3507:
3475:
3460:
2215:
2157:
2047:
1661:"Developmental enhancement of adenylate kinase-AMPK metabolic signaling axis supports stem cell cardiac differentiation"
802:
1779:"Defective metabolic signaling in adenylate kinase AK1 gene knock-out hearts compromises post-ischemic coronary reflow"
3142:
2269:
910:
Enhanced growth and elevated photosynthetic amino acid is associated with plastidial adenylate kinase deficiency in
2443:
2438:
2242:
2108:
1834:"Human adenylate kinase 2 deficiency causes a profound hematopoietic defect associated with sensorineural deafness"
3937:
1932:
Carrari F, Coll-Garcia D, Schauer N, Lytovchenko A, Palacios-Rojas N, Balbo I, Rosso M, Fernie AR (January 2005).
846:, adenylate kinase performed dual enzymatic functions. ADK complements nucleoside diphosphate kinase deficiency.
4053:
4040:
4027:
4014:
4001:
3988:
3975:
3740:
3718:
3693:
3668:
3588:
3196:
3156:
3101:
3078:
3050:
3018:
2884:
2168:
1607:"Adenylate kinase and AMP signaling networks: metabolic monitoring, signal communication and body energy sensing"
672:
644:
596:
588:
3947:
2971:
1883:"Adenylate kinase 1 deficiency induces molecular and structural adaptations to support muscle energy metabolism"
3901:
3844:
3205:
3186:
3117:
2851:
2359:
2237:
1987:
478:
371:
176:
755:
and enhances catalytic efficiency. Local unfolding is the result of competing strain energies in the protein.
334:
148:
1254:
Reinstein J, Gilles AM, Rose T, Wittinghofer A, Saint Girons I, Bârzu O, Surewicz WK, Mantsch HH (May 1989).
3849:
3063:
2406:
2193:
707:
584:
565:
465:
3517:
3290:
3200:
2401:
2058:
872:
592:
576:
561:
3870:
3789:
3371:
3326:
3028:
3013:
2949:
2929:
2827:
2775:
2515:
2391:
2227:
2205:
580:
569:
529:
3942:
453:
330:
144:
2812:
742:
693:
of ADP by transfer of the γ-phosphoryl group to AMP. In the crystal structure of the ADK enzyme from
3285:
3280:
3160:
3023:
3003:
2912:
2616:
2522:
2379:
2328:
2274:
1731:
1672:
1554:
1543:"Overlap between folding and functional energy landscapes for adenylate kinase conformational change"
1408:
1212:
912:
876:
856:
287:
101:
658:
3906:
3548:
3540:
3321:
2988:
2924:
2888:
1038:
Panayiotou C, Solaroli N, Karlsson A (April 2014). "The many isoforms of human adenylate kinases".
833:
388:
383:
481:
33:
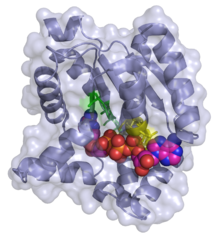
3D ribbon/surface model of adenylate kinase in complex with bis(adenosine)tetraphosphate (ADP-ADP)
3839:
3411:
3349:
3300:
3035:
2862:
2546:
2314:
2188:
1236:
1105:
699:
628:
405:
191:
1720:"Adenylate kinase complements nucleoside diphosphate kinase deficiency in nucleotide metabolism"
1306:
and the inhibitor Ap5A refined at 1.9 A resolution. A model for a catalytic transition state".
4102:
3426:
3416:
3406:
3331:
3315:
3274:
3260:
3255:
2993:
2956:
2536:
2339:
2120:
1963:
1914:
1863:
1810:
1759:
1700:
1638:
1572:
1523:
1485:
1436:
1377:
1323:
1281:
1228:
1181:
1146:
1097:
1055:
1017:
967:
472:
321:
135:
3885:
3880:
3854:
3782:
3759:
3421:
3401:
3343:
3239:
3137:
3068:
2966:
2961:
2820:
2349:
1983:
1953:
1945:
1904:
1894:
1853:
1845:
1800:
1790:
1749:
1739:
1690:
1680:
1628:
1618:
1562:
1515:
1475:
1467:
1426:
1416:
1367:
1359:
1315:
1271:
1220:
1173:
1136:
1089:
1047:
1007:
999:
957:
880:
838:
752:
1302:
Müller CW, Schulz GE (March 1992). "Structure of the complex between adenylate kinase from
832:
Nucleoside diphosphate (NDP) kinase catalyzes in vivo ATP-dependent synthesis of ribo- and
313:
127:
3932:
3916:
3829:
3682:
3672:
3494:
3465:
3091:
2998:
2983:
2254:
2147:
939:
746:
Describes the generic kinetic cycle of the ADK enzyme family. Ternary complex is labeled.
608:
441:
1881:
Janssen E, de Groof A, Wijers M, Fransen J, Dzeja PP, Terzic A, Wieringa B (April 2003).
887:
which indicates why individuals with an AK2 deficiency will have sensorineural deafness.
1735:
1676:
1558:
1456:"Many local motions cooperate to produce the adenylate kinase conformational transition"
1412:
1216:
946:"Thermodynamics of enzyme-catalyzed reactions--a database for quantitative biochemistry"
932:
417:
4081:
3970:
3911:
3650:
3489:
3396:
3388:
3265:
3228:
1858:
1833:
1805:
1778:
1695:
1660:
1633:
1606:
1480:
1455:
1431:
1396:
1372:
1347:
1141:
1125:"Mitochondrial GTP-AMP phosphotransferase. 2. Kinetic and equilibrium dialysis studies"
1124:
1012:
987:
815:
500:
376:
1958:
1933:
1276:
1255:
28:
4091:
3875:
3834:
2919:
2038:
1754:
1719:
1260:
adenylate kinase as evidenced by chemical modification and site-directed mutagenesis"
1177:
279:
93:
1109:
962:
945:
243:
57:
3824:
3008:
2978:
2907:
2671:
2646:
2304:
2183:
2096:
1240:
591:
is controlled by the availability of ADP. Thus, the mitochondrion attempts to keep
309:
123:
766:
has been shown to be significantly lower when compared with the CORE domain in ADK
2003:
1685:
1051:
680:
666:
652:
255:
69:
4048:
3983:
3819:
3574:
3564:
3165:
2902:
2843:
199:
4076:
1724:
Proceedings of the
National Academy of Sciences of the United States of America
1401:
Proceedings of the
National Academy of Sciences of the United States of America
595:
levels high due to the combined action of adenylate kinase and the controls on
3376:
3219:
3132:
2897:
2847:
2423:
2085:
1471:
1363:
896:
4022:
3996:
3655:
3568:
3105:
2871:
2354:
1744:
1421:
884:
736:
1967:
1918:
1899:
1882:
1867:
1814:
1795:
1704:
1642:
1576:
1527:
1489:
1440:
1381:
1232:
1101:
1059:
971:
541:
1949:
1763:
1327:
1319:
1285:
1185:
1021:
283:
214:
97:
3502:
3082:
2500:
2374:
2007:
1999:
1623:
1150:
1073:
Panayiotou C, Solaroli N, Xu Y, Johansson M, Karlsson A (February 2011).
675:
661:
647:
250:
64:
1224:
976:, gives equilibrium constants, search for adenylate kinase under enzymes
758:
The local (thermodynamic) stability of the substrate-binding domains ATP
3453:
3438:
3054:
2660:
2655:
2575:
1909:
1567:
1542:
1348:"Conformational transitions of adenylate kinase: switching by cracking"
1093:
899:
and mitochondrial metabolism. This supports muscle energy metabolism.
732:
612:
448:
429:
267:
262:
81:
76:
1519:
1003:
179:
4035:
3805:
3431:
2866:
2765:
2760:
2730:
2725:
2605:
2585:
2580:
2570:
2565:
2560:
2555:
512:
424:
412:
400:
341:
194:
155:
871:) deficiency in humans causes hematopoietic defects associated with
1849:
4009:
3448:
3443:
3295:
2785:
2780:
2770:
2755:
2750:
2745:
2740:
2735:
2720:
2715:
2710:
2705:
2700:
2695:
2690:
2685:
2680:
2476:
2463:
741:
718:
540:
3605:
CDP-diacylglycerol—glycerol-3-phosphate 3-phosphatidyltransferase
1659:
Dzeja PP, Chung S, Faustino RS, Behfar A, Terzic A (April 2011).
3359:
3354:
3250:
3245:
2944:
2939:
2934:
1995:
1777:
Dzeja PP, Bast P, Pucar D, Wieringa B, Terzic A (October 2007).
931:
The NIST Thermodynamics of Enzyme-Catalyzed
Reactions database,
436:
303:
238:
117:
52:
3778:
2816:
2011:
868:
770:. Furthermore, it has been shown that the two subdomains (ATP
1040:
1346:
Whitford PC, Miyashita O, Levy Y, Onuchic JN (March 2007).
3774:
1197:
1195:
702:
residues (Lys13, Arg123, Arg156, and Arg167 in ADK from
1994:
This article incorporates text from the public domain
4065:
3615:
CDP-diacylglycerol—inositol 3-phosphatidyltransferase
933:
http://xpdb.nist.gov/enzyme_thermodynamics/enzyme1.pl
731:
Flexibility and plasticity allow proteins to bind to
3620:
CDP-diacylglycerol—choline O-phosphatidyltransferase
3961:
3925:
3894:
3863:
3812:
3739:
3717:
3692:
3667:
3648:
3628:
3610:
CDP-diacylglycerol—serine O-phosphatidyltransferase
3597:
3587:
3557:
3539:
3516:
3488:
3387:
3227:
3218:
3195:
3155:
3100:
3077:
3049:
2883:
2859:
2669:
2644:
2614:
2594:
2544:
2535:
2489:
2452:
2420:
2388:
2336:
2327:
2283:
2251:
2224:
2202:
2165:
2156:
2133:
2105:
2082:
2055:
2046:
523:
511:
499:
494:
471:
459:
447:
435:
423:
411:
399:
394:
382:
370:
365:
360:
340:
320:
302:
297:
273:
261:
249:
237:
229:
224:
207:
154:
134:
116:
111:
87:
75:
63:
51:
43:
38:
21:
1256:"Structural and catalytic role of arginine 88 in
944:Goldberg RN, Tewari YB, Bhat TN (November 2004).
879:is an autosomal recessive form of human combined
1395:Schrank TP, Bolen DW, Hilser VJ (October 2009).
1826:
1824:
3790:
2828:
2023:
583:, and usually greater than 100 times that of
8:
219:Bacillus stearothermophilus adenylate kinase
3636:N-acetylglucosamine-1-phosphate transferase
3526:UTP—glucose-1-phosphate uridylyltransferase
1611:International Journal of Molecular Sciences
3797:
3783:
3775:
3664:
3594:
3224:
3215:
2880:
2835:
2821:
2813:
2541:
2511:Mitochondrial permeability transition pore
2493:
2333:
2162:
2052:
2030:
2016:
2008:
1454:Daily MD, Phillips GN, Cui Q (July 2010).
491:
294:
213:
108:
27:
3730:serine/threonine-specific protein kinases
3708:serine/threonine-specific protein kinases
3531:Galactose-1-phosphate uridylyltransferase
1986:at the U.S. National Library of Medicine
1957:
1908:
1898:
1857:
1804:
1794:
1753:
1743:
1694:
1684:
1632:
1622:
1566:
1479:
1430:
1420:
1371:
1275:
1140:
1011:
961:
2506:Mitochondrial membrane transport protein
828:Nucleoside diphosphate kinase deficiency
623:about a thousand times higher than the K
4072:
1541:Olsson U, Wolf-Watz M (November 2010).
1123:Tomasselli AG, Noda LH (January 1979).
924:
859:reflow following ischemia-reperfusion.
607:To date there have been nine human ADK
2265:Cholesterol side-chain cleavage enzyme
357:
204:
18:
1654:
1652:
1600:
1598:
1596:
1594:
1592:
1590:
1588:
1586:
1501:
1499:
1341:
1339:
1337:
986:Beis I, Newsholme EA (October 1975).
850:AK1 and post-ischemic coronary reflow
7:
1297:
1295:
1033:
1031:
2179:Coenzyme Q – cytochrome c reductase
1887:The Journal of Biological Chemistry
1783:The Journal of Biological Chemistry
1264:The Journal of Biological Chemistry
549:, crystallized with Ap5A inhibitor.
2365:Oxoglutarate dehydrogenase complex
2300:Glycerol-3-phosphate dehydrogenase
1142:10.1111/j.1432-1033.1979.tb12819.x
14:
3041:Glucose-1,6-bisphosphate synthase
2310:Carnitine palmitoyltransferase II
4075:
3182:Ribose-phosphate diphosphokinase
2434:Carbamoyl phosphate synthetase I
2074:Long-chain-fatty-acid—CoA ligase
2069:Carnitine palmitoyltransferase I
1605:Dzeja P, Terzic A (April 2009).
1129:European Journal of Biochemistry
579:is typically 7-10 times that of
2295:Glutamate aspartate transporter
2412:Pyruvate dehydrogenase complex
2370:Succinyl coenzyme A synthetase
727:involved in substrate binding
1:
3755:Protein-histidine tele-kinase
3750:Protein-histidine pros-kinase
3629:Glycosyl-1-phosphotransferase
1277:10.1016/S0021-9258(18)83156-8
963:10.1093/bioinformatics/bth314
903:Plastidial ADK deficiency in
844:nucleoside diphosphate kinase
298:Available protein structures:
112:Available protein structures:
3461:RNA-dependent RNA polymerase
2216:Dihydroorotate dehydrogenase
1718:Lu Q, Inouye M (June 1996).
1686:10.1371/journal.pone.0019300
1460:Journal of Molecular Biology
1352:Journal of Molecular Biology
1308:Journal of Molecular Biology
1178:10.1016/0378-1119(92)90721-Z
1052:10.1016/j.biocel.2014.01.014
3368:RNA-directed DNA polymerase
3236:DNA-directed DNA polymerase
2270:Steroid 11-beta-hydroxylase
836:triphosphates. In mutated
673:Adenylate kinase, isozyme 1
645:Adenylate kinase, subfamily
558:The reaction catalyzed is:
4119:
3721:: protein-dual-specificity
2444:N-Acetylglutamate synthase
2439:Ornithine transcarbamylase
2243:Glycerol phosphate shuttle
2109:monoamine neurotransmitter
1993:
3953:Michaelis–Menten kinetics
2798:
2496:
2472:
2169:oxidative phosphorylation
1472:10.1016/j.jmb.2010.05.015
1364:10.1016/j.jmb.2006.11.085
597:oxidative phosphorylation
589:oxidative phosphorylation
490:
293:
212:
107:
26:
3845:Diffusion-limited enzyme
3698:protein-serine/threonine
3598:Phosphatidyltransferases
3187:Thiamine diphosphokinase
2360:Isocitrate dehydrogenase
2238:Malate-aspartate shuttle
1988:Medical Subject Headings
2407:Glutamate dehydrogenase
2194:Succinate dehydrogenase
1745:10.1073/pnas.93.12.5720
1422:10.1073/pnas.0906510106
1082:The Biochemical Journal
992:The Biochemical Journal
3518:Nucleotidyltransferase
3201:nucleotidyltransferase
3128:Nucleoside-diphosphate
2803:mitochondrial diseases
2402:Aspartate transaminase
2059:fatty acid degradation
1900:10.1074/jbc.M211465200
1796:10.1074/jbc.M705268200
891:Structural adaptations
873:sensorineural deafness
747:
728:
554:Substrate and products
550:
3938:Eadie–Hofstee diagram
3871:Allosteric regulation
3372:Reverse transcriptase
2516:Mitochondrial carrier
2392:anaplerotic reactions
2228:mitochondrial shuttle
2206:pyrimidine metabolism
1950:10.1104/pp.104.056143
1547:Nature Communications
842:that had a disrupted
745:
722:
544:
4098:Cellular respiration
3948:Lineweaver–Burk plot
3161:diphosphotransferase
3143:Thiamine-diphosphate
2850:-containing groups (
2523:Translocator protein
2380:Malate dehydrogenase
2275:Aldosterone synthase
1624:10.3390/ijms10041729
913:Arabidopsis thaliana
905:Arabidopsis thaliana
877:Reticular dysgenesis
867:Adenylate Kinase 2 (
796:Metabolic monitoring
3743:: protein-histidine
3661:; protein acceptor)
3549:mRNA capping enzyme
3541:Guanylyltransferase
2135:Intermembrane space
1736:1996PNAS...93.5720L
1677:2011PLoSO...619300D
1559:2010NatCo...1..111O
1413:2009PNAS..10616984S
1320:10.2210/pdb1ake/pdb
1225:10.1038/nature06410
1217:2007Natur.450..838H
834:deoxyribonucleoside
3907:Enzyme superfamily
3840:Enzyme promiscuity
3019:Phosphoinositide 3
2863:phosphotransferase
2490:Other/to be sorted
2455:alcohol metabolism
2315:Uncoupling protein
2189:NADH dehydrogenase
1568:10.1038/ncomms1106
1094:10.1042/BJ20101443
938:2016-11-09 at the
748:
729:
629:targeting sequence
551:
192:phosphotransferase
4063:
4062:
3772:
3771:
3768:
3767:
3644:
3643:
3583:
3582:
3484:
3483:
3397:Template-directed
3151:
3150:
3118:Phosphomevalonate
2810:
2809:
2794:
2793:
2537:Mitochondrial DNA
2531:
2530:
2485:
2484:
2340:citric acid cycle
2323:
2322:
2129:
2128:
2121:Monoamine oxidase
1520:10.1021/bi8018042
1004:10.1042/bj1520023
823:Disease relevance
626:
621:
539:
538:
535:
534:
454:metabolic pathway
356:
355:
352:
351:
347:structure summary
182:) (also known as
170:
169:
166:
165:
161:structure summary
4110:
4080:
4079:
4071:
3943:Hanes–Woolf plot
3886:Enzyme activator
3881:Enzyme inhibitor
3855:Enzyme catalysis
3799:
3792:
3785:
3776:
3760:Histidine kinase
3683:tyrosine kinases
3673:protein-tyrosine
3665:
3595:
3402:RNA polymerase I
3225:
3216:
3069:Aspartate kinase
3064:Phosphoglycerate
2881:
2837:
2830:
2823:
2814:
2674:
2649:
2619:
2599:
2549:
2542:
2494:
2457:
2427:
2395:
2350:Citrate synthase
2343:
2334:
2288:
2258:
2231:
2209:
2172:
2163:
2143:Adenylate kinase
2114:
2090:
2062:
2053:
2032:
2025:
2018:
2009:
1984:Adenylate+kinase
1972:
1971:
1961:
1938:Plant Physiology
1929:
1923:
1922:
1912:
1902:
1893:(15): 12937–45.
1878:
1872:
1871:
1861:
1828:
1819:
1818:
1808:
1798:
1789:(43): 31366–72.
1774:
1768:
1767:
1757:
1747:
1715:
1709:
1708:
1698:
1688:
1656:
1647:
1646:
1636:
1626:
1602:
1581:
1580:
1570:
1538:
1532:
1531:
1503:
1494:
1493:
1483:
1451:
1445:
1444:
1434:
1424:
1392:
1386:
1385:
1375:
1343:
1332:
1331:
1304:Escherichia coli
1299:
1290:
1289:
1279:
1258:Escherichia coli
1251:
1245:
1244:
1211:(7171): 838–44.
1199:
1190:
1189:
1161:
1155:
1154:
1144:
1120:
1114:
1113:
1079:
1070:
1064:
1063:
1035:
1026:
1025:
1015:
983:
977:
975:
965:
929:
881:immunodeficiency
839:Escherichia coli
624:
619:
609:protein isoforms
492:
361:Adenylate kinase
358:
295:
217:
205:
173:Adenylate kinase
109:
31:
22:Adenylate kinase
19:
16:Class of enzymes
4118:
4117:
4113:
4112:
4111:
4109:
4108:
4107:
4088:
4087:
4086:
4074:
4066:
4064:
4059:
3971:Oxidoreductases
3957:
3933:Enzyme kinetics
3921:
3917:List of enzymes
3890:
3859:
3830:Catalytic triad
3808:
3803:
3773:
3764:
3735:
3713:
3688:
3659:
3653:
3649:2.7.10-2.7.13:
3640:
3624:
3591:: miscellaneous
3579:
3553:
3535:
3512:
3495:exoribonuclease
3492:
3480:
3466:Polyadenylation
3383:
3209:
3203:
3191:
3173:
3169:
3163:
3147:
3109:
3096:
3073:
3045:
2875:
2869:
2861:
2855:
2841:
2811:
2806:
2790:
2670:
2665:
2645:
2640:
2615:
2610:
2595:
2590:
2545:
2527:
2481:
2468:
2453:
2448:
2421:
2416:
2389:
2384:
2337:
2319:
2284:
2279:
2255:steroidogenesis
2252:
2247:
2225:
2220:
2203:
2198:
2166:
2152:
2148:Creatine kinase
2125:
2111:
2106:
2101:
2083:
2078:
2056:
2042:
2036:
2006:
1980:
1975:
1931:
1930:
1926:
1880:
1879:
1875:
1838:Nature Genetics
1830:
1829:
1822:
1776:
1775:
1771:
1717:
1716:
1712:
1658:
1657:
1650:
1604:
1603:
1584:
1540:
1539:
1535:
1505:
1504:
1497:
1453:
1452:
1448:
1407:(40): 16984–9.
1394:
1393:
1389:
1345:
1344:
1335:
1301:
1300:
1293:
1270:(14): 8107–12.
1253:
1252:
1248:
1201:
1200:
1193:
1163:
1162:
1158:
1122:
1121:
1117:
1077:
1072:
1071:
1067:
1037:
1036:
1029:
985:
984:
980:
943:
940:Wayback Machine
930:
926:
922:
908:
893:
865:
863:ADK2 deficiency
852:
830:
825:
811:
798:
793:
785:
781:
777:
773:
769:
765:
761:
726:
723:Residues of ADK
717:
690:
641:
605:
556:
220:
34:
17:
12:
11:
5:
4116:
4114:
4106:
4105:
4100:
4090:
4089:
4085:
4084:
4061:
4060:
4058:
4057:
4044:
4031:
4018:
4005:
3992:
3979:
3965:
3963:
3959:
3958:
3956:
3955:
3950:
3945:
3940:
3935:
3929:
3927:
3923:
3922:
3920:
3919:
3914:
3909:
3904:
3898:
3896:
3895:Classification
3892:
3891:
3889:
3888:
3883:
3878:
3873:
3867:
3865:
3861:
3860:
3858:
3857:
3852:
3847:
3842:
3837:
3832:
3827:
3822:
3816:
3814:
3810:
3809:
3804:
3802:
3801:
3794:
3787:
3779:
3770:
3769:
3766:
3765:
3763:
3762:
3757:
3752:
3746:
3744:
3737:
3736:
3734:
3733:
3724:
3722:
3715:
3714:
3712:
3711:
3702:
3700:
3690:
3689:
3687:
3686:
3677:
3675:
3662:
3657:
3651:protein kinase
3646:
3645:
3642:
3641:
3639:
3638:
3632:
3630:
3626:
3625:
3623:
3622:
3617:
3612:
3607:
3601:
3599:
3592:
3585:
3584:
3581:
3580:
3578:
3577:
3572:
3561:
3559:
3555:
3554:
3552:
3551:
3545:
3543:
3537:
3536:
3534:
3533:
3528:
3522:
3520:
3514:
3513:
3511:
3510:
3505:
3499:
3497:
3490:Phosphorolytic
3486:
3485:
3482:
3481:
3479:
3478:
3473:
3468:
3463:
3458:
3457:
3456:
3451:
3446:
3436:
3435:
3434:
3424:
3419:
3414:
3409:
3404:
3399:
3393:
3391:
3389:RNA polymerase
3385:
3384:
3382:
3381:
3380:
3379:
3369:
3365:
3364:
3363:
3362:
3357:
3352:
3341:
3340:
3339:
3334:
3329:
3324:
3313:
3307:
3306:
3305:
3298:
3293:
3288:
3283:
3272:
3271:
3270:
3263:
3258:
3253:
3248:
3237:
3233:
3231:
3229:DNA polymerase
3222:
3213:
3207:
3193:
3192:
3190:
3189:
3184:
3178:
3176:
3171:
3167:
3153:
3152:
3149:
3148:
3146:
3145:
3140:
3135:
3130:
3125:
3120:
3114:
3112:
3107:
3098:
3097:
3095:
3094:
3088:
3086:
3075:
3074:
3072:
3071:
3066:
3060:
3058:
3047:
3046:
3044:
3043:
3038:
3033:
3032:
3031:
3026:
3016:
3014:Diacylglycerol
3011:
3006:
3001:
2996:
2991:
2986:
2981:
2976:
2975:
2974:
2964:
2959:
2954:
2953:
2952:
2947:
2942:
2937:
2932:
2925:Phosphofructo-
2922:
2917:
2916:
2915:
2905:
2900:
2894:
2892:
2878:
2873:
2857:
2856:
2842:
2840:
2839:
2832:
2825:
2817:
2808:
2807:
2799:
2796:
2795:
2792:
2791:
2789:
2788:
2783:
2778:
2773:
2768:
2763:
2758:
2753:
2748:
2743:
2738:
2733:
2728:
2723:
2718:
2713:
2708:
2703:
2698:
2693:
2688:
2683:
2677:
2675:
2667:
2666:
2664:
2663:
2658:
2652:
2650:
2642:
2641:
2639:
2638:
2633:
2628:
2622:
2620:
2612:
2611:
2609:
2608:
2602:
2600:
2592:
2591:
2589:
2588:
2583:
2578:
2573:
2568:
2563:
2558:
2552:
2550:
2539:
2533:
2532:
2529:
2528:
2526:
2525:
2520:
2519:
2518:
2513:
2503:
2497:
2491:
2487:
2486:
2483:
2482:
2480:
2479:
2473:
2470:
2469:
2467:
2466:
2460:
2458:
2450:
2449:
2447:
2446:
2441:
2436:
2430:
2428:
2418:
2417:
2415:
2414:
2409:
2404:
2398:
2396:
2386:
2385:
2383:
2382:
2377:
2372:
2367:
2362:
2357:
2352:
2346:
2344:
2331:
2325:
2324:
2321:
2320:
2318:
2317:
2312:
2307:
2302:
2297:
2291:
2289:
2281:
2280:
2278:
2277:
2272:
2267:
2261:
2259:
2249:
2248:
2246:
2245:
2240:
2234:
2232:
2222:
2221:
2219:
2218:
2212:
2210:
2200:
2199:
2197:
2196:
2191:
2186:
2181:
2175:
2173:
2160:
2158:Inner membrane
2154:
2153:
2151:
2150:
2145:
2139:
2137:
2131:
2130:
2127:
2126:
2124:
2123:
2117:
2115:
2103:
2102:
2100:
2099:
2093:
2091:
2080:
2079:
2077:
2076:
2071:
2065:
2063:
2050:
2048:Outer membrane
2044:
2043:
2037:
2035:
2034:
2027:
2020:
2012:
1992:
1991:
1979:
1978:External links
1976:
1974:
1973:
1924:
1873:
1850:10.1038/ng.278
1820:
1769:
1730:(12): 5720–5.
1710:
1648:
1617:(4): 1729–72.
1582:
1533:
1514:(9): 1911–27.
1495:
1446:
1387:
1358:(5): 1661–71.
1333:
1291:
1246:
1191:
1156:
1115:
1065:
1027:
978:
956:(16): 2874–7.
950:Bioinformatics
923:
921:
918:
907:
901:
892:
889:
864:
861:
851:
848:
829:
826:
824:
821:
816:bucket brigade
810:
807:
797:
794:
792:
789:
783:
779:
775:
771:
767:
763:
759:
724:
716:
713:
689:
686:
685:
684:
670:
659:UMP-CMP kinase
656:
640:
637:
604:
601:
587:. The rate of
555:
552:
537:
536:
533:
532:
527:
521:
520:
515:
509:
508:
503:
497:
496:
488:
487:
476:
469:
468:
463:
457:
456:
451:
445:
444:
439:
433:
432:
427:
421:
420:
415:
409:
408:
403:
397:
396:
392:
391:
386:
380:
379:
374:
368:
367:
363:
362:
354:
353:
350:
349:
344:
338:
337:
324:
318:
317:
307:
300:
299:
291:
290:
277:
271:
270:
265:
259:
258:
253:
247:
246:
241:
235:
234:
231:
227:
226:
222:
221:
218:
210:
209:
168:
167:
164:
163:
158:
152:
151:
138:
132:
131:
121:
114:
113:
105:
104:
91:
85:
84:
79:
73:
72:
67:
61:
60:
55:
49:
48:
45:
41:
40:
36:
35:
32:
24:
23:
15:
13:
10:
9:
6:
4:
3:
2:
4115:
4104:
4101:
4099:
4096:
4095:
4093:
4083:
4078:
4073:
4069:
4055:
4051:
4050:
4045:
4042:
4038:
4037:
4032:
4029:
4025:
4024:
4019:
4016:
4012:
4011:
4006:
4003:
3999:
3998:
3993:
3990:
3986:
3985:
3980:
3977:
3973:
3972:
3967:
3966:
3964:
3960:
3954:
3951:
3949:
3946:
3944:
3941:
3939:
3936:
3934:
3931:
3930:
3928:
3924:
3918:
3915:
3913:
3912:Enzyme family
3910:
3908:
3905:
3903:
3900:
3899:
3897:
3893:
3887:
3884:
3882:
3879:
3877:
3876:Cooperativity
3874:
3872:
3869:
3868:
3866:
3862:
3856:
3853:
3851:
3848:
3846:
3843:
3841:
3838:
3836:
3835:Oxyanion hole
3833:
3831:
3828:
3826:
3823:
3821:
3818:
3817:
3815:
3811:
3807:
3800:
3795:
3793:
3788:
3786:
3781:
3780:
3777:
3761:
3758:
3756:
3753:
3751:
3748:
3747:
3745:
3742:
3738:
3732:
3731:
3726:
3725:
3723:
3720:
3716:
3710:
3709:
3704:
3703:
3701:
3699:
3695:
3691:
3685:
3684:
3679:
3678:
3676:
3674:
3670:
3666:
3663:
3660:
3652:
3647:
3637:
3634:
3633:
3631:
3627:
3621:
3618:
3616:
3613:
3611:
3608:
3606:
3603:
3602:
3600:
3596:
3593:
3590:
3586:
3576:
3573:
3570:
3566:
3563:
3562:
3560:
3556:
3550:
3547:
3546:
3544:
3542:
3538:
3532:
3529:
3527:
3524:
3523:
3521:
3519:
3515:
3509:
3506:
3504:
3501:
3500:
3498:
3496:
3491:
3487:
3477:
3474:
3472:
3469:
3467:
3464:
3462:
3459:
3455:
3452:
3450:
3447:
3445:
3442:
3441:
3440:
3437:
3433:
3430:
3429:
3428:
3425:
3423:
3420:
3418:
3415:
3413:
3410:
3408:
3405:
3403:
3400:
3398:
3395:
3394:
3392:
3390:
3386:
3378:
3375:
3374:
3373:
3370:
3367:
3366:
3361:
3358:
3356:
3353:
3351:
3348:
3347:
3345:
3342:
3338:
3335:
3333:
3330:
3328:
3325:
3323:
3320:
3319:
3317:
3314:
3311:
3308:
3304:
3303:
3299:
3297:
3294:
3292:
3289:
3287:
3284:
3282:
3279:
3278:
3276:
3273:
3269:
3268:
3264:
3262:
3259:
3257:
3254:
3252:
3249:
3247:
3244:
3243:
3241:
3238:
3235:
3234:
3232:
3230:
3226:
3223:
3221:
3217:
3214:
3211:
3202:
3198:
3194:
3188:
3185:
3183:
3180:
3179:
3177:
3174:
3162:
3158:
3154:
3144:
3141:
3139:
3136:
3134:
3131:
3129:
3126:
3124:
3121:
3119:
3116:
3115:
3113:
3110:
3103:
3099:
3093:
3090:
3089:
3087:
3084:
3080:
3076:
3070:
3067:
3065:
3062:
3061:
3059:
3056:
3052:
3048:
3042:
3039:
3037:
3034:
3030:
3029:Class II PI 3
3027:
3025:
3022:
3021:
3020:
3017:
3015:
3012:
3010:
3007:
3005:
3004:Deoxycytidine
3002:
3000:
2997:
2995:
2992:
2990:
2987:
2985:
2982:
2980:
2977:
2973:
2972:ADP-thymidine
2970:
2969:
2968:
2965:
2963:
2960:
2958:
2955:
2951:
2948:
2946:
2943:
2941:
2938:
2936:
2933:
2931:
2928:
2927:
2926:
2923:
2921:
2918:
2914:
2911:
2910:
2909:
2906:
2904:
2901:
2899:
2896:
2895:
2893:
2890:
2886:
2882:
2879:
2876:
2868:
2864:
2858:
2853:
2849:
2845:
2838:
2833:
2831:
2826:
2824:
2819:
2818:
2815:
2805:
2804:
2797:
2787:
2784:
2782:
2779:
2777:
2774:
2772:
2769:
2767:
2764:
2762:
2759:
2757:
2754:
2752:
2749:
2747:
2744:
2742:
2739:
2737:
2734:
2732:
2729:
2727:
2724:
2722:
2719:
2717:
2714:
2712:
2709:
2707:
2704:
2702:
2699:
2697:
2694:
2692:
2689:
2687:
2684:
2682:
2679:
2678:
2676:
2673:
2668:
2662:
2659:
2657:
2654:
2653:
2651:
2648:
2643:
2637:
2634:
2632:
2629:
2627:
2624:
2623:
2621:
2618:
2613:
2607:
2604:
2603:
2601:
2598:
2593:
2587:
2584:
2582:
2579:
2577:
2574:
2572:
2569:
2567:
2564:
2562:
2559:
2557:
2554:
2553:
2551:
2548:
2543:
2540:
2538:
2534:
2524:
2521:
2517:
2514:
2512:
2509:
2508:
2507:
2504:
2502:
2499:
2498:
2495:
2492:
2488:
2478:
2475:
2474:
2471:
2465:
2462:
2461:
2459:
2456:
2451:
2445:
2442:
2440:
2437:
2435:
2432:
2431:
2429:
2426:
2425:
2419:
2413:
2410:
2408:
2405:
2403:
2400:
2399:
2397:
2394:
2393:
2387:
2381:
2378:
2376:
2373:
2371:
2368:
2366:
2363:
2361:
2358:
2356:
2353:
2351:
2348:
2347:
2345:
2342:
2341:
2335:
2332:
2330:
2326:
2316:
2313:
2311:
2308:
2306:
2303:
2301:
2298:
2296:
2293:
2292:
2290:
2287:
2282:
2276:
2273:
2271:
2268:
2266:
2263:
2262:
2260:
2257:
2256:
2250:
2244:
2241:
2239:
2236:
2235:
2233:
2230:
2229:
2223:
2217:
2214:
2213:
2211:
2208:
2207:
2201:
2195:
2192:
2190:
2187:
2185:
2182:
2180:
2177:
2176:
2174:
2171:
2170:
2164:
2161:
2159:
2155:
2149:
2146:
2144:
2141:
2140:
2138:
2136:
2132:
2122:
2119:
2118:
2116:
2113:
2110:
2104:
2098:
2095:
2094:
2092:
2089:
2087:
2081:
2075:
2072:
2070:
2067:
2066:
2064:
2061:
2060:
2054:
2051:
2049:
2045:
2040:
2039:Mitochondrial
2033:
2028:
2026:
2021:
2019:
2014:
2013:
2010:
2005:
2001:
1997:
1989:
1985:
1982:
1981:
1977:
1969:
1965:
1960:
1955:
1951:
1947:
1943:
1939:
1935:
1928:
1925:
1920:
1916:
1911:
1906:
1901:
1896:
1892:
1888:
1884:
1877:
1874:
1869:
1865:
1860:
1855:
1851:
1847:
1844:(1): 106–11.
1843:
1839:
1835:
1827:
1825:
1821:
1816:
1812:
1807:
1802:
1797:
1792:
1788:
1784:
1780:
1773:
1770:
1765:
1761:
1756:
1751:
1746:
1741:
1737:
1733:
1729:
1725:
1721:
1714:
1711:
1706:
1702:
1697:
1692:
1687:
1682:
1678:
1674:
1671:(4): e19300.
1670:
1666:
1662:
1655:
1653:
1649:
1644:
1640:
1635:
1630:
1625:
1620:
1616:
1612:
1608:
1601:
1599:
1597:
1595:
1593:
1591:
1589:
1587:
1583:
1578:
1574:
1569:
1564:
1560:
1556:
1552:
1548:
1544:
1537:
1534:
1529:
1525:
1521:
1517:
1513:
1509:
1502:
1500:
1496:
1491:
1487:
1482:
1477:
1473:
1469:
1466:(3): 618–31.
1465:
1461:
1457:
1450:
1447:
1442:
1438:
1433:
1428:
1423:
1418:
1414:
1410:
1406:
1402:
1398:
1391:
1388:
1383:
1379:
1374:
1369:
1365:
1361:
1357:
1353:
1349:
1342:
1340:
1338:
1334:
1329:
1325:
1321:
1317:
1314:(1): 159–77.
1313:
1309:
1305:
1298:
1296:
1292:
1287:
1283:
1278:
1273:
1269:
1265:
1261:
1259:
1250:
1247:
1242:
1238:
1234:
1230:
1226:
1222:
1218:
1214:
1210:
1206:
1198:
1196:
1192:
1187:
1183:
1179:
1175:
1171:
1167:
1160:
1157:
1152:
1148:
1143:
1138:
1134:
1130:
1126:
1119:
1116:
1111:
1107:
1103:
1099:
1095:
1091:
1088:(3): 527–34.
1087:
1083:
1076:
1069:
1066:
1061:
1057:
1053:
1049:
1045:
1041:
1034:
1032:
1028:
1023:
1019:
1014:
1009:
1005:
1001:
997:
993:
989:
982:
979:
973:
969:
964:
959:
955:
951:
947:
941:
937:
934:
928:
925:
919:
917:
915:
914:
906:
902:
900:
898:
890:
888:
886:
882:
878:
874:
870:
862:
860:
858:
849:
847:
845:
841:
840:
835:
827:
822:
820:
817:
808:
806:
804:
795:
790:
788:
756:
754:
744:
740:
738:
734:
721:
714:
712:
709:
705:
701:
696:
687:
683:
682:
677:
674:
671:
669:
668:
663:
660:
657:
655:
654:
649:
646:
643:
642:
638:
636:
632:
630:
622:
614:
610:
602:
600:
598:
594:
590:
586:
582:
578:
572:
571:
567:
563:
559:
553:
548:
543:
531:
528:
526:
522:
519:
516:
514:
510:
507:
504:
502:
498:
493:
489:
486:
483:
480:
477:
474:
470:
467:
464:
462:
458:
455:
452:
450:
446:
443:
440:
438:
434:
431:
430:NiceZyme view
428:
426:
422:
419:
416:
414:
410:
407:
404:
402:
398:
393:
390:
387:
385:
381:
378:
375:
373:
369:
364:
359:
348:
345:
343:
339:
336:
332:
328:
325:
323:
319:
315:
311:
308:
305:
301:
296:
292:
289:
285:
281:
278:
276:
272:
269:
266:
264:
260:
257:
254:
252:
248:
245:
242:
240:
236:
232:
228:
223:
216:
211:
206:
203:
201:
196:
193:
189:
185:
181:
178:
174:
162:
159:
157:
153:
150:
146:
142:
139:
137:
133:
129:
125:
122:
119:
115:
110:
106:
103:
99:
95:
92:
90:
86:
83:
80:
78:
74:
71:
68:
66:
62:
59:
56:
54:
50:
46:
42:
37:
30:
25:
20:
4049:Translocases
4046:
4033:
4020:
4007:
3994:
3984:Transferases
3981:
3968:
3825:Binding site
3727:
3705:
3680:
3301:
3266:
3122:
3024:Class I PI 3
2989:Pantothenate
2860:2.7.1-2.7.4:
2844:Transferases
2800:
2647:ATP synthase
2454:
2422:
2390:
2338:
2305:ATP synthase
2285:
2253:
2226:
2204:
2184:Cytochrome c
2167:
2142:
2107:
2097:Kynureninase
2084:
2057:
1944:(1): 70–82.
1941:
1937:
1927:
1890:
1886:
1876:
1841:
1837:
1786:
1782:
1772:
1727:
1723:
1713:
1668:
1664:
1614:
1610:
1550:
1546:
1536:
1511:
1508:Biochemistry
1507:
1463:
1459:
1449:
1404:
1400:
1390:
1355:
1351:
1311:
1307:
1303:
1267:
1263:
1257:
1249:
1208:
1204:
1172:(1): 145–8.
1169:
1165:
1159:
1135:(2): 263–7.
1132:
1128:
1118:
1085:
1081:
1068:
1043:
1039:
998:(1): 23–32.
995:
991:
981:
953:
949:
927:
911:
909:
904:
894:
866:
853:
837:
831:
812:
799:
757:
749:
730:
703:
694:
691:
679:
665:
651:
633:
606:
573:
560:
557:
546:
418:BRENDA entry
187:
183:
172:
171:
3820:Active site
3575:Transposase
3565:Recombinase
3210:-nucleoside
3036:Sphingosine
2597:Complex III
1910:2066/186091
809:ADK shuttle
639:Subfamilies
406:IntEnz view
366:Identifiers
225:Identifiers
200:homeostasis
39:Identifiers
4092:Categories
4023:Isomerases
3997:Hydrolases
3864:Regulation
3377:Telomerase
3220:Polymerase
2994:Mevalonate
2957:Riboflavin
2848:phosphorus
2617:Complex IV
2424:urea cycle
2112:metabolism
2088:metabolism
2086:tryptophan
1553:(8): 111.
920:References
897:glycolysis
475:structures
442:KEGG entry
310:structures
124:structures
3902:EC number
3569:Integrase
3493:3' to 5'
3138:Guanylate
3133:Uridylate
3123:Adenylate
2967:Thymidine
2962:Shikimate
2801:see also
2547:Complex I
2355:Aconitase
2004:IPR000850
1046:: 75–83.
885:inner ear
737:oligomers
715:Structure
700:conserved
688:Mechanism
681:IPR006267
667:IPR006266
653:IPR006259
395:Databases
268:PDOC00104
256:IPR007862
188:myokinase
82:PDOC00104
70:IPR000850
4103:EC 2.7.4
3926:Kinetics
3850:Cofactor
3813:Activity
3503:RNase PH
3111:acceptor
3092:Creatine
3085:acceptor
3057:acceptor
2999:Pyruvate
2984:Glycerol
2945:Platelet
2920:Galacto-
2891:acceptor
2501:Frataxin
2375:Fumarase
2041:proteins
2000:InterPro
1968:15618410
1919:12562761
1868:19043416
1815:17704060
1705:21556322
1665:PLOS ONE
1643:19468337
1577:21081909
1528:19219996
1490:20471396
1441:19805185
1382:17217965
1233:18026086
1110:33249169
1102:21080915
1060:24495878
972:15145806
936:Archived
857:coronary
791:Function
708:cofactor
676:InterPro
662:InterPro
648:InterPro
603:Isozymes
530:proteins
518:articles
506:articles
479:RCSB PDB
389:2598011
327:RCSB PDB
251:InterPro
141:RCSB PDB
65:InterPro
4082:Biology
4036:Ligases
3806:Enzymes
3454:PrimPol
3439:Primase
2913:Hepatic
2908:Fructo-
2661:MT-ATP8
2656:MT-ATP6
2576:MT-ND4L
1859:2612090
1806:3232003
1764:8650159
1732:Bibcode
1696:3083437
1673:Bibcode
1634:2680645
1555:Bibcode
1481:2902635
1432:2761315
1409:Bibcode
1373:2561047
1328:1548697
1286:2542263
1241:4406037
1213:Bibcode
1186:1587477
1022:1212224
1013:1172435
782:and AMP
774:and AMP
768:E. coli
762:and AMP
735:, form
733:ligands
725:E. coli
704:E. coli
695:E. coli
678::
664::
650::
613:cytosol
547:E. coli
466:profile
449:MetaCyc
384:CAS no.
377:2.7.4.3
263:PROSITE
244:PF05191
233:ADK_lid
208:ADK_lid
190:) is a
180:2.7.4.3
77:PROSITE
58:PF00406
4068:Portal
4010:Lyases
3741:2.7.13
3719:2.7.12
3694:2.7.11
3669:2.7.10
3508:PNPase
3476:PNPase
3432:POLRMT
3427:ssRNAP
2940:Muscle
2903:Gluco-
2867:kinase
2766:MT-TS2
2761:MT-TS1
2731:MT-TL2
2726:MT-TL1
2636:MT-CO3
2631:MT-CO2
2626:MT-CO1
2606:MT-CYB
2586:MT-ND6
2581:MT-ND5
2571:MT-ND4
2566:MT-ND3
2561:MT-ND2
2556:MT-ND1
2329:Matrix
1990:(MeSH)
1966:
1959:548839
1956:
1917:
1866:
1856:
1813:
1803:
1762:
1752:
1703:
1693:
1641:
1631:
1575:
1526:
1488:
1478:
1439:
1429:
1380:
1370:
1326:
1284:
1239:
1231:
1205:Nature
1184:
1151:218813
1149:
1108:
1100:
1058:
1020:
1010:
970:
753:strain
513:PubMed
495:Search
485:PDBsum
425:ExPASy
413:BRENDA
401:IntEnz
372:EC no.
342:PDBsum
316:
306:
288:SUPFAM
230:Symbol
195:enzyme
156:PDBsum
130:
120:
102:SUPFAM
44:Symbol
3962:Types
3589:2.7.8
3558:Other
3197:2.7.7
3157:2.7.6
3102:2.7.4
3079:2.7.3
3051:2.7.2
2935:Liver
2898:Hexo-
2885:2.7.1
2786:MT-TY
2781:MT-TW
2776:MT-TV
2771:MT-TT
2756:MT-TR
2751:MT-TQ
2746:MT-TP
2741:MT-TN
2736:MT-TM
2721:MT-TK
2716:MT-TI
2711:MT-TH
2706:MT-TG
2701:MT-TF
2696:MT-TE
2691:MT-TD
2686:MT-TC
2681:MT-TA
2477:PMPCB
2464:ALDH2
2286:other
1755:39127
1237:S2CID
1106:S2CID
1078:(PDF)
461:PRIAM
284:SCOPe
275:SCOP2
98:SCOPe
89:SCOP2
4054:list
4047:EC7
4041:list
4034:EC6
4028:list
4021:EC5
4015:list
4008:EC4
4002:list
3995:EC3
3989:list
3982:EC2
3976:list
3969:EC1
3728:see
3706:see
3681:see
3055:COOH
2854:2.7)
2672:tRNA
1998:and
1996:Pfam
1964:PMID
1915:PMID
1864:PMID
1811:PMID
1760:PMID
1701:PMID
1639:PMID
1573:PMID
1524:PMID
1486:PMID
1437:PMID
1378:PMID
1324:PMID
1282:PMID
1229:PMID
1182:PMID
1166:Gene
1147:PMID
1098:PMID
1056:PMID
1018:PMID
968:PMID
803:AMPK
568:⇔ 2
525:NCBI
482:PDBe
437:KEGG
335:PDBj
331:PDBe
314:ECOD
304:Pfam
280:1ake
239:Pfam
149:PDBj
145:PDBe
128:ECOD
118:Pfam
94:1ake
53:Pfam
3471:PAP
3412:III
3346:/Y
3337:TDT
3318:/X
3310:III
3302:Pfu
3277:/B
3267:Taq
3242:/A
3009:PFP
2979:NAD
1954:PMC
1946:doi
1942:137
1905:hdl
1895:doi
1891:278
1854:PMC
1846:doi
1801:PMC
1791:doi
1787:282
1750:PMC
1740:doi
1691:PMC
1681:doi
1629:PMC
1619:doi
1563:doi
1516:doi
1476:PMC
1468:doi
1464:400
1427:PMC
1417:doi
1405:106
1368:PMC
1360:doi
1356:366
1316:doi
1312:224
1272:doi
1268:264
1221:doi
1209:450
1174:doi
1170:114
1137:doi
1090:doi
1086:433
1048:doi
1008:PMC
1000:doi
996:152
958:doi
875:.
869:AK2
784:lid
780:lid
776:lid
772:lid
764:lid
760:lid
593:ATP
585:AMP
581:ADP
577:ATP
570:ADP
566:AMP
562:ATP
501:PMC
473:PDB
322:PDB
186:or
184:ADK
136:PDB
47:ADK
4094::
3696::
3671::
3656:PO
3417:IV
3407:II
3316:IV
3312:/C
3275:II
3261:T7
3206:PO
3199::
3159::
3106:PO
3104::
3081::
3053::
2889:OH
2887::
2872:PO
2852:EC
2846::
2002::
1962:.
1952:.
1940:.
1936:.
1913:.
1903:.
1889:.
1885:.
1862:.
1852:.
1842:41
1840:.
1836:.
1823:^
1809:.
1799:.
1785:.
1781:.
1758:.
1748:.
1738:.
1728:93
1726:.
1722:.
1699:.
1689:.
1679:.
1667:.
1663:.
1651:^
1637:.
1627:.
1615:10
1613:.
1609:.
1585:^
1571:.
1561:.
1549:.
1545:.
1522:.
1512:48
1510:.
1498:^
1484:.
1474:.
1462:.
1458:.
1435:.
1425:.
1415:.
1403:.
1399:.
1376:.
1366:.
1354:.
1350:.
1336:^
1322:.
1310:.
1294:^
1280:.
1266:.
1262:.
1235:.
1227:.
1219:.
1207:.
1194:^
1180:.
1168:.
1145:.
1133:93
1131:.
1127:.
1104:.
1096:.
1084:.
1080:.
1054:.
1044:49
1042:.
1030:^
1016:.
1006:.
994:.
990:.
966:.
954:20
952:.
948:.
942:,
916:.
599:.
564:+
333:;
329:;
312:/
286:/
282:/
177:EC
147:;
143:;
126:/
100:/
96:/
4070::
4056:)
4052:(
4043:)
4039:(
4030:)
4026:(
4017:)
4013:(
4004:)
4000:(
3991:)
3987:(
3978:)
3974:(
3798:e
3791:t
3784:v
3658:4
3654:(
3571:)
3567:(
3449:2
3444:1
3422:V
3360:κ
3355:ι
3350:η
3344:V
3332:μ
3327:λ
3322:β
3296:ζ
3291:ε
3286:δ
3281:α
3256:ν
3251:θ
3246:γ
3240:I
3212:)
3208:4
3204:(
3175:)
3172:7
3170:O
3168:2
3166:P
3164:(
3108:4
3083:N
2950:2
2930:1
2877:)
2874:4
2870:(
2865:/
2836:e
2829:t
2822:v
2031:e
2024:t
2017:v
1970:.
1948::
1921:.
1907::
1897::
1870:.
1848::
1817:.
1793::
1766:.
1742::
1734::
1707:.
1683::
1675::
1669:6
1645:.
1621::
1579:.
1565::
1557::
1551:1
1530:.
1518::
1492:.
1470::
1443:.
1419::
1411::
1384:.
1362::
1330:.
1318::
1288:.
1274::
1243:.
1223::
1215::
1188:.
1176::
1153:.
1139::
1112:.
1092::
1062:.
1050::
1024:.
1002::
974:.
960::
625:m
620:m
618:K
202:.
175:(
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.