285:
37:
662:
967:, a major polysaccharide found in the cell walls of red seaweeds. The κ-carrageenase containing vesicles can be exploited for bioethanol production since they convert carbohydrate-rich biomass to sugars. Biomolecules present in the red seaweeds, such as vitamins and carotenoids, are also extracted for commercial use in tandem with the bioethanol production process.
730:
Content of genomic islands differs greatly between strains, especially those coding for polysaccharides that present on the flagellum and the outer surface of the cell, with possible roles in phage avoidance. Some strains have acquired heavy-metal tolerance and other important functional genes from
930:
contained in the Earth's crust with desirable optic and electronic properties. However, current industrial production of tellurium requires the usage of substances harmful to both humans and the environment. As a result, extraction of metalloids by biotechnological applications involving bacterial
255:
is able to grow on glucose-only solid medium, forming colonies up to 0.9 cm in diameter with irregular edges. As a result of phenotypic variability and differences in genomic content among strains, competitiveness in culture varies both between cultures of the same strain and between strains
705:
is globally distributed in the surface ocean at 0-50m depth, these strains are highly variable functionally despite sharing 97-99% nucleotide identity. Functional differences between surface strains are conferred by horizontally transferred genes, and are reflective of the variable conditions of
299:
are ubiquitous in the global oceans, typically adhering to small organic particles in the upper 50 metres of the water column. They constitute a significant proportion of the bacterial abundance in the North
Atlantic and Mediterranean at up to 9 and 23 percent of total particle-attached bacteria
888:
strains are likely involved in phage interactions. Some genomic islands encode specific surface receptors recognised by phages, increasing the susceptibility to phage infections. Some components of GIs are lysogenic or defective phages; one of these widespread GIs encodes virus-derived
818:. Specific subsets of the DGC genes are highly expressed in some strains, enhancing biofilm development by amplifying the transduction of signals that promote biofilm formation. This process changes the structure of the microbial community, affecting both the microenvironment and
986:, making it an alternative to other viscous polymers currently used in food and cosmetics. As of 2012, "deepsane" is also commercially available in the cosmetics industry and is referred to as Abyssine®, used in skincare products to reduce skin irritation from sunburns.
766:
generally has genomic features which confer very high tolerance to copper and other heavy metals. Strains with high copper tolerance all had at least one genomic island with metal tolerance genes, including several copies of the key cytoplasmic detoxifying factor
860:
for degrading recalcitrant DOM such as urea, molecular chaperones for protein folding at lower temperatures and hydrogenases associated with heavy-metal tolerance, located with other tolerance genes on a single GI. These sets of genes are not exclusive to
568:
in seawater and are then consumed by higher trophic levels, acting as a gateway for carbon into ecosystems. While natural ecosystems consist of a variety of heterotrophs contributing to the carbon cycle, it has been found in laboratory settings that
685:
strains have distinct genomic content associated with different lifestyles and geographical locations. Small differences in overall nucleotide identity between strains can be functionally substantial; many important functional genes are found on
275:
is able to use glucose as its sole carbon and energy source and blooms under high nutrient and sodium concentrations where it is able to outcompete other organisms. Low temperatures and low carbon availability generally impede growth.
1751:
Ivars-Martínez E, D'Auria G, Rodríguez-Valera F, Sânchez-Porro C, Ventosa A, Joint I, Mühling M (September 2008). "Biogeography of the ubiquitous marine bacterium
Alteromonas macleodii determined by multilocus sequence analysis".
539:
Increased exposure of bacteria to copper may occur in several ways, such as nutrient leaching, metals from ship hulls, or natural mineral deposits. Under conditions of increased copper concentrations, biofilm production of
544:
significantly increases as a defensive response to copper induced stress. These bacteria are able to colonise areas of very high copper concentration, giving them an advantage over other bacteria under these conditions.
2660:
Mitulla M, Dinasquet J, Guillemette R, Simon M, Azam F, Wietz M (December 2016). "Response of bacterial communities from
California coastal waters to alginate particles and an alginolytic Alteromonas macleodii strain".
624:
at different times to degrade the respective substrates. Laminarin is the first polysaccharide that is degraded, followed by alginate and pectin. This temporal variation in carbon utilisation is a result of a shift in
652:
plays a particularly relevant role in ecological carbon cycling. The degradation of algal polysaccharides and proteins is crucial for nutrient acquisition, and has the effect of preventing overgrowth of red algae.
942:
into elemental tellurium. The nanoparticles of the reduced tellurium are diffused into the cytoplasm, or into the extracellular space in the form of both electron-dense globules and metalloid crystals. This makes
698:
is vast, there is significant variation in functional genetic content between strains from different geographical regions. Additionally, closely related strains vary in functional genes found on genomic islands.
2160:
Neumann AM, Balmonte JP, Berger M, Giebel HA, Arnosti C, Voget S, et al. (October 2015). "Different utilization of alginate and other algal polysaccharides by marine
Alteromonas macleodii ecotypes".
378:
during growth. An ATPase-independent mechanism is involved in the transport of the siderophores which scavenge iron for the bacterium, iron which is then used by enzymes to facilitate carbon metabolism.
1910:
Ivanova, Elena P.; López-Pérez, Mario; Zabalos, Mila; Nguyen, Song Ha; Webb, Hayden K.; Ryan, Jason; Lagutin, Kiril; Vyssotski, Mikhail; Crawford, Russell J.; Rodriguez-Valera, Francisco (2015-01-01).
1409:
Raguénès G, Cambon-Bonavita MA, Lohier JF, Boisset C, Guezennec J (June 2003). "A novel, highly viscous polysaccharide excreted by an alteromonas isolated from a deep-sea hydrothermal vent shrimp".
1852:
Ivanova, Elena P.; López-Pérez, Mario; Zabalos, Mila; Nguyen, Song Ha; Webb, Hayden K.; Ryan, Jason; Lagutin, Kiril; Vyssotski, Mikhail; Crawford, Russell J.; Rodriguez-Valera, Francisco (2015).
2276:"Comparative genomics of two ecotypes of the marine planktonic copiotroph Alteromonas macleodii suggests alternative lifestyles associated with different kinds of particulate organic matter"
3829:
1577:
García-Martínez J, Acinas SG, Massana R, Rodríguez-Valera F (January 2002). "Prevalence and microdiversity of
Alteromonas macleodii-like microorganisms in different oceanic regions".
2122:
Rattanaphan P, Mittraparp-Arthorn P, Srinoun K, Vuddhakul V, Tansila N (July 2020). "Indole signaling decreases biofilm formation and related virulence of
Listeria monocytogenes".
315:
environments and it sinks rapidly into the deeper pelagic zones, relying on a different spectrum of carbon sources. Recently, the deep ecotype strains have been reclassified as
997:, when grown in glucose-supplemented media, secreted an unexpectedly high-molecular-weight polymer that changed carbohydrate composition. The polymer was found to be rich in
1627:"Description of a new polymer-secreting bacterium from a deep-sea hydrothermal vent, Alteromonas macleodii subsp. fijiensis, and preliminary characterization of the polymer"
1151:"Captured diversity in a culture collection: case study of the geographic and habitat distributions of environmental isolates held at the american type culture collection"
897:
of the bacteria may associate with the receptor binding proteins present on the
Alterophage R8W, allowing attachment and entry. Just over a dozen Alterophages infecting
366:(DOM) production in the oceans. These bacteria utilise unique Ton-B dependent transporters to acquire iron as well as carbon substrates. As a result, some strains of
3803:
531:
is therefore reduced when iron is limited, although the growth rate of strains from coastal populations is reduced more so than those from mid-oceanic populations.
978:
secretes "deepsane", an exopolysaccharide now used in cosmetics. Studied properties of "deepsane" include high viscosity possibly due to the interaction between
3842:
604:
exhibits two distinct strategies for carbon uptake, depending on the type of polysaccharide present in their habitat. When degrading the common polysaccharides
690:(GIs) exchanged between populations. Strains associated with surface waters such as ATCC 21726 have a single circular genome of about 4.6 million base pairs.
2508:
Cusick KD, Dale JR, Fitzgerald LA, Little BJ, Biffinger JC (July 2017). "Adaptation to copper stress influences biofilm formation in
Alteromonas macleodii".
452:. These bacteria are generally attached to small particles, but can also be free-living and are able to utilise a number of different substrates for growth.
355:
are also strain-specific, with different nutrient acquisition and cellular communication strategies between strains under different ecological conditions.
636:
Alginate is a gel textured polysaccharide that is a common component of macroalgal cell walls, and is a nutrient and carbon source for many organisms.
3790:
3816:
3694:"Alteromonas macleodii 107 - Type strain - DSM 6062, ATCC 27126, CCUG 16128, CIP 103198, JCM 20772, LMG 2843, NBRC 102226, IAM 12920, NCIMB 1963"
2945:
Manck, Lauren E.; Park, Jiwoon; Tully, Benjamin J.; Poire, Alfonso M.; Bundy, Randelle M.; Dupont, Christopher L.; Barbeau, Katherine A. (2022).
2709:"Draft Genome Sequence of Alteromonas macleodii Strain MIT1002, Isolated from an Enrichment Culture of the Marine Cyanobacterium Prochlorococcus"
2079:
Zhang Z, Li Z, Jiao N (June 2014). "Effects of D-amino acids on the EPS production and cell aggregation of
Alteromonas macleodii strain JL2069".
2769:"Why Close a Bacterial Genome? The Plasmid of Alteromonas Macleodii HOT1A3 is a Vector for Inter-Specific Transfer of a Flexible Genomic Island"
2882:"Differing Growth Responses of Major Phylogenetic Groups of Marine Bacteria to Natural Phytoplankton Blooms in the Western North Pacific Ocean"
1457:
Naval P, Chandra TS (June 2019). "Characterization of membrane vesicles secreted by seaweed associated bacterium
Alteromonas macleodii KS62".
3855:
869:
species along with other sets of functional genes such as enzymes for sugar and amino acid degradation, allowing for niche specialisation.
3012:"Draft Genome Sequences of Four Alteromonas macleodii Strains Isolated from Copper Coupons and Grown Long-Term at Elevated Copper Levels"
3914:
1349:"Genomic, metabolic and phenotypic variability shapes ecological differentiation and intraspecies interactions of Alteromonas macleodii"
762:
is an early coloniser of copper-based antifouling paint on ships, where it forms biofilms. While there is variability between strains,
284:
3517:"Genetic diversity of the causative agent of ice-ice disease of the seaweed Kappaphycus alvarezii from Karimunjawa island, Indonesia"
810:
strains are still able to induce growth of biofilms under elevated copper concentrations. These strains possess an alteration in the
213:
strains are currently being explored for their industrial uses, including in cosmetics, bioethanol production and rare earth mining.
1974:"Transcriptomic Study of Substrate-Specific Transport Mechanisms for Iron and Carbon in the Marine Copiotroph Alteromonas macleodii"
629:
activity of CAZymes and polysaccharide utilisation gene fragments. The biphasic nature of these cellular adaptations indicates that
391:
573:
is capable of drawing down the complete pool of labile DOC present in coastal waters. This indicates that the relationship between
3881:
2605:"Biphasic cellular adaptations and ecological implications of Alteromonas macleodii degrading a mixture of algal polysaccharides"
582:
481:
is unable to use its essential photosynthetic pigments, but is able to survive for an extended period of time in the presence of
267:; large cells with high nucleic-acid content are commonly seen, with high dividing frequencies and carbon production rates. As a
311:
had caused two distinct strains of the bacterium to occupy different water depth profiles. The “deep ecotype” is more suited to
3067:
Cusick, Kathleen; Iturbide, Ane; Gautam, Pratima; Price, Amelia; Polson, Shawn; MacDonald, Madolyn; Erill, Ivan (2021-09-28).
3919:
3821:
1912:"Ecophysiological diversity of a novel member of the genus Alteromonas, and description of Alteromonas mediterranea sp. nov"
1854:"Ecophysiological diversity of a novel member of the genus Alteromonas, and description of Alteromonas mediterranea sp. nov"
402:
impedes the production of EPS, but encourages the formation of biofilms by promoting other independent aggregation factors.
3069:"Enhanced copper-resistance gene repertoire in Alteromonas macleodii strains isolated from copper-treated marine coatings"
2831:"RNA sequencing provides evidence for functional variability between naturally co-existing Alteromonas macleodii lineages"
2880:
Tada, Yuya; Taniguchi, Akito; Nagao, Ippei; Miki, Takeshi; Uematsu, Mitsuo; Tsuda, Atsushi; Hamasaki, Koji (2011-06-15).
1210:
2204:
Mitulla, Maximilian; Dinasquet, Julie; Guillemette, Ryan; Simon, Meinhard; Azam, Farooq; Wietz, Matthias (2016-05-30).
633:’s role in the drawdown of polysaccharide DOC is adaptable to changing community structures of macroalgal communities.
2396:"The Trichodesmium consortium: conserved heterotrophic co-occurrence and genomic signatures of potential interactions"
2337:
Cells Rely on Microbial Interactions Rather than on Chlorotic Resting Stages To Survive Long-Term Nutrient Starvation"
3309:
Gonzaga A, Martin-Cuadrado AB, López-Pérez M, Megumi Mizuno C, García-Heredia I, Kimes NE, et al. (2012-12-01).
2767:
Fadeev, Eduard; De Pascale, Fabio; Vezzi, Alessandro; Hübner, Sariel; Aharonovich, Dikla; Sher, Daniel (2016-03-08).
409:
play a crucial role in algae degradation and habitat colonisation. These vesicles contain hydrolytic enzymes such as
343:
degradation, while others have a unique capacity to metabolise sugars from specific algae species. The production of
36:
515:
is significantly reduced. Iron metal is associated with several key processes for bacterial metabolism, such as the
711:
209:
confer functional diversity to closely related strains and facilitate different lifestyles and strategies. Certain
3847:
710:
also have a higher number of genes associated with utilising different sugar and amino acid substrates as well as
3755:
3468:"Bacterial recovery and recycling of tellurium from tellurium-containing compounds by Pseudoalteromonas sp. EPR3"
1269:
Gonzaga, Aitor; López-Pérez, Mario; Martin-Cuadrado, Ana-Belen; Ghai, Rohit; Rodriguez-Valera, Francisco (2012).
524:
3243:
Cusick, Kathleen D.; Dale, Jason R.; Fitzgerald, Lisa A.; Little, Brenda J.; Biffinger, Justin C. (2017-07-03).
2331:
Roth-Rosenberg D, Aharonovich D, Luzzatto-Knaan T, Vogts A, Zoccarato L, Eigemann F, et al. (August 2020).
1045:"Genomes of surface isolates of Alteromonas macleodii: the life of a widespread marine opportunistic copiotroph"
1001:
and therefore expected to have a heavy-metal-binding ability that could be used and applied in the treatment of
3909:
2205:
938:
contains a plasmid that houses genes allowing for resistance to multiple metals, and has the ability to reduce
565:
456:
is a copiotroph flexible in its use of substrates, growing rapidly at high carbon and nutrient concentrations.
363:
3621:
Le Costaouëc T, Cérantola S, Ropartz D, Ratiskol J, Sinquin C, Colliec-Jouault S, Boisset C (September 2012).
2947:"Petrobactin, a siderophore produced by Alteromonas, mediates community iron acquisition in the global ocean"
2274:
Ivars-Martinez E, Martin-Cuadrado AB, D'Auria G, Mira A, Ferriera S, Johnson J, et al. (December 2008).
390:
such as D-alanine, D-serine and D-glutamic acid reduce metabolic activity, also inhibiting the production of
1043:
López-Pérez M, Gonzaga A, Martin-Cuadrado AB, Onyshchenko O, Ghavidel A, Ghai R, Rodriguez-Valera F (2012).
626:
421:. These enzymes are responsible for the degradation of cell walls and inner components of red algae such as
335:, cellular communication, and nutrient acquisition. Some strains are specialised in their associations with
2603:
Koch H, Dürwald A, Schweder T, Noriega-Ortega B, Vidal-Melgosa S, Hehemann JH, et al. (January 2019).
3717:
2453:"Effects of iron limitation on growth and carbon metabolism in oceanic and coastal heterotrophic bacteria"
2206:"Response of bacterial communities from California coastal waters to alginate particles and an alginolytic
783:
found in particularly metal-tolerant strains contain multiple copies of metal detoxification systems with
226:
1211:"Prevalence and microdiversity of Alteromonas macleodii-like microorganisms in different oceanic regions"
308:
121:
3566:"A novel polymer produced by a bacterium isolated from a deep-sea hydrothermal vent polychaete annelid"
1271:"Complete Genome Sequence of the Copiotrophic Marine Bacterium Alteromonas macleodii Strain ATCC 27126"
718:
to grow rapidly and take advantage of increases in available organic matter. Key genes associated with
648:
has been found to outcompete other species of bacteria in the degradation of alginate, indicating that
3623:"Structural data on a bacterial exopolysaccharide produced by a deep-sea Alteromonas macleodii strain"
2565:
503:
metabolism, allowing for the catabolism of methanol and the detoxification of radical oxygen species.
3886:
3528:
3256:
3147:
3080:
2958:
2893:
2670:
2616:
2517:
2464:
2407:
2348:
2287:
2221:
2170:
1816:
1761:
1698:
1638:
1586:
1526:
1360:
1222:
1162:
1056:
819:
527:, all of which are functionally limited when iron availability is not sufficient. The growth rate of
264:
2023:"Production and Reutilization of Fluorescent Dissolved Organic Matter by a Marine Bacterial Strain,
1687:"Single bacterial strain capable of significant contribution to carbon cycling in the surface ocean"
1347:
Koch H, Germscheid N, Freese HM, Noriega-Ortega B, Lücking D, Berger M, et al. (January 2020).
931:
biosynthesis of nanoparticles from various uncommon and rare metals are increasingly being studied.
947:
a candidate for facilitating the extraction of tellurium with reduced reliance on toxic chemicals.
939:
890:
811:
232:
68:
3603:
3497:
3288:
2541:
2490:
2451:
Fourquez M, Devez A, Schaumann A, Gueneugues A, Jouenne T, Obernosterer I, Blain S (2014-01-27).
2313:
2104:
1947:
1889:
1785:
1482:
1434:
956:
894:
248:
147:
31:
747:
strains that enhance heavy-metal tolerance are found in genomic islands in other members of the
3808:
1625:
Raguenes G, Pignet P, Gauthier G, Peres A, Christen R, Rougeaux H, et al. (January 1996).
1209:
Garcia-Martinez, Jesus; Acinas, Silvia G.; Massana, Ramon; Rodriguez-Valera, Francisco (2002).
181:
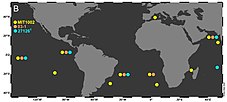
found in surface waters across temperate and tropical regions. First discovered in a survey of
3868:
3777:
3642:
3595:
3587:
3546:
3489:
3448:
3393:
3340:
3280:
3272:
3222:
3181:
3163:
3136:"Multiple Megaplasmids Confer Extremely High Levels of Metal Tolerance in Alteromonas Strains"
3116:
3098:
3049:
3031:
2992:
2974:
2927:
2909:
2862:
2808:
2790:
2746:
2728:
2686:
2642:
2585:
2533:
2482:
2433:
2376:
2305:
2245:
2237:
2186:
2139:
2096:
2058:
2003:
1969:
1939:
1931:
1881:
1873:
1834:
1777:
1726:
1664:
1602:
1554:
1474:
1426:
1386:
1308:
1290:
1246:
1238:
1188:
1131:
1082:
784:
516:
492:
1805:"Widespread Archaea and novel Bacteria from the deep sea as shown by 16S rRNA gene sequences"
3873:
3634:
3577:
3536:
3532:
3479:
3438:
3428:
3383:
3375:
3330:
3322:
3264:
3212:
3171:
3155:
3106:
3088:
3039:
3023:
2982:
2966:
2917:
2901:
2852:
2842:
2798:
2780:
2736:
2720:
2678:
2632:
2624:
2577:
2525:
2472:
2423:
2415:
2366:
2358:
2295:
2229:
2178:
2131:
2088:
2048:
2038:
1993:
1985:
1923:
1865:
1824:
1769:
1716:
1706:
1654:
1646:
1594:
1544:
1534:
1466:
1418:
1376:
1368:
1298:
1282:
1230:
1178:
1170:
1121:
1113:
1072:
1064:
182:
178:
88:
3010:
Cusick, Kathleen D.; Dale, Jason R.; Little, Brenda J.; Biffinger, Justin C. (2016-12-29).
2394:
Lee MD, Walworth NG, McParland EL, Fu FX, Mincer TJ, Levine NM, et al. (August 2017).
661:
748:
644:, in that they degrade alginate, increasing DOC drawdown in marine environments. Further,
469:
336:
244:
78:
3199:
Colin, M.; Carré, G.; Klingelschmitt, F.; Reffuveille, F.; Gangloff, S. C. (2021-12-15).
2707:
Biller, Steven J.; Coe, Allison; Martin-Cuadrado, Ana-Belen; Chisholm, Sallie W. (2015).
735:
GIs containing important functional genes are exchanged between different populations of
714:
for plasticity in changing conditions. This plasticity is associated with the ability of
394:(EPS). Exopolysaccharide production contributes to cell aggregation and the formation of
3541:
3516:
3260:
3151:
3084:
2962:
2897:
2674:
2620:
2521:
2468:
2411:
2291:
2225:
2174:
1820:
1765:
1702:
1642:
1590:
1530:
1364:
1226:
1166:
1060:
3443:
3412:
3388:
3359:
3335:
3310:
3176:
3135:
3111:
3068:
3044:
3011:
2987:
2946:
2922:
2881:
2857:
2830:
2803:
2768:
2741:
2708:
2637:
2604:
2428:
2395:
2371:
2332:
2053:
2022:
1998:
1973:
1804:
1721:
1686:
1549:
1506:
1381:
1348:
1303:
1270:
1183:
1150:
1077:
1044:
687:
593:
578:
312:
190:
58:
1659:
1626:
1126:
1101:
300:
respectively, and are also present in the Northeast Pacific and subtropical Atlantic.
193:
and is recognised as a prominent component of surface waters between 0 and 50 metres.
3903:
3834:
3665:
3582:
3565:
1853:
1773:
1598:
1486:
1234:
803:
487:
3607:
3501:
3292:
2564:
Chen, Chia-Lung; Maki, James S.; Rittschof, Dan; Teo, Serena Lay-Ming (2013-09-01).
2545:
2494:
2108:
1951:
1893:
1789:
1438:
1174:
3782:
3672:. Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures GmbH
3564:
Cambon-Bonavita, M.-A.; Raguenes, G.; Jean, J.; Vincent, P.; Guezennec, J. (2002).
3245:"Adaptation to copper stress influences biofilm formation in Alteromonas macleodii"
983:
641:
609:
430:
387:
3638:
3268:
2529:
2317:
1539:
1505:
Beleneva IA, Efimova KV, Eliseikina MG, Svetashev VI, Orlova TY (September 2019).
1117:
722:
production and degradation of algal substrates are also transferred horizontally.
429:
is also very efficient at degrading alginate, expressing as many as five separate
3217:
3200:
3093:
589:
is capable of carbon cycling to the same extent as entire microbial communities.
3749:
3244:
3134:
Cusick, Kathleen D.; Polson, Shawn W.; Duran, Gabriel; Hill, Russell T. (2020).
2581:
1650:
998:
964:
857:
795:
789:
719:
554:
422:
352:
229:
98:
3740:
3622:
2970:
1691:
Proceedings of the National Academy of Sciences of the United States of America
1470:
1372:
2628:
2477:
2452:
2092:
1989:
1927:
1869:
1422:
1002:
695:
621:
520:
449:
371:
344:
332:
268:
198:
3591:
3550:
3276:
3226:
3167:
3102:
3035:
3027:
2978:
2913:
2847:
2794:
2785:
2732:
2724:
2589:
2486:
2241:
2135:
2043:
1935:
1877:
1838:
1294:
1242:
2829:
Kimes NE, López-Pérez M, Ausó E, Ghai R, Rodriguez-Valera F (October 2014).
2682:
2233:
2182:
1911:
1711:
927:
923:
605:
474:
236:
3646:
3599:
3493:
3452:
3397:
3344:
3284:
3185:
3120:
3053:
2996:
2931:
2866:
2812:
2750:
2690:
2646:
2537:
2437:
2380:
2309:
2249:
2190:
2143:
2100:
2062:
2007:
1943:
1885:
1781:
1730:
1606:
1558:
1478:
1430:
1390:
1312:
1250:
1192:
1086:
398:
in marine bacterial species. Paradoxically, the uptake of D-amino acids by
3311:"Polyclonality of concurrent natural populations of Alteromonas macleodii"
2419:
2362:
2300:
2275:
1668:
1135:
243:
are between 0.6 to 0.8 μm width and 1.4 to 2.0 μm length, and are neither
3734:
3326:
3159:
2905:
833:
A closely related set of strains previously considered "deep-ecotype" of
779:
the ability to grow at metal levels lethal to most other marine species.
418:
414:
375:
48:
3201:"Copper alloys to prevent bacterial biofilm formation on touch surfaces"
1286:
3795:
3764:
3379:
3358:
Garcia-Heredia I, Rodriguez-Valera F, Martin-Cuadrado AB (April 2013).
1829:
979:
780:
740:
395:
348:
3484:
3467:
3433:
1068:
934:
Tellurium has toxic effects on bacteria through an unknown mechanism.
436:
and outcompeting other bacterial groups when grown on this substrate.
3769:
3693:
3411:
Feng X, Yan W, Wang A, Ma R, Chen X, Lin TH, et al. (May 2021).
881:
677:
in black. The grey colour indicates genes not of functional interest.
613:
410:
340:
202:
3711:
2566:"Early marine bacterial biofilm on a copper-based antifouling paint"
2353:
660:
433:
331:
leads to specific adaptive strategies in terms of carbon and iron
694:
has an estimated 4400 total genes with about 47% GC content. Its
673:
genes in blue, distinct A. macleodii genes in red, core genes of
3860:
806:
properties, minimising the formation of biofilms. However, some
3715:
3515:
Syafitri, E; Prayitno, S B; Ma’ruf, W F; Radjasa, O K (2017).
485:. The marine heterotroph has also been found associated with
370:
are able to more efficiently regulate the uptake of glucose,
235:. It is aerobic and motile, with a singular unsheathed polar
3670:
List of Prokaryotic names with Standing in Nomenclature LPSN
802:
Copper prevents bacterial growth due to its intrinsic
600:, and are a major source of carbon into marine ecosystems.
3360:"Novel group of podovirus infecting the marine bacterium '
901:
have been characterised to date, including members of the
511:
Under iron replete conditions, the rate of respiration in
448:
is well-suited to the degradation of a variety sugars and
292:
strains MIT1002, 83-1 and 27126 in TARA Ocean metagenomes.
814:(DGC) genes, which control the expression of biofilms in
1572:
1570:
1568:
1100:
Baumann L, Baumann P, Mandel M, Allen RD (April 1972).
918:
Reduction of potassium tellurite to elemental tellurium
865:
as they are exchanged between different populations of
596:
secreted by macroalga are degraded by microbes such as
3521:
IOP Conference Series: Earth and Environmental Science
2269:
2267:
2265:
2263:
2261:
2259:
3724:
2570:
International Biodeterioration & Biodegradation
1459:
Biochemical and Biophysical Research Communications
1038:
362:can influence iron concentrations and recalcitrant
201:of 4.6 million base pairs. Variable regions in the
1036:
1034:
1032:
1030:
1028:
1026:
1024:
1022:
1020:
1018:
893:and RNA chaperone genes. For example, the O-chain
1149:Floyd MM, Tang J, Kane M, Emerson D (June 2005).
1746:
1744:
1742:
1740:
1620:
1618:
1616:
841:as they share only 81% overall sequence identity
3304:
3302:
2155:
2153:
1500:
1498:
1496:
1404:
1402:
1400:
990:Additional future prospects in water treatments
669:strain HOT1A3 and its plasmid pAM1A3. Distinct
2021:Goto S, Tada Y, Suzuki K, Yamashita Y (2017).
1680:
1678:
256:from different geographical areas and depths.
2824:
2822:
2074:
2072:
1963:
1961:
8:
1685:Pedler BE, Aluwihare LI, Azam F (May 2014).
1452:
1450:
1448:
1342:
951:Extraction of biomolecules from red seaweeds
386:depend on the type of amino acids taken up.
1511:from a culture of the toxic dinoflagellate
1340:
1338:
1336:
1334:
1332:
1330:
1328:
1326:
1324:
1322:
3712:
3413:"A Novel Broad Host Range Phage Infecting
477:due to nutrient stress. During chlorosis,
20:
3581:
3540:
3483:
3466:Bonificio WD, Clarke DR (November 2014).
3442:
3432:
3387:
3334:
3216:
3175:
3110:
3092:
3043:
2986:
2921:
2856:
2846:
2802:
2784:
2740:
2636:
2476:
2427:
2370:
2352:
2299:
2052:
2042:
1997:
1828:
1720:
1710:
1658:
1548:
1538:
1380:
1302:
1182:
1125:
1076:
283:
189:has since been placed within the phylum
1102:"Taxonomy of aerobic marine eubacteria"
1014:
880:Genes such as phase integrases and the
405:The extracellular membrane vesicles in
3140:Applied and Environmental Microbiology
2886:Applied and Environmental Microbiology
1631:Applied and Environmental Microbiology
1155:Applied and Environmental Microbiology
3238:
3236:
2762:
2760:
2702:
2700:
2559:
2557:
2555:
1905:
1903:
739:, increasing functional flexibility.
7:
1264:
1262:
1260:
1204:
1202:
845:There are 3200 genes shared between
837:have since been reclassified under
706:surface waters. Surface strains of
491:, a filamentous cyanobacteria that
1968:Manck LE, Espinoza JL, Dupont CL,
1507:"The tellurite-reducing bacterium
771:and its transcriptional regulator
14:
847:A. macleodii and A. mediterranea,
3666:"Species: Alteromonas macleodii"
3583:10.1046/j.1365-2672.2002.01689.x
1774:10.1111/j.1365-294X.2008.03883.x
1599:10.1046/j.1462-2920.2002.00255.x
1235:10.1046/j.1462-2920.2002.00255.x
35:
3570:Journal of Applied Microbiology
3472:Journal of Applied Microbiology
1175:10.1128/aem.71.6.2813-2823.2005
971:EPS deepsane usage in cosmetics
382:The physiological responses of
3692:Podstawka, Adam (2021-12-21).
1809:Marine Ecology Progress Series
849:with 1200-1600 unique to each.
1:
3639:10.1016/j.carbpol.2012.04.059
3542:10.1088/1755-1315/55/1/012044
3269:10.1080/08927014.2017.1329423
2530:10.1080/08927014.2017.1329423
1540:10.1016/j.heliyon.2019.e02435
1118:10.1128/jb.110.1.402-429.1972
963:, which allows it to degrade
955:Membrane vesicles containing
288:Location and distribution of
3315:Genome Biology and Evolution
3218:10.1016/j.matlet.2021.130712
3094:10.1371/journal.pone.0257800
640:are a key components of the
581:of coastal waters is one of
2582:10.1016/j.ibiod.2013.04.012
1803:Fuhrman J, Davis A (1997).
1651:10.1128/aem.62.1.67-73.1996
327:Physiological variation in
303:Initially, two ecotypes of
177:is a species of widespread
3936:
3915:Bacteria described in 1972
2971:10.1038/s41396-021-01065-y
2663:Environmental Microbiology
2457:Limnology and Oceanography
2214:Environmental Microbiology
2163:Environmental Microbiology
1579:Environmental Microbiology
1471:10.1016/j.bbrc.2019.04.148
1373:10.1038/s41598-020-57526-5
1215:Environmental Microbiology
712:transcriptional regulators
577:and other bacteria in the
225:refers to an encapsulated
2773:Frontiers in Microbiology
2629:10.1038/s41396-018-0252-4
2478:10.4319/lo.2014.59.2.0349
2124:FEMS Microbiology Letters
2093:10.1007/s00284-014-0520-0
2031:Frontiers in Microbiology
1990:10.1128/msystems.00070-20
1928:10.1007/s10482-014-0309-y
1870:10.1007/s10482-014-0309-y
1423:10.1007/s00284-002-3922-3
828:Alteromonas medditerranea
525:oxidative phosphorylation
460:Interspecies interactions
339:species through enhanced
153:
146:
127:
120:
32:Scientific classification
30:
23:
3028:10.1128/genomea.01311-16
2848:10.1186/1471-2164-15-938
2786:10.3389/fmicb.2016.00248
2725:10.1128/genomea.00967-15
2044:10.3389/fmicb.2017.00507
1513:Prorocentrum foraminosum
566:dissolved organic carbon
364:dissolved organic matter
317:Alteromonas mediterranea
3533:2017E&ES...55a2044S
3362:Alteromonas macleodii.'
2683:10.1111/1462-2920.13314
2234:10.1111/1462-2920.13314
2183:10.1111/1462-2920.12862
1916:Antonie van Leeuwenhoek
1858:Antonie van Leeuwenhoek
1712:10.1073/pnas.1401887111
1275:Journal of Bacteriology
1106:Journal of Bacteriology
259:Bacteria classified as
2136:10.1093/femsle/fnaa116
884:cluster found in some
820:biogeochemical cycling
678:
293:
197:has a single circular
3920:Marine microorganisms
3835:alteromonas-macleodii
3756:Alteromonas macleodii
3726:Alteromonas macleodii
3627:Carbohydrate Polymers
2420:10.1038/ismej.2017.49
2363:10.1128/mbio.01846-20
2301:10.1038/ismej.2008.74
2208:Alteromonas macleodii
2025:Alteromonas macleodii
1509:Alteromonas macleodii
995:Alteromonas macleodii
976:Alteromonas macleodii
936:Alteromonas macleodii
926:is an extremely rare
856:strains contain more
760:Alteromonas macleodii
755:Heavy metal tolerance
703:Alteromonas macleodii
692:Alteromonas macleodii
664:
602:Alteromonas macleodii
583:functional redundancy
497:Alteromonas macleodii
465:Alteromonas macleodii
454:Alteromonas macleodii
444:The surface-dwelling
427:Alteromonas macleodii
360:Alteromonas macleodii
329:Alteromonas macleodii
309:niche differentiation
305:Alteromonas macleodii
297:Alteromonas macleodii
290:Alteromonas macleodii
287:
253:Alteromonas macleodii
223:Alteromonas macleodii
195:Alteromonas macleodii
174:Alteromonas macleodii
156:Alteromonas macleodii
131:Alteromonas macleodii
25:Alteromonas macleodii
3160:10.1128/aem.01831-19
3016:Genome Announcements
2906:10.1128/aem.02952-10
2713:Genome Announcements
2081:Current Microbiology
1411:Current Microbiology
549:Role in carbon cycle
16:Species of bacterium
3261:2017Biofo..33..505C
3152:2020ApEnM..86E1831C
3085:2021PLoSO..1657800C
2963:2022ISMEJ..16..358M
2898:2011ApEnM..77.4055T
2675:2016EnvMi..18.4369M
2621:2019ISMEJ..13...92K
2522:2017Biofo..33..505C
2469:2014LimOc..59..349F
2412:2017ISMEJ..11.1813L
2292:2008ISMEJ...2.1194I
2226:2016EnvMi..18.4369M
2175:2015EnvMi..17.3857N
1821:1997MEPS..150..275F
1766:2008MolEc..17.4092I
1703:2014PNAS..111.7202P
1643:1996ApEnM..62...67R
1591:2002EnvMi...4...42G
1531:2019Heliy...502435B
1365:2020NatSR..10..809K
1287:10.1128/jb.01565-12
1227:2002EnvMi...4...42G
1167:2005ApEnM..71.2813F
1061:2012NatSR...2E.696L
940:potassium tellurite
895:lipopolysaccharides
852:The "deep-ecotype"
812:diguanylate cyclase
775:These factors give
620:releases different
467:is able to sustain
307:were described, as
69:Gammaproteobacteria
3380:10.4161/bact.24766
3327:10.1093/gbe/evs112
1830:10.3354/meps150275
1353:Scientific Reports
1049:Scientific Reports
679:
358:The physiology of
294:
185:bacteria in 1972,
3897:
3896:
3869:Open Tree of Life
3718:Taxon identifiers
3485:10.1111/jam.12629
3434:10.3390/v13060987
3321:(12): 1360–1374.
3205:Materials Letters
2892:(12): 4055–4065.
2669:(12): 4369–4377.
2286:(12): 1194–1212.
2220:(12): 4369–4377.
2169:(10): 3857–3868.
1760:(18): 4092–4106.
1754:Molecular Ecology
1697:(20): 7202–7207.
1069:10.1038/srep00696
907:Autographiviridae
517:citric acid cycle
473:cells undergoing
392:exopolysaccharide
233:γ-proteobacterium
170:
169:
165:
113:A. macleodii
3927:
3890:
3889:
3877:
3876:
3864:
3863:
3851:
3850:
3838:
3837:
3825:
3824:
3812:
3811:
3799:
3798:
3786:
3785:
3773:
3772:
3760:
3759:
3758:
3745:
3744:
3743:
3713:
3708:
3706:
3705:
3680:
3678:
3677:
3651:
3650:
3618:
3612:
3611:
3585:
3561:
3555:
3554:
3544:
3512:
3506:
3505:
3487:
3478:(5): 1293–1304.
3463:
3457:
3456:
3446:
3436:
3408:
3402:
3401:
3391:
3355:
3349:
3348:
3338:
3306:
3297:
3296:
3240:
3231:
3230:
3220:
3196:
3190:
3189:
3179:
3131:
3125:
3124:
3114:
3096:
3064:
3058:
3057:
3047:
3007:
3001:
3000:
2990:
2951:The ISME Journal
2942:
2936:
2935:
2925:
2877:
2871:
2870:
2860:
2850:
2826:
2817:
2816:
2806:
2788:
2764:
2755:
2754:
2744:
2704:
2695:
2694:
2657:
2651:
2650:
2640:
2609:The ISME Journal
2600:
2594:
2593:
2561:
2550:
2549:
2505:
2499:
2498:
2480:
2448:
2442:
2441:
2431:
2406:(8): 1813–1824.
2400:The ISME Journal
2391:
2385:
2384:
2374:
2356:
2328:
2322:
2321:
2303:
2280:The ISME Journal
2271:
2254:
2253:
2201:
2195:
2194:
2157:
2148:
2147:
2119:
2113:
2112:
2076:
2067:
2066:
2056:
2046:
2018:
2012:
2011:
2001:
1965:
1956:
1955:
1907:
1898:
1897:
1849:
1843:
1842:
1832:
1800:
1794:
1793:
1748:
1735:
1734:
1724:
1714:
1682:
1673:
1672:
1662:
1622:
1611:
1610:
1574:
1563:
1562:
1552:
1542:
1502:
1491:
1490:
1454:
1443:
1442:
1406:
1395:
1394:
1384:
1344:
1317:
1316:
1306:
1266:
1255:
1254:
1206:
1197:
1196:
1186:
1161:(6): 2813–2823.
1146:
1140:
1139:
1129:
1097:
1091:
1090:
1080:
1040:
959:are produced by
873:Phage infecting
863:A. mediterranea,
826:Relationship to
743:carried by some
733:A. mediterranea,
499:might influence
179:marine bacterium
159:
133:
89:Alteromonadaceae
40:
39:
21:
3935:
3934:
3930:
3929:
3928:
3926:
3925:
3924:
3910:Alteromonadales
3900:
3899:
3898:
3893:
3885:
3880:
3872:
3867:
3859:
3854:
3846:
3841:
3833:
3828:
3820:
3815:
3807:
3802:
3794:
3789:
3781:
3776:
3768:
3763:
3754:
3753:
3748:
3739:
3738:
3733:
3720:
3703:
3701:
3700:. BacDiveID:444
3691:
3688:
3683:
3675:
3673:
3664:
3660:
3658:Further reading
3655:
3654:
3620:
3619:
3615:
3563:
3562:
3558:
3514:
3513:
3509:
3465:
3464:
3460:
3410:
3409:
3405:
3357:
3356:
3352:
3308:
3307:
3300:
3242:
3241:
3234:
3198:
3197:
3193:
3133:
3132:
3128:
3079:(9): e0257800.
3066:
3065:
3061:
3009:
3008:
3004:
2944:
2943:
2939:
2879:
2878:
2874:
2828:
2827:
2820:
2766:
2765:
2758:
2706:
2705:
2698:
2659:
2658:
2654:
2602:
2601:
2597:
2563:
2562:
2553:
2507:
2506:
2502:
2450:
2449:
2445:
2393:
2392:
2388:
2335:Prochlorococcus
2330:
2329:
2325:
2273:
2272:
2257:
2203:
2202:
2198:
2159:
2158:
2151:
2130:(14): fnaa116.
2121:
2120:
2116:
2078:
2077:
2070:
2020:
2019:
2015:
1967:
1966:
1959:
1909:
1908:
1901:
1851:
1850:
1846:
1802:
1801:
1797:
1750:
1749:
1738:
1684:
1683:
1676:
1624:
1623:
1614:
1576:
1575:
1566:
1504:
1503:
1494:
1456:
1455:
1446:
1408:
1407:
1398:
1346:
1345:
1320:
1268:
1267:
1258:
1208:
1207:
1200:
1148:
1147:
1143:
1099:
1098:
1094:
1042:
1041:
1016:
1011:
992:
973:
953:
920:
915:
913:Industrial uses
891:mismatch repair
878:
854:A. mediterranea
839:A. mediterranea
831:
757:
749:Alteromonadales
728:
726:Genomic Islands
688:genomic islands
681:The genomes of
671:A. mediterranea
659:
627:transcriptional
594:polysaccharides
551:
537:
509:
507:Iron limitation
495:in the oceans.
479:Prochlorococcus
470:Prochlorococcus
462:
442:
337:prochlorococcus
325:
313:microaerophilic
282:
219:
158:
142:
135:
129:
116:
79:Alteromonadales
34:
17:
12:
11:
5:
3933:
3931:
3923:
3922:
3917:
3912:
3902:
3901:
3895:
3894:
3892:
3891:
3878:
3865:
3852:
3839:
3826:
3813:
3800:
3787:
3774:
3761:
3746:
3730:
3728:
3722:
3721:
3716:
3710:
3709:
3687:
3686:External links
3684:
3682:
3681:
3661:
3659:
3656:
3653:
3652:
3613:
3576:(2): 310–315.
3556:
3507:
3458:
3403:
3350:
3298:
3255:(6): 505–519.
3232:
3191:
3126:
3059:
3002:
2957:(2): 358–369.
2937:
2872:
2818:
2756:
2696:
2652:
2595:
2551:
2516:(6): 505–519.
2500:
2463:(2): 349–360.
2443:
2386:
2354:10.1101/657627
2323:
2255:
2196:
2149:
2114:
2087:(6): 751–755.
2068:
2013:
1972:(April 2020).
1957:
1922:(1): 119–132.
1899:
1864:(1): 119–132.
1844:
1795:
1736:
1674:
1612:
1564:
1492:
1465:(2): 422–427.
1444:
1417:(6): 448–452.
1396:
1318:
1256:
1198:
1141:
1112:(1): 402–429.
1092:
1013:
1012:
1010:
1007:
991:
988:
972:
969:
957:κ-carrageenase
952:
949:
919:
916:
914:
911:
877:
871:
830:
824:
756:
753:
727:
724:
665:The genome of
658:
655:
579:microbial loop
550:
547:
536:
533:
508:
505:
493:fixes nitrogen
461:
458:
441:
438:
324:
321:
281:
278:
239:. Isolates of
218:
215:
199:DNA chromosome
191:Pseudomonadota
168:
167:
151:
150:
144:
143:
136:
125:
124:
118:
117:
110:
108:
104:
103:
96:
92:
91:
86:
82:
81:
76:
72:
71:
66:
62:
61:
59:Pseudomonadota
56:
52:
51:
46:
42:
41:
28:
27:
15:
13:
10:
9:
6:
4:
3:
2:
3932:
3921:
3918:
3916:
3913:
3911:
3908:
3907:
3905:
3888:
3883:
3879:
3875:
3870:
3866:
3862:
3857:
3853:
3849:
3844:
3840:
3836:
3831:
3827:
3823:
3818:
3814:
3810:
3805:
3801:
3797:
3792:
3788:
3784:
3779:
3775:
3771:
3766:
3762:
3757:
3751:
3747:
3742:
3736:
3732:
3731:
3729:
3727:
3723:
3719:
3714:
3699:
3695:
3690:
3689:
3685:
3671:
3667:
3663:
3662:
3657:
3648:
3644:
3640:
3636:
3632:
3628:
3624:
3617:
3614:
3609:
3605:
3601:
3597:
3593:
3589:
3584:
3579:
3575:
3571:
3567:
3560:
3557:
3552:
3548:
3543:
3538:
3534:
3530:
3527:(1): 012044.
3526:
3522:
3518:
3511:
3508:
3503:
3499:
3495:
3491:
3486:
3481:
3477:
3473:
3469:
3462:
3459:
3454:
3450:
3445:
3440:
3435:
3430:
3426:
3422:
3418:
3416:
3407:
3404:
3399:
3395:
3390:
3385:
3381:
3377:
3374:(2): e24766.
3373:
3369:
3368:Bacteriophage
3365:
3363:
3354:
3351:
3346:
3342:
3337:
3332:
3328:
3324:
3320:
3316:
3312:
3305:
3303:
3299:
3294:
3290:
3286:
3282:
3278:
3274:
3270:
3266:
3262:
3258:
3254:
3250:
3246:
3239:
3237:
3233:
3228:
3224:
3219:
3214:
3210:
3206:
3202:
3195:
3192:
3187:
3183:
3178:
3173:
3169:
3165:
3161:
3157:
3153:
3149:
3145:
3141:
3137:
3130:
3127:
3122:
3118:
3113:
3108:
3104:
3100:
3095:
3090:
3086:
3082:
3078:
3074:
3070:
3063:
3060:
3055:
3051:
3046:
3041:
3037:
3033:
3029:
3025:
3021:
3017:
3013:
3006:
3003:
2998:
2994:
2989:
2984:
2980:
2976:
2972:
2968:
2964:
2960:
2956:
2952:
2948:
2941:
2938:
2933:
2929:
2924:
2919:
2915:
2911:
2907:
2903:
2899:
2895:
2891:
2887:
2883:
2876:
2873:
2868:
2864:
2859:
2854:
2849:
2844:
2840:
2836:
2832:
2825:
2823:
2819:
2814:
2810:
2805:
2800:
2796:
2792:
2787:
2782:
2778:
2774:
2770:
2763:
2761:
2757:
2752:
2748:
2743:
2738:
2734:
2730:
2726:
2722:
2718:
2714:
2710:
2703:
2701:
2697:
2692:
2688:
2684:
2680:
2676:
2672:
2668:
2664:
2656:
2653:
2648:
2644:
2639:
2634:
2630:
2626:
2622:
2618:
2615:(1): 92–103.
2614:
2610:
2606:
2599:
2596:
2591:
2587:
2583:
2579:
2575:
2571:
2567:
2560:
2558:
2556:
2552:
2547:
2543:
2539:
2535:
2531:
2527:
2523:
2519:
2515:
2511:
2504:
2501:
2496:
2492:
2488:
2484:
2479:
2474:
2470:
2466:
2462:
2458:
2454:
2447:
2444:
2439:
2435:
2430:
2425:
2421:
2417:
2413:
2409:
2405:
2401:
2397:
2390:
2387:
2382:
2378:
2373:
2368:
2364:
2360:
2355:
2350:
2347:(4): 657627.
2346:
2342:
2338:
2336:
2327:
2324:
2319:
2315:
2311:
2307:
2302:
2297:
2293:
2289:
2285:
2281:
2277:
2270:
2268:
2266:
2264:
2262:
2260:
2256:
2251:
2247:
2243:
2239:
2235:
2231:
2227:
2223:
2219:
2215:
2211:
2209:
2200:
2197:
2192:
2188:
2184:
2180:
2176:
2172:
2168:
2164:
2156:
2154:
2150:
2145:
2141:
2137:
2133:
2129:
2125:
2118:
2115:
2110:
2106:
2102:
2098:
2094:
2090:
2086:
2082:
2075:
2073:
2069:
2064:
2060:
2055:
2050:
2045:
2040:
2036:
2032:
2028:
2026:
2017:
2014:
2009:
2005:
2000:
1995:
1991:
1987:
1983:
1979:
1975:
1971:
1964:
1962:
1958:
1953:
1949:
1945:
1941:
1937:
1933:
1929:
1925:
1921:
1917:
1913:
1906:
1904:
1900:
1895:
1891:
1887:
1883:
1879:
1875:
1871:
1867:
1863:
1859:
1855:
1848:
1845:
1840:
1836:
1831:
1826:
1822:
1818:
1814:
1810:
1806:
1799:
1796:
1791:
1787:
1783:
1779:
1775:
1771:
1767:
1763:
1759:
1755:
1747:
1745:
1743:
1741:
1737:
1732:
1728:
1723:
1718:
1713:
1708:
1704:
1700:
1696:
1692:
1688:
1681:
1679:
1675:
1670:
1666:
1661:
1656:
1652:
1648:
1644:
1640:
1636:
1632:
1628:
1621:
1619:
1617:
1613:
1608:
1604:
1600:
1596:
1592:
1588:
1584:
1580:
1573:
1571:
1569:
1565:
1560:
1556:
1551:
1546:
1541:
1536:
1532:
1528:
1525:(9): e02435.
1524:
1520:
1516:
1514:
1510:
1501:
1499:
1497:
1493:
1488:
1484:
1480:
1476:
1472:
1468:
1464:
1460:
1453:
1451:
1449:
1445:
1440:
1436:
1432:
1428:
1424:
1420:
1416:
1412:
1405:
1403:
1401:
1397:
1392:
1388:
1383:
1378:
1374:
1370:
1366:
1362:
1358:
1354:
1350:
1343:
1341:
1339:
1337:
1335:
1333:
1331:
1329:
1327:
1325:
1323:
1319:
1314:
1310:
1305:
1300:
1296:
1292:
1288:
1284:
1280:
1276:
1272:
1265:
1263:
1261:
1257:
1252:
1248:
1244:
1240:
1236:
1232:
1228:
1224:
1220:
1216:
1212:
1205:
1203:
1199:
1194:
1190:
1185:
1180:
1176:
1172:
1168:
1164:
1160:
1156:
1152:
1145:
1142:
1137:
1133:
1128:
1123:
1119:
1115:
1111:
1107:
1103:
1096:
1093:
1088:
1084:
1079:
1074:
1070:
1066:
1062:
1058:
1054:
1050:
1046:
1039:
1037:
1035:
1033:
1031:
1029:
1027:
1025:
1023:
1021:
1019:
1015:
1008:
1006:
1004:
1000:
996:
989:
987:
985:
981:
977:
970:
968:
966:
962:
958:
950:
948:
946:
941:
937:
932:
929:
925:
917:
912:
910:
908:
904:
900:
896:
892:
887:
883:
876:
872:
870:
868:
864:
859:
855:
851:
848:
844:
840:
836:
829:
825:
823:
821:
817:
813:
809:
805:
804:antimicrobial
800:
799:
797:
792:
791:
786:
782:
778:
774:
770:
765:
761:
754:
752:
750:
746:
742:
738:
734:
731:GIs found in
725:
723:
721:
717:
713:
709:
704:
700:
697:
693:
689:
684:
676:
672:
668:
663:
656:
654:
651:
647:
643:
639:
634:
632:
628:
623:
619:
615:
611:
607:
603:
599:
595:
590:
588:
584:
580:
576:
572:
567:
563:
560:
556:
555:heterotrophic
548:
546:
543:
535:Copper stress
534:
532:
530:
526:
522:
518:
514:
506:
504:
502:
501:Trichodesmium
498:
494:
490:
489:
488:Trichodesmium
484:
480:
476:
472:
471:
466:
459:
457:
455:
451:
447:
439:
437:
435:
432:
428:
424:
420:
416:
412:
408:
403:
401:
397:
393:
389:
388:D-amino acids
385:
380:
377:
373:
369:
365:
361:
356:
354:
350:
346:
342:
338:
334:
330:
322:
320:
318:
314:
310:
306:
301:
298:
291:
286:
279:
277:
274:
270:
266:
265:r-strategists
262:
257:
254:
250:
246:
242:
238:
234:
231:
230:heterotrophic
228:
227:gram-negative
224:
216:
214:
212:
208:
204:
200:
196:
192:
188:
184:
180:
176:
175:
166:
163:
157:
152:
149:
145:
140:
134:
132:
126:
123:
122:Binomial name
119:
115:
114:
109:
106:
105:
102:
101:
97:
94:
93:
90:
87:
84:
83:
80:
77:
74:
73:
70:
67:
64:
63:
60:
57:
54:
53:
50:
47:
44:
43:
38:
33:
29:
26:
22:
19:
3725:
3702:. Retrieved
3697:
3674:. Retrieved
3669:
3633:(1): 49–59.
3630:
3626:
3616:
3573:
3569:
3559:
3524:
3520:
3510:
3475:
3471:
3461:
3424:
3420:
3414:
3406:
3371:
3367:
3361:
3353:
3318:
3314:
3252:
3248:
3208:
3204:
3194:
3143:
3139:
3129:
3076:
3072:
3062:
3019:
3015:
3005:
2954:
2950:
2940:
2889:
2885:
2875:
2838:
2835:BMC Genomics
2834:
2776:
2772:
2716:
2712:
2666:
2662:
2655:
2612:
2608:
2598:
2573:
2569:
2513:
2509:
2503:
2460:
2456:
2446:
2403:
2399:
2389:
2344:
2340:
2334:
2326:
2283:
2279:
2217:
2213:
2207:
2199:
2166:
2162:
2127:
2123:
2117:
2084:
2080:
2034:
2030:
2024:
2016:
1981:
1977:
1919:
1915:
1861:
1857:
1847:
1812:
1808:
1798:
1757:
1753:
1694:
1690:
1637:(1): 67–73.
1634:
1630:
1585:(1): 42–50.
1582:
1578:
1522:
1518:
1512:
1508:
1462:
1458:
1414:
1410:
1356:
1352:
1281:(24): 6998.
1278:
1274:
1221:(1): 42–50.
1218:
1214:
1158:
1154:
1144:
1109:
1105:
1095:
1052:
1048:
999:uronic acids
994:
993:
975:
974:
961:A. macleodii
960:
954:
945:A. macleodii
944:
935:
933:
921:
906:
902:
899:A. macleodii
898:
886:A. macleodii
885:
879:
875:A. macleodii
874:
866:
862:
858:dioxygenases
853:
850:
846:
842:
838:
835:A. macleodii
834:
832:
827:
816:A. macleodii
815:
808:A. macleodii
807:
801:
794:
788:
781:Megaplasmids
777:A. macleodii
776:
772:
768:
764:A. macleodii
763:
759:
758:
745:A. macleodii
744:
737:A. macleodii
736:
732:
729:
715:
708:A. macleodii
707:
702:
701:
691:
683:A. macleodii
682:
680:
674:
670:
666:
650:A. macleodii
649:
646:A. macleodii
645:
642:carbon cycle
638:A. macleodii
637:
635:
631:A. macleodii
630:
618:A. macleodii
617:
601:
597:
591:
586:
575:A. macleodii
574:
571:A. macleodii
570:
561:
558:
552:
542:A. macleodii
541:
538:
529:A. macleodii
528:
513:A. macleodii
512:
510:
500:
496:
486:
483:A. macleodii
482:
478:
468:
464:
463:
453:
446:A. macleodii
445:
443:
426:
407:A. macleodii
406:
404:
400:A. macleodii
399:
384:A. macleodii
383:
381:
368:A. macleodii
367:
359:
357:
353:siderophores
328:
326:
316:
304:
302:
296:
295:
289:
280:Distribution
273:A. macleodii
272:
261:A. macleodii
260:
258:
252:
241:A. macleodii
240:
222:
221:The species
220:
211:A. macleodii
210:
207:A. macleodii
206:
194:
187:A. macleodii
186:
173:
172:
171:
161:
155:
154:
138:
130:
128:
112:
111:
99:
24:
18:
3750:Wikispecies
3415:Alteromonas
1815:: 275–285.
965:carrageenan
903:Podoviridae
867:Alteromonas
796:Pseudomonas
790:Escherichia
720:siderophore
716:Alteromonas
675:Alteromonas
667:Alteromonas
622:catabolites
598:Alteromonas
587:Alteromonas
450:amino acids
423:Kappaphycus
245:luminescent
100:Alteromonas
3904:Categories
3704:2022-04-04
3676:2022-04-04
3427:(6): 987.
3249:Biofouling
3211:: 130712.
2841:(1): 938.
2510:Biofouling
1970:Barbeau KA
1359:(1): 809.
1055:(1): 696.
1009:References
1003:wastewater
922:Elemental
592:Cell wall
557:bacteria,
521:glycolysis
372:tryptophan
345:homoserine
333:metabolism
323:Physiology
269:copiotroph
217:Morphology
3592:1364-5072
3551:1755-1307
3277:0892-7014
3227:0167-577X
3168:0099-2240
3103:1932-6203
3036:2169-8287
2979:1751-7370
2914:0099-2240
2795:1664-302X
2733:2169-8287
2590:0964-8305
2576:: 71–76.
2487:0024-3590
2242:1462-2912
1936:1572-9699
1878:0003-6072
1839:0171-8630
1487:145022909
1295:0021-9193
1243:1462-2912
928:metalloid
924:tellurium
785:orthologs
696:pangenome
606:laminarin
562:macleodii
475:chlorosis
419:nucleases
415:proteases
249:pigmented
237:flagellum
107:Species:
3809:10034789
3741:Q4736627
3735:Wikidata
3647:24751009
3608:23606744
3600:12147080
3502:18504786
3494:25175548
3453:34073246
3398:24228219
3345:23212172
3293:46739077
3285:28604167
3186:31757820
3121:34582496
3073:PLOS ONE
3054:27881542
2997:34341506
2932:21515719
2867:25344729
2813:27014193
2751:26316635
2691:27059936
2647:30116038
2546:46739077
2538:28604167
2495:85813253
2438:28440800
2381:32788385
2310:18670397
2250:27059936
2191:25847866
2144:32658271
2109:14922228
2101:24526245
2063:28400762
2008:32345736
1978:mSystems
1952:18595072
1944:25326795
1894:18595072
1886:25326795
1790:38830049
1782:19238708
1731:24733921
1607:11966824
1559:31687549
1479:31053303
1439:23929160
1431:12732953
1391:31964928
1313:23209244
1251:11966824
1193:15932972
1087:23019517
984:pyruvate
741:Plasmids
610:alginate
564:consume
431:alginate
396:biofilms
376:tyrosine
349:lactones
160:Baumann
148:Synonyms
137:Baumann
85:Family:
55:Phylum:
49:Bacteria
45:Domain:
3796:3222676
3765:BacDive
3698:BacDive
3529:Bibcode
3444:8228385
3421:Viruses
3389:3821669
3336:3542563
3257:Bibcode
3177:6974629
3148:Bibcode
3112:8478169
3081:Bibcode
3045:5122684
2988:8776838
2959:Bibcode
2923:3131633
2894:Bibcode
2858:4223743
2804:4781885
2779:: 248.
2742:4551879
2671:Bibcode
2638:6298977
2617:Bibcode
2518:Bibcode
2465:Bibcode
2429:5520037
2408:Bibcode
2372:7439483
2349:bioRxiv
2288:Bibcode
2222:Bibcode
2210:strain"
2171:Bibcode
2054:5368175
2037:: 507.
1999:7190382
1817:Bibcode
1762:Bibcode
1722:4034236
1699:Bibcode
1669:8572714
1639:Bibcode
1587:Bibcode
1550:6819836
1527:Bibcode
1519:Heliyon
1382:6972757
1361:Bibcode
1304:3510622
1223:Bibcode
1184:1151842
1163:Bibcode
1136:4552999
1078:3458243
1057:Bibcode
980:acetate
440:Ecology
411:lipases
183:aerobic
95:Genus:
75:Order:
65:Class:
3887:742978
3874:726602
3861:742978
3822:959258
3645:
3606:
3598:
3590:
3549:
3500:
3492:
3451:
3441:
3396:
3386:
3343:
3333:
3291:
3283:
3275:
3225:
3184:
3174:
3166:
3119:
3109:
3101:
3052:
3042:
3034:
2995:
2985:
2977:
2930:
2920:
2912:
2865:
2855:
2811:
2801:
2793:
2749:
2739:
2731:
2689:
2645:
2635:
2588:
2544:
2536:
2493:
2485:
2436:
2426:
2379:
2369:
2351:
2318:520422
2316:
2308:
2248:
2240:
2189:
2142:
2107:
2099:
2061:
2051:
2006:
1996:
1950:
1942:
1934:
1892:
1884:
1876:
1837:
1788:
1780:
1729:
1719:
1667:
1660:167774
1657:
1605:
1557:
1547:
1485:
1477:
1437:
1429:
1389:
1379:
1311:
1301:
1293:
1249:
1241:
1191:
1181:
1134:
1127:247423
1124:
1085:
1075:
882:CRISPR
657:Genome
614:pectin
612:, and
523:, and
434:lyases
374:, and
341:phenol
203:genome
164:, 1972
162:et al.
141:, 1972
139:et al.
3882:WoRMS
3848:28108
3804:IRMNG
3783:65Y2Q
3604:S2CID
3498:S2CID
3289:S2CID
3146:(3).
3022:(6).
2719:(4).
2542:S2CID
2491:S2CID
2314:S2CID
2105:S2CID
1984:(2).
1948:S2CID
1890:S2CID
1786:S2CID
1483:S2CID
1435:S2CID
773:merR.
3856:OBIS
3843:NCBI
3830:LPSN
3817:ITIS
3791:GBIF
3643:PMID
3596:PMID
3588:ISSN
3547:ISSN
3490:PMID
3449:PMID
3394:PMID
3341:PMID
3281:PMID
3273:ISSN
3223:ISSN
3182:PMID
3164:ISSN
3117:PMID
3099:ISSN
3050:PMID
3032:ISSN
2993:PMID
2975:ISSN
2928:PMID
2910:ISSN
2863:PMID
2809:PMID
2791:ISSN
2747:PMID
2729:ISSN
2687:PMID
2643:PMID
2586:ISSN
2534:PMID
2483:ISSN
2434:PMID
2377:PMID
2341:mBio
2306:PMID
2246:PMID
2238:ISSN
2187:PMID
2140:PMID
2097:PMID
2059:PMID
2004:PMID
1940:PMID
1932:ISSN
1882:PMID
1874:ISSN
1835:ISSN
1778:PMID
1727:PMID
1665:PMID
1603:PMID
1555:PMID
1475:PMID
1427:PMID
1387:PMID
1309:PMID
1291:ISSN
1247:PMID
1239:ISSN
1189:PMID
1132:PMID
1083:PMID
982:and
905:and
793:and
769:copA
417:and
351:and
263:are
247:nor
3778:CoL
3770:444
3635:doi
3578:doi
3537:doi
3480:doi
3476:117
3439:PMC
3429:doi
3384:PMC
3376:doi
3331:PMC
3323:doi
3265:doi
3213:doi
3209:305
3172:PMC
3156:doi
3107:PMC
3089:doi
3040:PMC
3024:doi
2983:PMC
2967:doi
2918:PMC
2902:doi
2853:PMC
2843:doi
2799:PMC
2781:doi
2737:PMC
2721:doi
2679:doi
2633:PMC
2625:doi
2578:doi
2526:doi
2473:doi
2424:PMC
2416:doi
2367:PMC
2359:doi
2296:doi
2230:doi
2179:doi
2132:doi
2128:367
2089:doi
2049:PMC
2039:doi
1994:PMC
1986:doi
1924:doi
1920:107
1866:doi
1862:107
1825:doi
1813:150
1770:doi
1717:PMC
1707:doi
1695:111
1655:PMC
1647:doi
1595:doi
1545:PMC
1535:doi
1467:doi
1463:514
1419:doi
1377:PMC
1369:doi
1299:PMC
1283:doi
1279:194
1231:doi
1179:PMC
1171:doi
1122:PMC
1114:doi
1110:110
1073:PMC
1065:doi
787:in
553:As
205:of
3906::
3884::
3871::
3858::
3845::
3832::
3819::
3806::
3793::
3780::
3767::
3752::
3737::
3696:.
3668:.
3641:.
3631:90
3629:.
3625:.
3602:.
3594:.
3586:.
3574:93
3572:.
3568:.
3545:.
3535:.
3525:55
3523:.
3519:.
3496:.
3488:.
3474:.
3470:.
3447:.
3437:.
3425:13
3423:.
3419:.
3392:.
3382:.
3370:.
3366:.
3339:.
3329:.
3317:.
3313:.
3301:^
3287:.
3279:.
3271:.
3263:.
3253:33
3251:.
3247:.
3235:^
3221:.
3207:.
3203:.
3180:.
3170:.
3162:.
3154:.
3144:86
3142:.
3138:.
3115:.
3105:.
3097:.
3087:.
3077:16
3075:.
3071:.
3048:.
3038:.
3030:.
3018:.
3014:.
2991:.
2981:.
2973:.
2965:.
2955:16
2953:.
2949:.
2926:.
2916:.
2908:.
2900:.
2890:77
2888:.
2884:.
2861:.
2851:.
2839:15
2837:.
2833:.
2821:^
2807:.
2797:.
2789:.
2775:.
2771:.
2759:^
2745:.
2735:.
2727:.
2715:.
2711:.
2699:^
2685:.
2677:.
2667:18
2665:.
2641:.
2631:.
2623:.
2613:13
2611:.
2607:.
2584:.
2574:83
2572:.
2568:.
2554:^
2540:.
2532:.
2524:.
2514:33
2512:.
2489:.
2481:.
2471:.
2461:59
2459:.
2455:.
2432:.
2422:.
2414:.
2404:11
2402:.
2398:.
2375:.
2365:.
2357:.
2345:11
2343:.
2339:.
2312:.
2304:.
2294:.
2282:.
2278:.
2258:^
2244:.
2236:.
2228:.
2218:18
2216:.
2212:.
2185:.
2177:.
2167:17
2165:.
2152:^
2138:.
2126:.
2103:.
2095:.
2085:68
2083:.
2071:^
2057:.
2047:.
2033:.
2029:.
2002:.
1992:.
1980:.
1976:.
1960:^
1946:.
1938:.
1930:.
1918:.
1914:.
1902:^
1888:.
1880:.
1872:.
1860:.
1856:.
1833:.
1823:.
1811:.
1807:.
1784:.
1776:.
1768:.
1758:17
1756:.
1739:^
1725:.
1715:.
1705:.
1693:.
1689:.
1677:^
1663:.
1653:.
1645:.
1635:62
1633:.
1629:.
1615:^
1601:.
1593:.
1581:.
1567:^
1553:.
1543:.
1533:.
1521:.
1517:.
1495:^
1481:.
1473:.
1461:.
1447:^
1433:.
1425:.
1415:46
1413:.
1399:^
1385:.
1375:.
1367:.
1357:10
1355:.
1351:.
1321:^
1307:.
1297:.
1289:.
1277:.
1273:.
1259:^
1245:.
1237:.
1229:.
1217:.
1213:.
1201:^
1187:.
1177:.
1169:.
1159:71
1157:.
1153:.
1130:.
1120:.
1108:.
1104:.
1081:.
1071:.
1063:.
1051:.
1047:.
1017:^
1005:.
909:.
822:.
751:.
616:,
608:,
585::
559:A.
519:,
425:.
413:,
347:,
319:.
271:,
251:.
3707:.
3679:.
3649:.
3637::
3610:.
3580::
3553:.
3539::
3531::
3504:.
3482::
3455:.
3431::
3417:"
3400:.
3378::
3372:3
3364:"
3347:.
3325::
3319:4
3295:.
3267::
3259::
3229:.
3215::
3188:.
3158::
3150::
3123:.
3091::
3083::
3056:.
3026::
3020:4
2999:.
2969::
2961::
2934:.
2904::
2896::
2869:.
2845::
2815:.
2783::
2777:7
2753:.
2723::
2717:3
2693:.
2681::
2673::
2649:.
2627::
2619::
2592:.
2580::
2548:.
2528::
2520::
2497:.
2475::
2467::
2440:.
2418::
2410::
2383:.
2361::
2333:"
2320:.
2298::
2290::
2284:2
2252:.
2232::
2224::
2193:.
2181::
2173::
2146:.
2134::
2111:.
2091::
2065:.
2041::
2035:8
2027:"
2010:.
1988::
1982:5
1954:.
1926::
1896:.
1868::
1841:.
1827::
1819::
1792:.
1772::
1764::
1733:.
1709::
1701::
1671:.
1649::
1641::
1609:.
1597::
1589::
1583:4
1561:.
1537::
1529::
1523:5
1515:"
1489:.
1469::
1441:.
1421::
1393:.
1371::
1363::
1315:.
1285::
1253:.
1233::
1225::
1219:4
1195:.
1173::
1165::
1138:.
1116::
1089:.
1067::
1059::
1053:2
843:.
798:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.