360:
5′ phosphate group and the terminal 3′ hydroxyl group (at the ends of the strand or sequence in question), because these ends determine the direction of transcription and translation. A sequence written 5′-CGCTAT-3′ is equivalent to a sequence written 3′-TATCGC-5′ as long as the 5′ and 3′ ends are noted. If the ends are not labeled, convention is to assume that both sequences are written in the 5′-to-3′ direction. The "Watson strand" refers to 5′-to-3′ top strand (5′→3′), whereas the "Crick strand" refers to the 5′-to-3′ bottom strand (3′←5′). Both Watson and Crick strands can be either sense or antisense strands depending on the specific gene product made from them.
371:(ORF) from the centromere of the left arm (L) of Yeast (Y) chromosome number V (E), and that the expression coding strand is the Watson strand (W). "YKL074C" denotes the 74th ORF to the left of the centromere of chromosome XI and that the coding strand is the Crick strand (C). Another confusing term referring to "Plus" and "Minus" strand is also widely used. Whether the strand is sense (positive) or antisense (negative), the default query sequence in NCBI BLAST alignment is "Plus" strand.
200:
254:. The transcribed DNA strand is called the template strand, with antisense sequence, and the mRNA transcript produced from it is said to be sense sequence (the complement of antisense). The untranscribed DNA strand, complementary to the transcribed strand, is also said to have sense sequence; it has the same sense sequence as the mRNA transcript (though T bases in DNA are substituted with U bases in RNA).
208:
be used to make methionine because it will not be directly used to make mRNA. The DNA sense strand is called a "sense" strand not because it will be used to make protein (it won't be), but because it has a sequence that corresponds directly to the RNA codon sequence. By this logic, the RNA transcript itself is sometimes described as "sense".
538:
from it prior to translation. Like DNA, negative-sense RNA has a nucleotide sequence complementary to the mRNA that it encodes; also like DNA, this RNA cannot be translated into protein directly. Instead, it must first be transcribed into a positive-sense RNA that acts as an mRNA. Some viruses (e.g.
359:
you are writing the sequence that contains the information for proteins (the "sense" information), not on which strand is depicted as "on the top" or "on the bottom" (which is arbitrary). The only biological information that is important for labeling strands is the relative locations of the terminal
207:
Hence, a base triplet 3′-TAC-5′ in the DNA antisense strand (complementary to the 5′-ATG-3′ of the DNA sense strand) is used as the template which results in a 5′-AUG-3′ base triplet in the mRNA. The DNA sense strand will have the triplet ATG, which looks similar to the mRNA triplet AUG but will not
82:
between nucleic acid polymers, a double-stranded DNA molecule will be composed of two strands with sequences that are reverse complements of each other. To help molecular biologists specifically identify each strand individually, the two strands are usually differentiated as the "sense" strand and
507:
into viral proteins (e.g., those needed for viral replication). Therefore, in positive-sense RNA viruses, the viral RNA genome can be considered viral mRNA, and can be immediately translated by the host cell. Unlike negative-sense RNA, positive-sense RNA is of the same sense as mRNA. Some viruses
554:
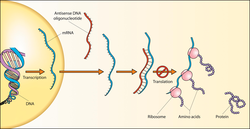
can be achieved by introducing into cells a short "antisense oligonucleotide" that is complementary to an RNA target. This experiment was first done by
Zamecnik and Stephenson in 1978 and continues to be a useful approach, both for laboratory experiments and potentially for clinical applications
155:
The terms "sense" and "antisense" are relative only to the particular RNA transcript in question, and not to the DNA strand as a whole. In other words, either DNA strand can serve as the sense or antisense strand. Most organisms with sufficiently large genomes make use of both strands, with each
589:
Other antisense mechanisms are not enzyme-dependent, but involve steric blocking of their target RNA (e.g. to prevent translation or to induce alternative splicing). Steric blocking antisense mechanisms often use oligonucleotides that are heavily modified. Since there is no need for RNase H
147:
are encountered in place of sense and antisense, respectively, and in the context of a double-stranded DNA molecule the usage of these terms is essentially equivalent. However, the coding/sense strand need not always contain a code that is used to make a protein; both protein-coding and
62:. Depending on the context, sense may have slightly different meanings. For example, the negative-sense strand of DNA is equivalent to the template strand, whereas the positive-sense strand is the non-template strand whose nucleotide sequence is equivalent to the sequence of the
135:
construct the RNA transcript, but the complementary base-pairing by which nucleic acid polymerization occurs means that the sequence of the RNA transcript will look identical to the positive-sense strand, apart from the RNA transcript's use of uracil instead of thymine.
192:. However, the DNA sense strand itself is not used as the template for the mRNA; it is the DNA antisense strand that serves as the source for the protein code, because, with bases complementary to the DNA sense strand, it is used as a template for the mRNA. Since
413:". In other words, it is a non-coding strand complementary to the coding sequence of RNA; this is similar to negative-sense viral RNA. When mRNA forms a duplex with a complementary antisense RNA sequence, translation is blocked. This process is related to
1153:
Kumar, B.; Khanna, Madhu; Kumar, P.; Sood, V.; Vyas, R.; Banerjea, A. C. (2011-07-09). "Nucleic Acid-Mediated
Cleavage of M1 Gene of Influenza A Virus Is Significantly Augmented by Antisense Molecules Targeted to Hybridize Close to the Cleavage Site".
477:, also known as a "minus-strand". In most cases, the terms "sense" and "strand" are used interchangeably, making terms such as "positive-strand" equivalent to "positive-sense", and "plus-strand" equivalent to "plus-sense". Whether a
1101:
Kumar, Prashant; Kumar, Binod; Rajput, Roopali; Saxena, Latika; Banerjea, Akhil C.; Khanna, Madhu (2013-06-02). "Cross-Protective Effect of
Antisense Oligonucleotide Developed Against the Common 3′ NCR of Influenza A Virus Genome".
156:
strand functioning as the template strand for different RNA transcripts in different places along the same DNA molecule. In some cases, RNA transcripts can be transcribed in both directions (i.e. on either strand) from a common
429:
coding for antisense RNA in order to block the expression of a gene of interest. Radioactively or fluorescently labelled antisense RNA can be used to show the level of transcription of genes in various cell types.
828:
512:) have positive-sense genomes that can act as mRNA and be used directly to synthesize proteins without the help of a complementary RNA intermediate. Because of this, these viruses do not need to have an
391:
are also ssRNA viruses with an ambisense genome, as they have three fragments that are mainly negative-sense except for part of the 5′ ends of the large and small segments of their genome.
184:(protein synthesis) to build an amino acid sequence and then a protein. For example, the sequence "ATG" within a DNA sense strand corresponds to an "AUG" codon in the mRNA, which
570:
to degrade the target RNA. This makes the mechanism of gene silencing catalytic. Double-stranded RNA can also act as a catalytic, enzyme-dependent antisense agent through the
131:), and is reverse complementary to both the positive-sense strand and the RNA transcript. It is actually the antisense strand that is used as the template from which
993:
Kumar, Binod; Khanna, Madhu; Meseko, Clement A.; Sanicas, Melvin; Kumar, Prashant; Asha, Kumari; Asha, Kumari; Kumar, Prashant; Sanicas, Melvin (January 2019).
364:
520:—the RNA polymerase will be one of the first proteins produced by the host cell, since it is needed in order for the virus's genome to be replicated.
529:
490:
586:
provides yet another example of an enzyme-dependent antisense regulation process through enzymatic degradation of the resulting RNA duplex.
310:
RNA strand that is transcribed from the noncoding (template/antisense) strand. Note: Except for the fact that all thymines are now uracils (
238:, which are usually instructions specifying the order in which amino acids are assembled to make proteins, as well as regulatory sequences,
337:
RNA strand that is transcribed from the coding (nontemplate/sense) strand. Note: Except for the fact that all thymines are now uracils (
75:
55:
616:
356:
566:
If the antisense oligonucleotide contains a stretch of DNA or a DNA mimic (phosphorothioate DNA, 2′F-ANA, or others) it can recruit
250:. For a cell to use this information, one strand of the DNA serves as a template for the synthesis of a complementary strand of
578:
pathway, involving target mRNA recognition through sense-antisense strand pairing followed by target mRNA degradation by the
387:
have three single-stranded RNA (ssRNA) fragments, some of them containing both positive-sense and negative-sense sections;
854:
579:
196:
results in an RNA product complementary to the DNA template strand, the mRNA is complementary to the DNA antisense strand.
534:
Negative-sense (3′-to-5′) viral RNA is complementary to the viral mRNA, thus a positive-sense RNA must be produced by an
535:
442:
1044:
Kumar, Binod; Asha, Kumari; Khanna, Madhu; Ronsard, Larance; Meseko, Clement
Adebajo; Sanicas, Melvin (2018-01-10).
1215:
631:
193:
563:(SARS-CoV), have been targeted using antisense oligonucleotides to inhibit their replication in host cells.
100:
697:"Ambisense segment 3 of rice stripe virus: the first instance of a virus containing two ambisense segments"
636:
434:
882:"Inhibition of Rous sarcoma Virus Replication and Cell Transformation by a Specific Oligodeoxynucleotide"
379:
A single-stranded genome that is used in both positive-sense and negative-sense capacities is said to be
504:
181:
104:
96:
425:. The concept has also been exploited as a molecular biology technique, by artificially introducing a
893:
591:
595:
119:
bases in the RNA sequence). The other strand of the double-stranded DNA molecule is referred to as
1187:
1135:
368:
157:
199:
1225:
1179:
1171:
1127:
1119:
1083:
1065:
1026:
970:
921:
810:
759:
718:
677:
611:
556:
438:
31:
1163:
1111:
1073:
1057:
1016:
1006:
960:
952:
911:
901:
800:
790:
749:
708:
669:
560:
543:
viruses) have negative-sense genomes and so must carry an RNA polymerase inside the virion.
414:
1046:"The emerging influenza virus threat: status and new prospects for its therapy and control"
738:"Complete nucleotide sequence of RNA 3 of rice stripe virus: an ambisense coding strategy"
621:
422:
897:
1078:
1045:
1021:
994:
965:
940:
805:
778:
583:
551:
513:
149:
132:
995:"Advancements in Nucleic Acid Based Therapeutics against Respiratory Viral Infections"
916:
881:
673:
1204:
509:
410:
400:
173:
1191:
1139:
559:). Several viruses, such as influenza viruses Respiratory syncytial virus (RSV) and
481:
is positive-sense or negative-sense can be used as a basis for classifying viruses.
1220:
1210:
384:
247:
239:
185:
79:
43:
27:
Property of nucleic acid strands with respect to their translatability into protein
203:
Schematic showing how antisense DNA strands can interfere with protein translation
754:
737:
713:
696:
1167:
1115:
1061:
599:
450:
446:
406:
388:
189:
108:
59:
1175:
1123:
1069:
626:
540:
466:
426:
1183:
1131:
1087:
1030:
974:
814:
681:
503:) viral RNA signifies that a particular viral RNA sequence may be directly
363:
For example, the notation "YEL021W", an alias of the URA3 gene used in the
906:
795:
763:
722:
462:
418:
35:
17:
925:
1011:
660:
Anne-Lise Haenni (2003). "Expression strategies of ambisense viruses".
567:
161:
112:
956:
500:
496:
465:, the term "sense" has a slightly different meaning. The genome of an
83:
the "antisense" strand. An individual strand of DNA is referred to as
517:
421:, which interact with complementary mRNA molecules and inhibit their
243:
116:
445:(FDA) has approved the phosphorothioate antisense oligonucleotides
641:
575:
478:
198:
177:
176:(mRNA) transcript, and can therefore be used to read the expected
571:
235:
63:
590:
recognition, this can include chemistries such as 2′-O-alkyl,
417:. Cells can produce antisense RNA molecules naturally, called
251:
51:
47:
941:"Silencing Disease Genes in the Laboratory and in the Clinic"
855:"FDA approves orphan drug for inherited cholesterol disorder"
234:
Some regions within a double-stranded DNA molecule code for
355:
The names assigned to each strand actually depend on which
54:, refers to the nature of the roles of the strand and its
779:"The multiple personalities of Watson and Crick strands"
367:(NCBI) database, denotes that this gene is in the 21st
318:
to the noncoding (template/antisense) DNA strand and
736:
Zhu Y; Hayakawa T; Toriyama S; Takahashi M. (1991).
695:Kakutani T; Hayano Y; Hayashi T; Minobe Y. (1991).
349:to the noncoding (template/antisense) DNA strand.
345:to the coding (nontemplate/sense) DNA strand and
99:corresponds directly to the sequence of an RNA
322:to the coding (nontemplate/sense) DNA strand.
365:National Center for Biotechnology Information
180:sequence that will ultimately be used during
8:
405:An RNA sequence that is complementary to an
115:bases in the DNA sequence are replaced with
777:Cartwright, Reed; Dan Graur (Feb 8, 2011).
164:on either strand (see "ambisense" below).
1077:
1020:
1010:
964:
915:
905:
880:Zamecnik, P.C.; Stephenson, M.L. (1978).
804:
794:
753:
712:
530:Negative-sense single-stranded RNA virus
491:Positive-sense single-stranded RNA virus
256:
652:
383:. Some viruses have ambisense genomes.
435:alternative antisense structural types
295:Complementary to the template strand.
275:Used as a template for transcription.
160:region, or be transcribed from within
46:molecule, particularly of a strand of
453:(Kynamro) for human therapeutic use.
409:mRNA transcript is sometimes called "
7:
988:
986:
984:
473:, also known as a "plus-strand", or
437:have been experimentally applied as
172:The DNA sense strand looks like the
107:or translatable into a sequence of
617:Directionality (molecular biology)
25:
939:Watts, J.K.; Corey, D.R. (2012).
829:"FDA approves fomivirsen for CMV"
212:Example with double-stranded DNA
218:DNA strand 1: antisense strand
1:
674:10.1016/S0168-1702(03)00094-7
580:RNA-induced silencing complex
999:Journal of Clinical Medicine
536:RNA-dependent RNA polymerase
443:Food and Drug Administration
441:. In the United States, the
58:in specifying a sequence of
1242:
886:Proc. Natl. Acad. Sci. USA
755:10.1099/0022-1317-72-4-763
714:10.1099/0022-1317-72-2-465
547:Antisense oligonucleotides
527:
488:
398:
229:DNA strand 2: sense strand
1168:10.1007/s12033-011-9437-z
1116:10.1007/s12033-013-9670-8
1062:10.1007/s00705-018-3708-y
469:can be said to be either
334:mRNA antisense transcript
632:Transcription (genetics)
1156:Molecular Biotechnology
1104:Molecular Biotechnology
582:(RISC). The R1 plasmid
329:3′CGCUAUAGCGUUU 5′
302:5′GCGAUAUCGCAAA 3′
283:5′GCGATATCGCAAA 3′
263:3′CGCTATAGCGTTT 5′
637:Translation (genetics)
204:
139:Sometimes the phrases
907:10.1073/pnas.75.1.280
796:10.1186/1745-6150-6-7
307:mRNA sense transcript
202:
1050:Archives of Virology
592:peptide nucleic acid
457:RNA sense in viruses
292:(nontemplate/coding)
272:(template/noncoding)
270:DNA antisense strand
152:may be transcribed.
898:1978PNAS...75..280Z
596:locked nucleic acid
220:(transcribed to) →
188:for the amino acid
111:(provided that any
97:nucleotide sequence
1012:10.3390/jcm8010006
516:packaged into the
369:open reading frame
242:sites, non-coding
222:RNA strand (sense)
205:
1216:Molecular biology
957:10.1002/path.2993
861:. 30 January 2013
642:Viral replication
612:Antisense therapy
557:antisense therapy
439:antisense therapy
353:
352:
32:molecular biology
16:(Redirected from
1233:
1196:
1195:
1150:
1144:
1143:
1098:
1092:
1091:
1081:
1041:
1035:
1034:
1024:
1014:
990:
979:
978:
968:
936:
930:
929:
919:
909:
877:
871:
870:
868:
866:
851:
845:
844:
842:
840:
835:. 1 October 1998
825:
819:
818:
808:
798:
774:
768:
767:
757:
733:
727:
726:
716:
692:
686:
685:
657:
561:SARS coronavirus
495:Positive-sense (
449:(Vitravene) and
415:RNA interference
340:
331:
330:
313:
304:
303:
291:
290:DNA sense strand
286:
284:
271:
266:
264:
257:
21:
1241:
1240:
1236:
1235:
1234:
1232:
1231:
1230:
1201:
1200:
1199:
1152:
1151:
1147:
1100:
1099:
1095:
1043:
1042:
1038:
992:
991:
982:
938:
937:
933:
879:
878:
874:
864:
862:
853:
852:
848:
838:
836:
827:
826:
822:
776:
775:
771:
735:
734:
730:
694:
693:
689:
659:
658:
654:
650:
622:DNA codon table
608:
549:
532:
526:
493:
487:
459:
403:
397:
377:
338:
328:
327:
311:
301:
300:
289:
282:
280:
269:
262:
260:
214:
170:
150:non-coding RNAs
145:template strand
133:RNA polymerases
74:Because of the
72:
28:
23:
22:
15:
12:
11:
5:
1239:
1237:
1229:
1228:
1223:
1218:
1213:
1203:
1202:
1198:
1197:
1145:
1110:(3): 203–211.
1093:
1056:(4): 831–844.
1036:
980:
951:(2): 365–379.
931:
892:(1): 280–284.
872:
846:
820:
783:Biology Direct
769:
728:
687:
668:(2): 141–150.
662:Virus Research
651:
649:
646:
645:
644:
639:
634:
629:
624:
619:
614:
607:
604:
584:hok/sok system
552:Gene silencing
548:
545:
528:Main article:
525:
524:Negative-sense
522:
514:RNA polymerase
489:Main article:
486:
485:Positive-sense
483:
475:negative-sense
471:positive-sense
458:
455:
399:Main article:
396:
393:
376:
373:
351:
350:
335:
332:
324:
323:
308:
305:
297:
296:
293:
287:
277:
276:
273:
267:
232:
231:
225:
224:
213:
210:
169:
166:
121:negative-sense
85:positive-sense
71:
68:
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
1238:
1227:
1224:
1222:
1219:
1217:
1214:
1212:
1209:
1208:
1206:
1193:
1189:
1185:
1181:
1177:
1173:
1169:
1165:
1161:
1157:
1149:
1146:
1141:
1137:
1133:
1129:
1125:
1121:
1117:
1113:
1109:
1105:
1097:
1094:
1089:
1085:
1080:
1075:
1071:
1067:
1063:
1059:
1055:
1051:
1047:
1040:
1037:
1032:
1028:
1023:
1018:
1013:
1008:
1004:
1000:
996:
989:
987:
985:
981:
976:
972:
967:
962:
958:
954:
950:
946:
942:
935:
932:
927:
923:
918:
913:
908:
903:
899:
895:
891:
887:
883:
876:
873:
860:
856:
850:
847:
834:
830:
824:
821:
816:
812:
807:
802:
797:
792:
788:
784:
780:
773:
770:
765:
761:
756:
751:
747:
743:
739:
732:
729:
724:
720:
715:
710:
706:
702:
698:
691:
688:
683:
679:
675:
671:
667:
663:
656:
653:
647:
643:
640:
638:
635:
633:
630:
628:
625:
623:
620:
618:
615:
613:
610:
609:
605:
603:
601:
597:
593:
587:
585:
581:
577:
573:
569:
564:
562:
558:
553:
546:
544:
542:
537:
531:
523:
521:
519:
515:
511:
510:Coronaviridae
506:
502:
498:
492:
484:
482:
480:
476:
472:
468:
464:
456:
454:
452:
448:
444:
440:
436:
431:
428:
424:
420:
416:
412:
411:antisense RNA
408:
402:
401:Antisense RNA
395:Antisense RNA
394:
392:
390:
386:
382:
374:
372:
370:
366:
361:
358:
348:
344:
343:complementary
336:
333:
326:
325:
321:
317:
316:complementary
309:
306:
299:
298:
294:
288:
285:
279:
278:
274:
268:
265:
259:
258:
255:
253:
249:
248:gene products
245:
241:
237:
230:
227:
226:
223:
219:
216:
215:
211:
209:
201:
197:
195:
194:transcription
191:
187:
183:
179:
175:
174:messenger RNA
167:
165:
163:
159:
153:
151:
146:
142:
141:coding strand
137:
134:
130:
126:
122:
118:
114:
110:
106:
102:
98:
94:
90:
86:
81:
77:
76:complementary
69:
67:
65:
61:
57:
53:
49:
45:
41:
37:
33:
19:
1162:(1): 27–36.
1159:
1155:
1148:
1107:
1103:
1096:
1053:
1049:
1039:
1002:
998:
948:
944:
934:
889:
885:
875:
865:18 September
863:. Retrieved
858:
849:
839:18 September
837:. Retrieved
832:
823:
786:
782:
772:
748:(4): 763–7.
745:
741:
731:
707:(2): 465–8.
704:
700:
690:
665:
661:
655:
588:
565:
550:
533:
494:
479:viral genome
474:
470:
460:
432:
404:
389:arenaviruses
385:Bunyaviruses
380:
378:
362:
354:
346:
342:
319:
315:
281:
261:
246:, and other
233:
228:
221:
217:
206:
171:
154:
144:
140:
138:
128:
125:negative (−)
124:
120:
92:
89:positive (+)
88:
84:
80:base-pairing
73:
66:transcript.
44:nucleic acid
39:
29:
859:Drug Topics
742:J Gen Virol
701:J Gen Virol
602:oligomers.
598:(LNA), and
182:translation
109:amino acids
60:amino acids
1205:Categories
648:References
600:Morpholino
505:translated
451:mipomersen
447:fomivirsen
423:expression
407:endogenous
190:methionine
105:translated
101:transcript
91:or simply
78:nature of
56:complement
1176:1073-6085
1124:1073-6085
1070:0304-8608
945:J. Pathol
627:RNA virus
541:influenza
467:RNA virus
427:transgene
419:microRNAs
381:ambisense
375:Ambisense
357:direction
347:identical
341:), it is
320:identical
314:), it is
168:Sense DNA
129:antisense
103:which is
95:) if its
70:DNA sense
18:Ambisense
1226:Virology
1192:45686564
1184:21744034
1140:24496875
1132:23729285
1088:29322273
1031:30577479
1005:(1): 6.
975:22069063
815:21303550
682:12782362
606:See also
463:virology
240:splicing
158:promoter
36:genetics
1079:7087104
1022:6351902
966:3916955
894:Bibcode
806:3055211
764:2016591
723:1993885
594:(PNA),
568:RNase H
244:introns
162:introns
113:thymine
1190:
1182:
1174:
1138:
1130:
1122:
1086:
1076:
1068:
1029:
1019:
973:
963:
924:
917:411230
914:
833:healio
813:
803:
762:
721:
680:
518:virion
508:(e.g.
123:(also
117:uracil
87:(also
38:, the
1188:S2CID
1136:S2CID
926:75545
789:: 7.
576:siRNA
433:Some
339:T → U
312:T → U
236:genes
186:codes
178:codon
93:sense
42:of a
40:sense
1180:PMID
1172:ISSN
1128:PMID
1120:ISSN
1084:PMID
1066:ISSN
1027:PMID
971:PMID
922:PMID
867:2020
841:2020
811:PMID
760:PMID
719:PMID
678:PMID
572:RNAi
499:-to-
143:and
64:mRNA
34:and
1221:RNA
1211:DNA
1164:doi
1112:doi
1074:PMC
1058:doi
1054:163
1017:PMC
1007:doi
961:PMC
953:doi
949:226
912:PMC
902:doi
801:PMC
791:doi
750:doi
709:doi
670:doi
461:In
252:RNA
127:or
52:RNA
50:or
48:DNA
30:In
1207::
1186:.
1178:.
1170:.
1160:51
1158:.
1134:.
1126:.
1118:.
1108:55
1106:.
1082:.
1072:.
1064:.
1052:.
1048:.
1025:.
1015:.
1001:.
997:.
983:^
969:.
959:.
947:.
943:.
920:.
910:.
900:.
890:75
888:.
884:.
857:.
831:.
809:.
799:.
785:.
781:.
758:.
746:72
744:.
740:.
717:.
705:72
703:.
699:.
676:.
666:93
664:.
501:3′
497:5′
1194:.
1166::
1142:.
1114::
1090:.
1060::
1033:.
1009::
1003:8
977:.
955::
928:.
904::
896::
869:.
843:.
817:.
793::
787:6
766:.
752::
725:.
711::
684:.
672::
574:/
555:(
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.