317:
294:
191:
216:
2277:
575:
568:
2292:
323:
222:
40:
1252:
However, newborn infants absorb lipids relatively well, considering the low level of CDL and BSDL they produce. This observation has led to the suggestion that BSDL produced by lactating mammary gland and present within milk, may compensate for the low levels of other TG-digesting enzymes and aid
66:
1561:"A prospective, randomized, double-blind crossover study comparing rhBSSL (recombinant human Bile Salt Stimulated Lipase) and placebo added to infant formula during one week of treatment in preterm infants born before 32 weeks of gestational age: preliminary results"
1611:
Lidberg U, Nilsson J, Strömberg K, et al. (1992). "Genomic organization, sequence analysis, and chromosomal localization of the human carboxyl ester lipase (CEL) gene and a CEL-like (CELL) gene".
1704:
Lindström MB, Persson J, Thurn L, Borgström B (1991). "Effect of pancreatic phospholipase A2 and gastric lipase on the action of pancreatic carboxyl ester lipase against lipid substrates in vitro".
1912:
Erlanson-Albertsson C, Sternby B, Johannesson U (1985). "The interaction between human pancreatic carboxylester hydrolase (bile-salt-stimulated lipase of human milk) and lactoferrin".
2205:
Landberg E, Påhlsson P, Krotkiewski H, et al. (1997). "Glycosylation of bile-salt-stimulated lipase from human milk: comparison of native and recombinant forms".
1182:. BSDL has been found in the pancreatic secretions of all species in which it has been looked for. BSSL, originally discovered in the milk of humans and various other
330:
229:
1253:
newborns in lipid absorption. The importance of BSSL in breast milk for the preterm infant nutrition was suggested at 2007. It was also directly shown recently.
2276:
2055:
Nilsson J, Hellquist M, Bjursell G (1993). "The human carboxyl ester lipase-like (CELL) gene is ubiquitously expressed and contains a hypervariable region".
888:
152:
869:
1764:"Human milk bile-salt stimulated lipase. Sequence similarity with rat lysophospholipase and homology with the active site region of cholinesterases"
2589:
2291:
2836:
2026:
Wang CS, Dashti A, Jackson KW, et al. (1995). "Isolation and characterization of human milk bile salt-activated lipase C-tail fragment".
2256:
1330:
1312:
3222:
2750:
2851:
2845:
2646:
2396:
1516:
Andersson Y, Sävman K, et al. (2007). "Pasteurization of mother's own milk reduces fat absorption and growth in preterm infants".
1088:
316:
2694:
2356:
2805:
2594:
2337:
2318:
1640:
Taylor AK, Zambaux JL, Klisak I, et al. (1991). "Carboxyl ester lipase: a highly polymorphic locus on human chromosome 9qter".
1095:
293:
1840:
Hui DY, Kissel JA (1991). "Sequence identity between human pancreatic cholesterol esterase and bile salt-stimulated milk lipase".
1879:"Purification and molecular characterization of FAP, a feto-acinar protein associated with the differentiation of human pancreas"
2912:
2800:
1671:"cDNA cloning of human-milk bile-salt-stimulated lipase and evidence for its identity to pancreatic carboxylic ester hydrolase"
1299:
1278:
2721:
2631:
2584:
2121:"Human fetoacinar pancreatic protein: an oncofetal glycoform of the normally secreted pancreatic bile-salt-dependent lipase"
1295:
1477:"Molecular cloning of the bile salt-dependent lipase of ferret lactating mammary gland: an overview of functional residues"
215:
190:
63:
3143:
3084:
2362:
2343:
2324:
1582:
Kumar BV, Aleman-Gomez JA, Colwell N, et al. (1992). "Structure of the human pancreatic cholesterol esterase gene".
1274:
3188:
2755:
2745:
1962:
Roudani S, Miralles F, Margotat A, et al. (1995). "Bile salt-dependent lipase transcripts in human fetal tissues".
132:
2086:"Chaperone function of a Grp 94-related protein for folding and transport of the pancreatic bile salt-dependent lipase"
3148:
3040:
2665:
2734:
2730:
2726:
2642:
329:
228:
2996:
2942:
2902:
2861:
2765:
2604:
2572:
2458:
2422:
2249:
2164:"Pancreatic carboxyl ester lipase: a circulating enzyme that modifies normal and oxidized lipoproteins in vitro"
322:
221:
2650:
2499:
2413:
1203:
933:
140:
2519:
2389:
1436:
Murasugi A, Asami Y, Mera-Kikuchi Y (2001). "Production of recombinant human bile-salt-stimulated lipase in
914:
3075:
2714:
123:, BAL, BSDL, BSSL, CELL, CEase, FAP, FAPP, LIPA, MODY8, Bile salt-dependent lipase, carboxyl ester lipase
3010:
2907:
2699:
2660:
2524:
2446:
1214:
1733:
Baba T, Downs D, Jackson KW, et al. (1991). "Structure of human milk bile salt activated lipase".
3193:
3028:
3023:
2956:
2616:
2441:
2242:
1807:"cDNA cloning of carboxyl ester lipase from human pancreas reveals a unique proline-rich repeat unit"
1242:
204:
119:
3048:
3018:
2822:
2817:
2741:
2677:
2463:
2368:
2349:
2330:
3181:
3033:
2514:
2504:
2382:
1865:
1793:
1560:
1541:
164:
1071:
1050:
1024:
1003:
2982:
2927:
2895:
2770:
2489:
2222:
2193:
2150:
2107:
2072:
2043:
2014:
1979:
1950:
1929:
1900:
1857:
1828:
1785:
1750:
1721:
1692:
1657:
1628:
1599:
1533:
1498:
1457:
1417:
1379:
112:
56:
3232:
3058:
2638:
2543:
2538:
2494:
2214:
2183:
2175:
2140:
2132:
2097:
2064:
2035:
2004:
1971:
1921:
1890:
1849:
1818:
1775:
1742:
1713:
1682:
1649:
1620:
1591:
1525:
1488:
1449:
1409:
1369:
1361:
1245:. BSDL production in the newborn pancreas is quite low when compared with production in the
1218:
1186:, has since been found in the milk of many animals including dogs, cats, rats, and rabbits.
409:
340:
284:
239:
1238:
574:
567:
3160:
2974:
2890:
2885:
2880:
2793:
2788:
2548:
2426:
1195:
384:
1993:"Association of bile-salt-dependent lipase with membranes of human pancreatic microsomes"
160:
1400:
Lombardo, D. (2001). "Bile salt-dependent lipase: its pathophysiological implications".
1194:
More than 95% of the fat present in human milk and in infant formulas is in the form of
3133:
3128:
3123:
2509:
2484:
2480:
2453:
2436:
2145:
2120:
1687:
1670:
1374:
1349:
2188:
2163:
1895:
1878:
1823:
1806:
1493:
1476:
1413:
1237:). In particular, they can hydrolyze esters of the essential fatty acids (n-3 and n-6
803:
798:
793:
788:
783:
778:
773:
768:
763:
758:
753:
748:
743:
738:
733:
717:
712:
707:
702:
697:
692:
687:
682:
666:
661:
656:
651:
646:
641:
636:
631:
621:
616:
39:
3227:
3216:
2874:
2783:
2533:
2009:
1992:
1975:
1925:
1853:
1780:
1763:
1717:
1653:
1624:
1529:
1246:
603:
1869:
1797:
3001:
2932:
2655:
2576:
1222:
402:
181:
1545:
144:
168:
3176:
2947:
2841:
2704:
2670:
2608:
2374:
1335:
National Center for
Biotechnology Information, U.S. National Library of Medicine
1317:
National Center for
Biotechnology Information, U.S. National Library of Medicine
1230:
1179:
485:
3153:
1199:
301:
198:
148:
2102:
2085:
3066:
2865:
2778:
2405:
833:
545:
423:
368:
355:
267:
254:
156:
2218:
2068:
1537:
1461:
1453:
1421:
2226:
2197:
2154:
2111:
2076:
2047:
2018:
1983:
1954:
1933:
1904:
1861:
1832:
1789:
1754:
1725:
1696:
1661:
1632:
1603:
1502:
1383:
1135:
1130:
3111:
3106:
3101:
2922:
2557:
2409:
1234:
1207:
1167:
1119:
978:
959:
2039:
1746:
1595:
626:
3118:
3096:
3091:
2709:
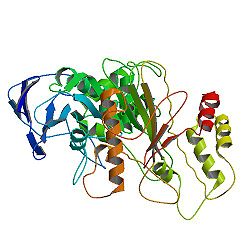
2285:: STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN BILE SALT ACTIVATED LIPASE
1183:
945:
900:
2179:
2136:
1365:
89:
85:
3138:
3071:
2964:
2562:
2470:
1163:
1103:
855:
17:
3080:
2831:
2827:
1226:
2162:
Shamir R, Johnson WJ, Morlock-Fitzpatrick K, et al. (1996).
1941:
Chekhranova MK, Il'ina EN, Shuvalova ER, et al. (1994). "".
818:
814:
2812:
2689:
2682:
2626:
2621:
136:
2378:
2238:
583:
2234:
2300:: Truncated Recombinant Human Bile Salt Stimulated Lipase
1991:
Bruneau N, de la Porte PL, Sbarra V, Lombardo D (1995).
1669:
Nilsson J, Bläckberg L, Carlsson P, et al. (1990).
392:
1198:(TG). In adults, TGs are thought to be broken down or
1350:"Two forms of human milk bile-salt-stimulated lipase"
557:
3169:
3057:
3009:
2995:
2973:
2955:
2941:
2921:
2860:
2764:
2603:
2571:
2421:
1064:
1043:
1017:
996:
2119:Mas E, Abouakil N, Roudani S, et al. (1993).
1291:
1289:
1287:
1270:
1268:
1266:
1206:(CDL) enzyme. In the newborn, CDL activity in the
2700:Fructose 6-P,2-kinase:fructose 2,6-bisphosphatase
339:
238:
1805:Reue K, Zambaux J, Wong H, et al. (1991).
1762:Christie DL, Cleverly DR, O'Connor CJ (1991).
1296:GRCm38: Ensembl release 89: ENSMUSG00000026818
2390:
2250:
8:
794:modulation of chemical synaptic transmission
1275:GRCh38: Ensembl release 89: ENSG00000170835
3006:
2952:
2938:
2397:
2383:
2375:
2257:
2243:
2235:
1395:
1393:
1217:and, like CDL, are capable of hydrolyzing
829:
599:
380:
279:
176:
74:
2187:
2144:
2101:
2008:
1894:
1822:
1779:
1686:
1492:
1475:Sbarra V, Bruneau N, et al. (1998).
1373:
1348:Swan JS, Hoffman MM, et al. (1992).
642:hydrolase activity, acting on ester bonds
1559:Maggio, L.; Bellagamba, M.; et al.
2590:Ubiquitin carboxy-terminal hydrolase L1
2272:
1262:
1178:) is an equivalent enzyme found within
27:Mammalian protein found in Homo sapiens
29:
3170:either deoxy- or ribo-
703:integral component of plasma membrane
344:
305:
300:
243:
202:
197:
7:
2751:Protein serine/threonine phosphatase
2852:Cyclic nucleotide phosphodiesterase
2846:Clostridium perfringens alpha toxin
2647:Tartrate-resistant acid phosphatase
1442:Protein Expression and Purification
1170:and aids in the digestion of fats.
647:carboxylic ester hydrolase activity
524:migratory enteric neural crest cell
2695:Pyruvate dehydrogenase phosphatase
1688:10.1111/j.1432-1033.1990.tb19259.x
1061:
1040:
1014:
993:
969:
950:
924:
905:
879:
860:
764:intestinal lipid catabolic process
562:
480:
418:
397:
25:
2595:4-hydroxybenzoyl-CoA thioesterase
1877:Escribano MJ, Imperial S (1990).
774:intestinal cholesterol absorption
2290:
2275:
2010:10.1111/j.1432-1033.1995.209_1.x
1530:10.1111/j.1651-2227.2007.00450.x
1213:Both BSDL and BSSL have a broad
573:
566:
328:
321:
315:
292:
227:
220:
214:
189:
38:
2913:N-acetylglucosamine-6-sulfatase
2801:Sphingomyelin phosphodiesterase
667:neurexin family protein binding
2722:Inositol-phosphate phosphatase
2585:Palmitoyl protein thioesterase
2084:Bruneau N, Lombardo D (1995).
799:postsynaptic membrane assembly
779:triglyceride metabolic process
584:More reference expression data
546:More reference expression data
462:right hemisphere of cerebellum
1:
3085:RNA-induced silencing complex
1896:10.1016/S0021-9258(20)88264-7
1824:10.1016/S0022-2275(20)42088-7
1494:10.1016/S0005-2760(98)00067-8
1481:Biochimica et Biophysica Acta
1414:10.1016/S1388-1981(01)00130-5
1402:Biochimica et Biophysica Acta
804:presynaptic membrane assembly
754:cholesterol catabolic process
313:
212:
3189:Serratia marcescens nuclease
2756:Dual-specificity phosphatase
2746:Protein tyrosine phosphatase
1976:10.1016/0167-4781(95)00141-3
1926:10.1016/0167-4838(85)90199-2
1854:10.1016/0014-5793(90)80525-N
1781:10.1016/0014-5793(91)80114-I
1718:10.1016/0005-2760(91)90220-C
1654:10.1016/0888-7543(91)90328-C
1625:10.1016/0888-7543(92)90134-E
789:synaptic vesicle endocytosis
734:fatty acid catabolic process
652:triglyceride lipase activity
622:acylglycerol lipase activity
3223:Genes on human chromosome 9
2666:Fructose 1,6-bisphosphatase
1566:. Swedish Orphan Biovitrum.
1172:Bile salt-stimulated lipase
662:signaling receptor activity
470:paraflocculus of cerebellum
3249:
2367:gene details page in the
2348:gene details page in the
2329:gene details page in the
1148:Bile salt-dependent lipase
759:pancreatic juice secretion
516:sexually immature organism
2903:Galactosamine-6 sulfatase
2459:6-phosphogluconolactonase
2270:
1331:"Mouse PubMed Reference:"
1313:"Human PubMed Reference:"
1210:is lower than in adults.
1204:colipase-dependent lipase
1134:
1129:
1125:
1118:
1102:
1089:Chr 9: 133.06 – 133.07 Mb
1083:
1068:
1047:
1036:
1021:
1000:
989:
976:
972:
957:
953:
944:
931:
927:
912:
908:
899:
886:
882:
867:
863:
854:
839:
832:
828:
812:
784:neuron cell-cell adhesion
602:
598:
581:
565:
556:
543:
492:
483:
430:
421:
391:
383:
379:
362:
349:
312:
291:
282:
278:
261:
248:
211:
188:
179:
175:
130:
127:
117:
110:
105:
82:
77:
60:
55:
50:
46:
37:
32:
2651:Purple acid phosphatases
2103:10.1074/jbc.270.22.13524
657:sterol esterase activity
1096:Chr 2: 28.45 – 28.45 Mb
769:lipid catabolic process
3076:Microprocessor complex
2715:Beta-propeller phytase
2219:10.1006/abbi.1997.0188
2207:Arch. Biochem. Biophys
2069:10.1006/geno.1993.1341
1964:Biochim. Biophys. Acta
1914:Biochim. Biophys. Acta
1706:Biochim. Biophys. Acta
1454:10.1006/prep.2001.1509
1235:lipid-soluble vitamins
1166:produced by the adult
744:protein esterification
520:mucous cell of stomach
3011:Endodeoxyribonuclease
2908:Iduronate-2-sulfatase
2661:Glucose 6-phosphatase
2447:Butyrylcholinesterase
1215:substrate specificity
1156:carboxyl ester lipase
693:extracellular exosome
532:tracheobronchial tree
508:epithelium of stomach
3194:Micrococcal nuclease
3029:Deoxyribonuclease IV
3024:Deoxyribonuclease II
2957:Exodeoxyribonuclease
2617:Alkaline phosphatase
2442:Acetylcholinesterase
2361:genome location and
2342:genome location and
2323:genome location and
688:extracellular region
346:2 A3|2 19.38 cM
307:Chromosome 2 (mouse)
205:Chromosome 9 (human)
78:List of PDB id codes
51:Available structures
3049:UvrABC endonuclease
3019:Deoxyribonuclease I
2742:Protein phosphatase
2678:Protein phosphatase
2476:Bile salt-dependent
2464:PAF acetylhydrolase
2369:UCSC Genome Browser
2350:UCSC Genome Browser
2331:UCSC Genome Browser
2040:10.1021/bi00033a039
1747:10.1021/bi00216a028
1596:10.1021/bi00141a017
1249:or adult pancreas.
698:extracellular space
500:islet of Langerhans
3182:Mung bean nuclease
3041:Restriction enzyme
3034:Restriction enzyme
1943:Mol. Biol. (Mosk.)
1190:Enzymatic activity
934:ENSMUSG00000026818
727:Biological process
676:Cellular component
637:hydrolase activity
627:catalytic activity
610:Molecular function
466:right uterine tube
446:anterior pituitary
3210:
3209:
3206:
3205:
3202:
3201:
2991:
2990:
2983:Oligonucleotidase
2928:deoxyribonuclease
2896:Steroid sulfatase
2771:Phosphodiesterase
2500:Hormone-sensitive
2308:
2307:
2180:10.1172/JCI118596
2137:10.1042/bj2890609
1524:(10): 1445–1449.
1366:10.1042/bj2830119
1219:triacylglycerides
1154:), also known as
1145:
1144:
1141:
1140:
1114:
1113:
1079:
1078:
1058:
1057:
1032:
1031:
1011:
1010:
985:
984:
966:
965:
940:
939:
921:
920:
895:
894:
876:
875:
824:
823:
594:
593:
590:
589:
552:
551:
539:
538:
504:lactiferous gland
477:
476:
450:corpus epididymis
375:
374:
274:
273:
101:
100:
97:
96:
61:Ortholog search:
16:(Redirected from
3240:
3059:Endoribonuclease
3045:
3039:
3007:
2953:
2939:
2639:Acid phosphatase
2520:Monoacylglycerol
2430:ester hydrolases
2399:
2392:
2385:
2376:
2294:
2279:
2259:
2252:
2245:
2236:
2230:
2201:
2191:
2158:
2148:
2115:
2105:
2096:(22): 13524–33.
2080:
2051:
2034:(33): 10639–44.
2022:
2012:
1987:
1958:
1937:
1908:
1898:
1889:(36): 21865–71.
1873:
1836:
1826:
1801:
1783:
1758:
1729:
1700:
1690:
1665:
1636:
1607:
1568:
1567:
1565:
1556:
1550:
1549:
1518:Acta Paediatrica
1513:
1507:
1506:
1496:
1472:
1466:
1465:
1432:
1426:
1425:
1397:
1388:
1387:
1377:
1345:
1339:
1338:
1327:
1321:
1320:
1309:
1303:
1293:
1282:
1272:
1221:(in addition to
1196:triacylglycerols
1127:
1126:
1098:
1091:
1074:
1062:
1053:
1041:
1037:RefSeq (protein)
1027:
1015:
1006:
994:
970:
951:
925:
906:
880:
861:
830:
749:lipid metabolism
600:
586:
577:
570:
563:
548:
488:
486:Top expressed in
481:
454:caput epididymis
434:body of pancreas
426:
424:Top expressed in
419:
398:
381:
371:
358:
347:
332:
325:
319:
308:
296:
280:
270:
257:
246:
231:
224:
218:
207:
193:
177:
171:
122:
115:
92:
75:
69:
48:
47:
42:
30:
21:
3248:
3247:
3243:
3242:
3241:
3239:
3238:
3237:
3213:
3212:
3211:
3198:
3165:
3053:
3043:
3037:
3000:
2987:
2975:Exoribonuclease
2969:
2946:
2930:
2926:
2917:
2891:Arylsulfatase L
2886:Arylsulfatase B
2881:Arylsulfatase A
2856:
2769:
2760:
2599:
2567:
2429:
2417:
2403:
2314:
2309:
2304:
2301:
2295:
2286:
2280:
2266:
2263:
2233:
2204:
2174:(7): 1696–704.
2168:J. Clin. Invest
2161:
2118:
2083:
2054:
2025:
1997:Eur. J. Biochem
1990:
1961:
1940:
1911:
1876:
1839:
1804:
1761:
1732:
1703:
1675:Eur. J. Biochem
1668:
1639:
1610:
1590:(26): 6077–81.
1581:
1577:
1575:Further reading
1572:
1571:
1563:
1558:
1557:
1553:
1515:
1514:
1510:
1474:
1473:
1469:
1438:Pichia pastoris
1435:
1433:
1429:
1399:
1398:
1391:
1347:
1346:
1342:
1329:
1328:
1324:
1311:
1310:
1306:
1294:
1285:
1273:
1264:
1259:
1192:
1136:View/Edit Mouse
1131:View/Edit Human
1094:
1087:
1084:Location (UCSC)
1070:
1049:
1023:
1002:
915:ENSG00000170835
808:
739:lipid digestion
722:
671:
632:protein binding
617:heparin binding
582:
572:
571:
544:
535:
530:
526:
522:
518:
514:
510:
506:
502:
498:
484:
473:
468:
464:
460:
456:
452:
448:
444:
442:pituitary gland
440:
436:
422:
366:
353:
345:
335:
334:
333:
326:
306:
283:Gene location (
265:
252:
244:
234:
233:
232:
225:
203:
180:Gene location (
169:CEL - orthologs
131:
118:
111:
84:
62:
28:
23:
22:
15:
12:
11:
5:
3246:
3244:
3236:
3235:
3230:
3225:
3215:
3214:
3208:
3207:
3204:
3203:
3200:
3199:
3197:
3196:
3191:
3186:
3185:
3184:
3173:
3171:
3167:
3166:
3164:
3163:
3158:
3157:
3156:
3151:
3146:
3141:
3131:
3126:
3121:
3116:
3115:
3114:
3109:
3104:
3099:
3089:
3088:
3087:
3078:
3063:
3061:
3055:
3054:
3052:
3051:
3046:
3031:
3026:
3021:
3015:
3013:
3004:
2993:
2992:
2989:
2988:
2986:
2985:
2979:
2977:
2971:
2970:
2968:
2967:
2961:
2959:
2950:
2936:
2919:
2918:
2916:
2915:
2910:
2905:
2900:
2899:
2898:
2893:
2888:
2883:
2870:
2868:
2858:
2857:
2855:
2854:
2849:
2839:
2834:
2825:
2820:
2815:
2810:
2809:
2808:
2798:
2797:
2796:
2791:
2781:
2775:
2773:
2762:
2761:
2759:
2758:
2753:
2748:
2739:
2738:
2737:
2719:
2718:
2717:
2707:
2702:
2697:
2692:
2687:
2686:
2685:
2675:
2674:
2673:
2663:
2658:
2653:
2636:
2635:
2634:
2629:
2624:
2613:
2611:
2601:
2600:
2598:
2597:
2592:
2587:
2581:
2579:
2569:
2568:
2566:
2565:
2560:
2554:
2553:
2552:
2551:
2546:
2541:
2530:
2529:
2528:
2527:
2525:Diacylglycerol
2522:
2517:
2512:
2507:
2502:
2497:
2492:
2487:
2478:
2467:
2466:
2461:
2456:
2454:Pectinesterase
2451:
2450:
2449:
2444:
2437:Cholinesterase
2433:
2431:
2419:
2418:
2404:
2402:
2401:
2394:
2387:
2379:
2373:
2372:
2353:
2334:
2313:
2312:External links
2310:
2306:
2305:
2303:
2302:
2296:
2289:
2287:
2281:
2274:
2271:
2268:
2267:
2264:
2262:
2261:
2254:
2247:
2239:
2232:
2231:
2202:
2159:
2116:
2081:
2052:
2023:
1988:
1959:
1938:
1909:
1874:
1848:(1–2): 131–4.
1837:
1802:
1759:
1730:
1701:
1666:
1637:
1608:
1578:
1576:
1573:
1570:
1569:
1551:
1508:
1467:
1448:(2): 282–288.
1427:
1389:
1360:(1): 119–122.
1340:
1322:
1304:
1283:
1261:
1260:
1258:
1255:
1202:mainly by the
1191:
1188:
1143:
1142:
1139:
1138:
1133:
1123:
1122:
1116:
1115:
1112:
1111:
1109:
1107:
1100:
1099:
1092:
1085:
1081:
1080:
1077:
1076:
1066:
1065:
1059:
1056:
1055:
1045:
1044:
1038:
1034:
1033:
1030:
1029:
1019:
1018:
1012:
1009:
1008:
998:
997:
991:
987:
986:
983:
982:
974:
973:
967:
964:
963:
955:
954:
948:
942:
941:
938:
937:
929:
928:
922:
919:
918:
910:
909:
903:
897:
896:
893:
892:
884:
883:
877:
874:
873:
865:
864:
858:
852:
851:
846:
841:
837:
836:
826:
825:
822:
821:
810:
809:
807:
806:
801:
796:
791:
786:
781:
776:
771:
766:
761:
756:
751:
746:
741:
736:
730:
728:
724:
723:
721:
720:
715:
710:
705:
700:
695:
690:
685:
679:
677:
673:
672:
670:
669:
664:
659:
654:
649:
644:
639:
634:
629:
624:
619:
613:
611:
607:
606:
596:
595:
592:
591:
588:
587:
579:
578:
560:
554:
553:
550:
549:
541:
540:
537:
536:
534:
533:
529:
525:
521:
517:
513:
509:
505:
501:
497:
496:pyloric antrum
493:
490:
489:
478:
475:
474:
472:
471:
467:
463:
459:
455:
451:
447:
443:
439:
435:
431:
428:
427:
415:
414:
406:
395:
389:
388:
385:RNA expression
377:
376:
373:
372:
364:
360:
359:
351:
348:
343:
337:
336:
327:
320:
314:
310:
309:
304:
298:
297:
289:
288:
276:
275:
272:
271:
263:
259:
258:
250:
247:
242:
236:
235:
226:
219:
213:
209:
208:
201:
195:
194:
186:
185:
173:
172:
129:
125:
124:
116:
108:
107:
103:
102:
99:
98:
95:
94:
80:
79:
71:
70:
59:
53:
52:
44:
43:
35:
34:
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
3245:
3234:
3231:
3229:
3226:
3224:
3221:
3220:
3218:
3195:
3192:
3190:
3187:
3183:
3180:
3179:
3178:
3175:
3174:
3172:
3168:
3162:
3159:
3155:
3152:
3150:
3147:
3145:
3142:
3140:
3137:
3136:
3135:
3132:
3130:
3127:
3125:
3122:
3120:
3117:
3113:
3110:
3108:
3105:
3103:
3100:
3098:
3095:
3094:
3093:
3090:
3086:
3082:
3079:
3077:
3073:
3070:
3069:
3068:
3065:
3064:
3062:
3060:
3056:
3050:
3047:
3042:
3035:
3032:
3030:
3027:
3025:
3022:
3020:
3017:
3016:
3014:
3012:
3008:
3005:
3003:
2998:
2994:
2984:
2981:
2980:
2978:
2976:
2972:
2966:
2963:
2962:
2960:
2958:
2954:
2951:
2949:
2944:
2940:
2937:
2934:
2929:
2924:
2920:
2914:
2911:
2909:
2906:
2904:
2901:
2897:
2894:
2892:
2889:
2887:
2884:
2882:
2879:
2878:
2877:
2876:
2875:arylsulfatase
2872:
2871:
2869:
2867:
2863:
2859:
2853:
2850:
2847:
2843:
2840:
2838:
2835:
2833:
2829:
2826:
2824:
2821:
2819:
2816:
2814:
2811:
2807:
2804:
2803:
2802:
2799:
2795:
2792:
2790:
2787:
2786:
2785:
2784:Phospholipase
2782:
2780:
2777:
2776:
2774:
2772:
2767:
2763:
2757:
2754:
2752:
2749:
2747:
2743:
2740:
2736:
2732:
2728:
2725:
2724:
2723:
2720:
2716:
2713:
2712:
2711:
2708:
2706:
2703:
2701:
2698:
2696:
2693:
2691:
2688:
2684:
2681:
2680:
2679:
2676:
2672:
2669:
2668:
2667:
2664:
2662:
2659:
2657:
2654:
2652:
2648:
2644:
2640:
2637:
2633:
2630:
2628:
2625:
2623:
2620:
2619:
2618:
2615:
2614:
2612:
2610:
2606:
2602:
2596:
2593:
2591:
2588:
2586:
2583:
2582:
2580:
2578:
2574:
2570:
2564:
2561:
2559:
2556:
2555:
2550:
2547:
2545:
2542:
2540:
2537:
2536:
2535:
2534:Phospholipase
2532:
2531:
2526:
2523:
2521:
2518:
2516:
2513:
2511:
2508:
2506:
2503:
2501:
2498:
2496:
2493:
2491:
2488:
2486:
2482:
2479:
2477:
2474:
2473:
2472:
2469:
2468:
2465:
2462:
2460:
2457:
2455:
2452:
2448:
2445:
2443:
2440:
2439:
2438:
2435:
2434:
2432:
2428:
2424:
2420:
2415:
2411:
2407:
2400:
2395:
2393:
2388:
2386:
2381:
2380:
2377:
2370:
2366:
2365:
2360:
2359:
2354:
2351:
2347:
2346:
2341:
2340:
2335:
2332:
2328:
2327:
2322:
2321:
2316:
2315:
2311:
2299:
2293:
2288:
2284:
2278:
2273:
2269:
2260:
2255:
2253:
2248:
2246:
2241:
2240:
2237:
2228:
2224:
2220:
2216:
2213:(1): 94–102.
2212:
2208:
2203:
2199:
2195:
2190:
2185:
2181:
2177:
2173:
2169:
2165:
2160:
2156:
2152:
2147:
2142:
2138:
2134:
2131:(2): 609–15.
2130:
2126:
2122:
2117:
2113:
2109:
2104:
2099:
2095:
2091:
2090:J. Biol. Chem
2087:
2082:
2078:
2074:
2070:
2066:
2063:(2): 416–22.
2062:
2058:
2053:
2049:
2045:
2041:
2037:
2033:
2029:
2024:
2020:
2016:
2011:
2006:
2003:(1): 209–18.
2002:
1998:
1994:
1989:
1985:
1981:
1977:
1973:
1970:(1): 141–50.
1969:
1965:
1960:
1956:
1952:
1948:
1944:
1939:
1935:
1931:
1927:
1923:
1919:
1915:
1910:
1906:
1902:
1897:
1892:
1888:
1884:
1883:J. Biol. Chem
1880:
1875:
1871:
1867:
1863:
1859:
1855:
1851:
1847:
1843:
1838:
1834:
1830:
1825:
1820:
1817:(2): 267–76.
1816:
1812:
1808:
1803:
1799:
1795:
1791:
1787:
1782:
1777:
1773:
1769:
1765:
1760:
1756:
1752:
1748:
1744:
1741:(2): 500–10.
1740:
1736:
1731:
1727:
1723:
1719:
1715:
1711:
1707:
1702:
1698:
1694:
1689:
1684:
1681:(2): 543–50.
1680:
1676:
1672:
1667:
1663:
1659:
1655:
1651:
1648:(2): 425–31.
1647:
1643:
1638:
1634:
1630:
1626:
1622:
1619:(3): 630–40.
1618:
1614:
1609:
1605:
1601:
1597:
1593:
1589:
1585:
1580:
1579:
1574:
1562:
1555:
1552:
1547:
1543:
1539:
1535:
1531:
1527:
1523:
1519:
1512:
1509:
1504:
1500:
1495:
1490:
1486:
1482:
1478:
1471:
1468:
1463:
1459:
1455:
1451:
1447:
1443:
1439:
1431:
1428:
1423:
1419:
1415:
1411:
1407:
1403:
1396:
1394:
1390:
1385:
1381:
1376:
1371:
1367:
1363:
1359:
1355:
1351:
1344:
1341:
1336:
1332:
1326:
1323:
1318:
1314:
1308:
1305:
1301:
1297:
1292:
1290:
1288:
1284:
1280:
1276:
1271:
1269:
1267:
1263:
1256:
1254:
1250:
1248:
1247:mammary gland
1244:
1240:
1236:
1232:
1228:
1224:
1223:phospholipids
1220:
1216:
1211:
1209:
1205:
1201:
1197:
1189:
1187:
1185:
1181:
1177:
1173:
1169:
1165:
1161:
1157:
1153:
1149:
1137:
1132:
1128:
1124:
1121:
1117:
1110:
1108:
1105:
1101:
1097:
1093:
1090:
1086:
1082:
1075:
1073:
1067:
1063:
1060:
1054:
1052:
1046:
1042:
1039:
1035:
1028:
1026:
1020:
1016:
1013:
1007:
1005:
999:
995:
992:
990:RefSeq (mRNA)
988:
981:
980:
975:
971:
968:
962:
961:
956:
952:
949:
947:
943:
936:
935:
930:
926:
923:
917:
916:
911:
907:
904:
902:
898:
891:
890:
885:
881:
878:
872:
871:
866:
862:
859:
857:
853:
850:
847:
845:
842:
838:
835:
831:
827:
820:
816:
811:
805:
802:
800:
797:
795:
792:
790:
787:
785:
782:
780:
777:
775:
772:
770:
767:
765:
762:
760:
757:
755:
752:
750:
747:
745:
742:
740:
737:
735:
732:
731:
729:
726:
725:
719:
716:
714:
711:
709:
706:
704:
701:
699:
696:
694:
691:
689:
686:
684:
681:
680:
678:
675:
674:
668:
665:
663:
660:
658:
655:
653:
650:
648:
645:
643:
640:
638:
635:
633:
630:
628:
625:
623:
620:
618:
615:
614:
612:
609:
608:
605:
604:Gene ontology
601:
597:
585:
580:
576:
569:
564:
561:
559:
555:
547:
542:
531:
527:
523:
519:
515:
511:
507:
503:
499:
495:
494:
491:
487:
482:
479:
469:
465:
461:
457:
453:
449:
445:
441:
437:
433:
432:
429:
425:
420:
417:
416:
413:
411:
407:
405:
404:
400:
399:
396:
394:
390:
386:
382:
378:
370:
365:
361:
357:
352:
342:
338:
331:
324:
318:
311:
303:
299:
295:
290:
286:
281:
277:
269:
264:
260:
256:
251:
241:
237:
230:
223:
217:
210:
206:
200:
196:
192:
187:
183:
178:
174:
170:
166:
162:
158:
154:
150:
146:
142:
138:
134:
126:
121:
114:
109:
104:
93:
91:
87:
81:
76:
73:
72:
68:
65:
58:
54:
49:
45:
41:
36:
31:
19:
3044:}}
3038:{{
3002:Endonuclease
2933:ribonuclease
2873:
2656:Nucleotidase
2577:Thioesterase
2475:
2363:
2357:
2344:
2338:
2325:
2319:
2297:
2282:
2210:
2206:
2171:
2167:
2128:
2124:
2093:
2089:
2060:
2056:
2031:
2028:Biochemistry
2027:
2000:
1996:
1967:
1963:
1949:(2): 464–7.
1946:
1942:
1920:(2): 282–7.
1917:
1913:
1886:
1882:
1845:
1841:
1814:
1811:J. Lipid Res
1810:
1774:(2): 190–4.
1771:
1767:
1738:
1735:Biochemistry
1734:
1712:(2): 194–7.
1709:
1705:
1678:
1674:
1645:
1641:
1616:
1612:
1587:
1584:Biochemistry
1583:
1554:
1521:
1517:
1511:
1487:(1): 80–89.
1484:
1480:
1470:
1445:
1441:
1437:
1430:
1405:
1401:
1357:
1353:
1343:
1334:
1325:
1316:
1307:
1251:
1212:
1193:
1175:
1171:
1159:
1155:
1151:
1147:
1146:
1069:
1048:
1022:
1001:
977:
958:
932:
913:
887:
868:
848:
843:
708:cell surface
408:
401:
266:133,071,861
253:133,061,981
128:External IDs
83:
3177:Nuclease S1
2948:Exonuclease
2842:Lecithinase
2671:Calcineurin
2609:Phosphatase
2515:Lipoprotein
2505:Endothelial
2265:PDB gallery
1408:(1): 1–28.
1231:cholesterol
1180:breast milk
367:28,453,415
354:28,445,807
106:Identifiers
3217:Categories
2490:Pancreatic
2427:Carboxylic
2125:Biochem. J
1354:Biochem. J
1302:, May 2017
1281:, May 2017
1257:References
1200:hydrolyzed
718:presynapse
412:(ortholog)
149:HomoloGene
3067:RNase III
2925:(includes
2866:Sulfatase
2779:Autotaxin
2643:Prostatic
2495:Lysosomal
2410:esterases
2406:Hydrolase
1842:FEBS Lett
1768:FEBS Lett
1072:NP_034015
1051:NP_001798
1025:NM_009885
1004:NM_001807
834:Orthologs
683:cytoplasm
438:beta cell
157:GeneCards
3161:RNase T1
2923:Nuclease
2558:Cutinase
2057:Genomics
1870:10716446
1798:28123400
1642:Genomics
1613:Genomics
1538:17714541
1462:11676603
1422:11514232
1298:–
1277:–
1208:duodenum
1184:primates
1168:pancreas
1162:) is an
1120:Wikidata
813:Sources:
512:duodenum
458:duodenum
3233:Enzymes
3134:RNase E
3129:RNase Z
3124:RNase A
3119:RNase P
3092:RNase H
2710:Phytase
2510:Hepatic
2485:Lingual
2481:Gastric
2227:9244386
2198:8601635
2155:8424803
2146:1132213
2112:7768954
2077:7691717
2048:7654718
2019:7588748
1984:7578248
1955:7514266
1934:3995055
1905:2600091
1862:2265692
1833:2066663
1790:1991511
1755:1988041
1726:1854805
1697:1698625
1662:1676983
1633:1639390
1604:1627550
1503:9714751
1384:1567358
1375:1131002
1300:Ensembl
1279:Ensembl
946:UniProt
901:Ensembl
840:Species
819:QuickGO
713:synapse
387:pattern
245:9q34.13
113:Aliases
3072:Drosha
2997:3.1.21
2965:RecBCD
2943:3.1.11
2563:PETase
2471:Lipase
2355:Human
2336:Human
2317:Human
2225:
2196:
2189:507234
2186:
2153:
2143:
2110:
2075:
2046:
2017:
1982:
1953:
1932:
1903:
1868:
1860:
1831:
1796:
1788:
1753:
1724:
1695:
1660:
1631:
1602:
1546:995002
1544:
1536:
1501:
1460:
1420:
1382:
1372:
1241:) and
1233:, and
1227:esters
1164:enzyme
1106:search
1104:PubMed
979:Q64285
960:P19835
856:Entrez
558:BioGPS
528:embryo
137:114840
3081:Dicer
3036:;see
2862:3.1.6
2832:PDE4B
2828:PDE4A
2766:3.1.4
2735:IMPA3
2731:IMPA2
2727:IMPA1
2605:3.1.3
2573:3.1.2
2423:3.1.1
1866:S2CID
1794:S2CID
1564:(PDF)
1542:S2CID
1239:PUFAs
889:12613
849:Mouse
844:Human
815:Amigo
410:Mouse
403:Human
350:Start
285:Mouse
249:Start
182:Human
153:37529
145:88374
3228:Milk
2999:-31:
2945:-16:
2931:and
2837:PDE5
2823:PDE3
2818:PDE2
2813:PDE1
2705:PTEN
2690:OCRL
2683:PP2A
2632:ALPP
2627:ALPL
2622:ALPI
2416:3.1)
2364:LIPA
2358:LIPA
2298:1jmy
2283:1f6w
2223:PMID
2194:PMID
2151:PMID
2108:PMID
2073:PMID
2044:PMID
2015:PMID
1980:PMID
1968:1264
1951:PMID
1930:PMID
1901:PMID
1858:PMID
1829:PMID
1786:PMID
1751:PMID
1722:PMID
1710:1084
1693:PMID
1658:PMID
1629:PMID
1600:PMID
1534:PMID
1499:PMID
1485:1393
1458:PMID
1418:PMID
1406:1533
1380:PMID
1176:BSSL
1174:(or
1158:(or
1152:BSDL
1150:(or
870:1056
393:Bgee
341:Band
302:Chr.
240:Band
199:Chr.
133:OMIM
90:1JMY
86:1F6W
67:RCSB
64:PDBe
18:BSSL
3154:4/5
2345:FAP
2339:FAP
2326:CEL
2320:CEL
2215:doi
2211:344
2184:PMC
2176:doi
2141:PMC
2133:doi
2129:289
2098:doi
2094:270
2065:doi
2036:doi
2005:doi
2001:233
1972:doi
1922:doi
1918:829
1891:doi
1887:264
1850:doi
1846:276
1819:doi
1776:doi
1772:278
1743:doi
1714:doi
1683:doi
1679:192
1650:doi
1621:doi
1592:doi
1526:doi
1489:doi
1450:doi
1440:".
1410:doi
1370:PMC
1362:doi
1358:283
1243:DHA
1229:of
1160:CEL
363:End
262:End
165:OMA
161:CEL
141:MGI
120:CEL
57:PDB
33:CEL
3219::
3112:2C
3107:2B
3102:2A
3083::
3074::
2864::
2744::
2733:,
2729:,
2645:)/
2607::
2575::
2544:A2
2539:A1
2425::
2414:EC
2408::
2221:.
2209:.
2192:.
2182:.
2172:97
2170:.
2166:.
2149:.
2139:.
2127:.
2123:.
2106:.
2092:.
2088:.
2071:.
2061:17
2059:.
2042:.
2032:34
2030:.
2013:.
1999:.
1995:.
1978:.
1966:.
1947:28
1945:.
1928:.
1916:.
1899:.
1885:.
1881:.
1864:.
1856:.
1844:.
1827:.
1815:32
1813:.
1809:.
1792:.
1784:.
1770:.
1766:.
1749:.
1739:30
1737:.
1720:.
1708:.
1691:.
1677:.
1673:.
1656:.
1646:10
1644:.
1627:.
1617:13
1615:.
1598:.
1588:31
1586:.
1540:.
1532:.
1522:96
1520:.
1497:.
1483:.
1479:.
1456:.
1446:23
1444:.
1416:.
1404:.
1392:^
1378:.
1368:.
1356:.
1352:.
1333:.
1315:.
1286:^
1265:^
1225:,
817:/
369:bp
356:bp
268:bp
255:bp
163:;
159::
155:;
151::
147:;
143::
139:;
135::
88:,
3149:3
3144:2
3139:1
3097:1
2935:)
2848:)
2844:(
2830:/
2806:1
2794:D
2789:C
2768::
2649:/
2641:(
2549:B
2483:/
2412:(
2398:e
2391:t
2384:v
2371:.
2352:.
2333:.
2258:e
2251:t
2244:v
2229:.
2217::
2200:.
2178::
2157:.
2135::
2114:.
2100::
2079:.
2067::
2050:.
2038::
2021:.
2007::
1986:.
1974::
1957:.
1936:.
1924::
1907:.
1893::
1872:.
1852::
1835:.
1821::
1800:.
1778::
1757:.
1745::
1728:.
1716::
1699:.
1685::
1664:.
1652::
1635:.
1623::
1606:.
1594::
1548:.
1528::
1505:.
1491::
1464:.
1452::
1434:*
1424:.
1412::
1386:.
1364::
1337:.
1319:.
287:)
184:)
167::
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.