49:
170:
473:
31:
327:
463:
of the packaging motor's three-way junction. When pRNA is in this tetramer ring form, it works as a part of the DNA packaging motor to transport DNA molecules to their destination location within the prohead capsule. Specifically, the functional domains of pRNA bind to the gp16 packaging enzyme and
511:
direction. This replication process also employs a sliding-back mechanism towards the 3’ end of the genome that uses a repeating TTT motif to move the replication complex backward without altering the template sequence. This allows the initiation of
1011:
556:, as most industries are currently unequipped to handle industrial pRNA synthesis. This is primarily because RNA nanotechnology is still an emerging field that lacks industrial application and manufacturing optimization of small RNAs.
1568:
Simpson, Alan A.; Tao, Yizhi; Leiman, Petr G.; Badasso, Mohammed O.; He, Yongning; Jardine, Paul J.; Olson, Norman H.; Morais, Marc C.; Grimes, Shelley; Anderson, Dwight L.; Baker, Timothy S.; Rossmann, Michael G. (2000).
342:(p8), the head or capsid fiber protein (p8.5), the distal tail knob (p9), the portal or connector protein (p10), the tail tube or lower collar proteins (p11), and the tail fibers or appendage proteins (p12*).
525:
464:
the structural connector molecule to aid in the translocation of DNA through the prohead channel. After DNA packaging is complete, the pRNA dissociates and is degraded.
2193:
507:
in other organisms. Φ29 forms a replication complex involving the p3 terminal protein, the dAMP nucleotide, and its own DNA polymerase to synthesize DNA in a 5’ to
357:
during viral replication. The Φ29 packaging motor is structurally composed of the procapsid and the connector proteins, which interact with the pRNA, the packaging
1117:
Zhang, Long; Mu, Chaofeng; Zhang, Tinghong; Yang, Dejun; Wang, Chenou; Chen, Qiong; Tang, Lin; Fan, Luhui; Liu, Cong; Shen, Jianliang; Li, Huaqiong (2021-01-07).
2152:
1119:"Development of targeted therapy therapeutics to sensitize triple-negative breast cancer chemosensitivity utilizing bacteriophage phi29 derived packaging RNA"
884:
Camacho, Ana; Jimenez, Fernando; Torre, Javier; Carrascosa, Jose L.; Mellado, Rafael P.; Vinuela, Eladio; Salas, Margarita; Vasquez, Cesar (February 1977).
1637:
Ding, Fang; Lu, Changrui; Zhao, Wei; Rajashankar, Kanagalaghatta R.; Anderson, Dwight L.; Jardine, Paul J.; Grimes, Shelley; Ke, Ailong (2011-05-03).
429:. Early studies such as Anderson (1990) and Trottier (1998) hypothesized that pRNA formed intermolecular hexamers, but these studies had a solely
1877:
De Vega, Miguel; Salas, Margarita (2011-09-26). "Chapter 9: Protein-Primed
Replication of Bacteriophage Φ29 DNA". In Kusic-Tisma, Jelena (ed.).
595:
Nanoparticles need to be stabilized as delivery mechanisms in order to adapt to microenvironments that may result in loss of therapeutic cargo.
1923:
1888:
1031:
846:
794:
213:. They are in the same order as phages PZA, Φ15, BS32, B103, M2Y (M2), Nf, and GA-1. First discovered in 1965, the Φ29 phage is the smallest
1063:"Current advances in Phi29 pRNA biology and its application in drug delivery: Current advances in Phi29 pRNA biology and its application"
1422:"Computer Modeling of Three-dimensional Structure of DNA-packaging RNA (pRNA) Monomer, Dimer, and Hexamer of Phi29 DNA Packaging Motor*"
2009:
Shu, Yi; Pi, Fengmei; Sharma, Ashwani; Rajabi, Mehdi; Haque, Farzin; Shu, Dan; Leggas, Markos; Evers, B. Mark; Guo, Peixuan (2014).
524:
2206:
2066:
629:
drug delivery mechanism to inhibit TNBC growth and volume. This treatment can also be combined with anti-cancer drugs like
603:
438:
516:
to be more accurate by having the polymerase complex check a specific sequence before beginning the elongation process.
447:, only a pentamer or smaller polymer could spatially fit in the virus. Ultimately, single isomorphous replacement with
459:
structure is a tetramer ring. This discovery aligned with what was known about the structural geometry and necessary
345:
The main difference between Φ29's structure and that of other phages is its use of pRNA in its DNA packaging motor.
48:
870:
260:
2157:
614:
is the only viable current treatment for TNBC because the loss of target receptors inherent to the disease causes
302:. Due to its small size and complex morphology, it has become an ideal model for the study of many processes in
2244:
2234:
2130:
549:. The pRNA in bacteriophage Φ29 can use its three-way junction in order to self-assemble into nanoparticles.
1344:
950:"Construction of Bacteriophage Phi29 DNA Packaging Motor and its Applications in Nanotechnology and Therapy"
625:
The three-way junction in the Φ29 DNA packaging motor can help sensitize TNBC cells to chemotherapy using a
564:Φ29’s DNA packaging system, using pRNA, incorporates a motor for the delivery of therapeutic molecules like
315:
299:
2092:
886:"Assembly of Bacillus subtilis Phage Phi29. 1. Mutants in the Cistrons Coding for the Structural Proteins"
553:
370:
268:
256:
1012:"Fabrication Methods for RNA Nanoparticle Assembly Based on Bacteriophage Phi29 pRNA Structural Features"
452:
362:
1178:"Microbe Profile: Bacillus subtilis: model organism for cellular development, and industrial workhorse"
652:
500:
1905:
1470:
2239:
2211:
1650:
1582:
448:
43:
133:
1817:"Bacteriophage-Encoded DNA Polymerases—Beyond the Traditional View of Polymerase Activities"
1706:"Inter-RNA Interaction of Phage φ29 pRNA to Form a Hexameric Complex for Viral DNA Transportation"
1518:"Inter-RNA Interaction of Phage φ29 pRNA to Form a Hexameric Complex for Viral DNA Transportation"
1092:
864:
647:
414:
223:
2198:
583:
The main difficulty in using aptamer-based drug delivery is sourcing unique aptamers and other
2139:
2048:
2030:
1991:
1973:
1929:
1919:
1884:
1856:
1838:
1792:
1774:
1735:
1727:
1686:
1668:
1616:
1598:
1547:
1539:
1498:
1490:
1451:
1443:
1402:
1394:
1326:
1318:
1274:
1256:
1217:
1199:
1158:
1140:
1084:
1027:
987:
969:
915:
907:
852:
842:
818:
800:
790:
748:
730:
410:
378:
303:
251:
231:
2038:
2022:
1981:
1965:
1911:
1846:
1828:
1782:
1766:
1717:
1676:
1658:
1606:
1590:
1529:
1482:
1433:
1386:
1308:
1264:
1248:
1207:
1189:
1148:
1130:
1074:
1019:
977:
961:
897:
808:
782:
738:
722:
679:
276:
1770:
1639:"Structure and assembly of the essential RNA ring component of a viral DNA packaging motor"
1390:
1313:
1296:
619:
513:
374:
1704:
Guo, Peixuan; Zhang, Chunlin; Chen, Chaoping; Garver, Kyle; Trottier, Mark (1998-07-01).
1516:
Guo, Peixuan; Zhang, Chunlin; Chen, Chaoping; Garver, Kyle; Trottier, Mark (1998-07-01).
1654:
1586:
1061:
Ye, Xin; Hemida, Maged; Zhang, Huifang M.; Hanson, Paul; Ye, Qiu; Yang, Decheng (2012).
472:
2043:
2010:
1986:
1953:
1851:
1816:
1787:
1754:
1681:
1638:
1611:
1570:
1212:
1177:
1153:
1118:
982:
902:
885:
813:
770:
493:
406:
382:
295:
235:
85:
1915:
1722:
1705:
1534:
1517:
1486:
1269:
1236:
786:
326:
2228:
1878:
743:
710:
642:
607:
489:
311:
307:
292:
239:
187:
73:
1096:
30:
841:. Stephen Mc Grath, Douwe van Sinderen. Norfolk, UK: Caister Academic Press. 2007.
726:
611:
577:
542:
205:
169:
121:
109:
97:
2144:
2115:
1252:
2124:
630:
615:
460:
280:
272:
194:
2184:
2026:
2011:"Stable RNA nanoparticles as potential new generation drugs for cancer therapy"
1135:
1952:
Jasinski, Daniel; Haque, Farzin; Binzel, Daniel W; Guo, Peixuan (2017-02-07).
1910:, Advances in Virus Research, vol. 58, Academic Press, pp. 255–294,
965:
504:
434:
386:
298:
Dr. Bernard Reilly discovered the Φ29 phage in Dr. John
Spizizen's lab at the
199:
145:
2034:
1977:
1842:
1778:
1731:
1672:
1602:
1543:
1494:
1447:
1398:
1322:
1260:
1203:
1144:
1023:
973:
911:
804:
734:
1969:
1663:
1374:
949:
856:
584:
354:
2052:
1995:
1933:
1860:
1796:
1690:
1620:
1455:
1438:
1421:
1406:
1330:
1278:
1237:"Bacteriophage Deoxyribonucleate Infection of Competent Bacillus subtilis1"
1221:
1162:
1088:
991:
752:
683:
485:
1739:
1551:
1502:
822:
38:
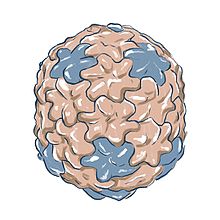
An illustration of Φ29's head based on electron microscopy data EMDB-2162
2178:
2109:
1833:
1815:
Morcinek-Orłowska, Joanna; Zdrojewska, Karolina; Węgrzyn, Alicja (2022).
1194:
919:
836:
565:
481:
430:
422:
418:
402:
215:
588:
569:
546:
443:
426:
335:
243:
191:
1079:
1062:
709:
Meijer, Wilfried J. J.; Horcajadas, José A.; Salas, Margarita (2001).
508:
330:
Schematic drawing of a Φ29 phage virion (cross section and side view).
1594:
610:
that accounts for ten to fifteen percent of all breast cancer cases.
572:. The small size of pRNA-derived nanoparticles also helps to deliver
503:
that has a different structure and function compared to standard DNA
437:
based approach. In the year 2000, a study by
Simpson et al. employed
366:
358:
339:
227:
219:
phage isolated to date and is among the smallest known dsDNA phages.
2086:
673:
1904:
Grimes, Shelley; Jardine, Paul J.; Anderson, Dwight (2002-01-01),
1755:"Mechanisms of DNA Packaging by Large Double-Stranded DNA Viruses"
626:
471:
390:
325:
168:
60:
1352:
591:
that potentially degrade therapeutic multimers and nanoparticles
573:
393:, making it one of the most powerful biomotors studied to date.
264:
2090:
2067:"Triple-negative Breast Cancer | Details, Diagnosis, and Signs"
385:. The Φ29 packaging motor is able to generate approximately 57
353:
The Φ29 DNA packaging motor packages the phage genome into the
234:. This novel structure system has inspired ongoing research in
1471:"RNA dependence of the bacteriophage φ29 DNA packaging ATPase"
538:
365:(genomic DNA-gp3). Because the process of genome packaging is
183:
480:
The Φ29 phage has a linear dsDNA genome consisting of 19,285
1010:
Shu, Yi; Wang, Hongzhi; Seremi, Bahar; Guo, Peixuan (2022),
222:Φ29 has a unique DNA packaging motor structure that employs
552:
One major challenge of using pRNA-derived nanoparticles is
1954:"Advancement of the Emerging Field of RNA Nanotechnology"
948:
Lee, Tae Jin; Schwartz, Chad; Guo, Peixuan (2009-10-01).
1571:"Structure of the bacteriophage φ29 DNA packaging motor"
1176:
Errington, Jeffery; van der Aart, Lizah T (2020-05-11).
541:
structure and function provides the ability to assemble
675:
Bacteriophage Φ29 structural model at atomic resolution
173:
Bacteriophage Φ29 structural model at atomic resolution
528:
Targeting of TNBC molecules by bacteriophage Φ29 pRNA
2168:
2099:
838:
Bacteriophage : genetics and molecular biology
1753:Rao, Venigalla B.; Feiss, Michael (2015-11-09).
1469:Grimes, Shelley; Anderson, Dwight (1990-10-20).
197:head and a short tail that belongs to the genus
1643:Proceedings of the National Academy of Sciences
771:"Tailed Bacteriophages: The Order Caudovirales"
334:The structure of Φ29 is composed of seven main
226:(pRNA) to guide the translocation of the phage
1880:DNA Replication and Related Cellular Processes
1420:Hoeprich, Stephen; Guo, Peixuan (2002-06-07).
476:The replication mechanism of bacteriophage Φ29
8:
1821:International Journal of Molecular Sciences
1235:Reilly, Bernard E.; Spizizen, John (1965).
2087:
1373:Rao, Venigalla B.; Feiss, Michael (2008).
715:Microbiology and Molecular Biology Reviews
492:terminal protein (p3) that complexes with
29:
18:
2042:
1985:
1850:
1832:
1786:
1721:
1680:
1662:
1610:
1533:
1437:
1312:
1268:
1211:
1193:
1152:
1134:
1078:
981:
901:
812:
742:
369:-intensive, it must be facilitated by an
338:: the terminal protein (p3), the head or
523:
1375:"The bacteriophage DNA packaging motor"
664:
599:Triple-negative breast cancer treatment
1771:10.1146/annurev-virology-100114-055212
1391:10.1146/annurev.genet.42.110807.091545
1314:10.1146/annurev.micro.61.080706.093415
862:
672:Padilla-Sanchez, Victor (2021-07-17),
1872:
1870:
1810:
1808:
1806:
1632:
1630:
1563:
1561:
1290:
1288:
764:
762:
7:
1112:
1110:
1108:
1106:
1067:Wiley Interdisciplinary Reviews: RNA
1056:
1054:
1052:
1050:
1048:
1005:
1003:
1001:
943:
941:
939:
937:
935:
933:
931:
929:
704:
702:
700:
698:
1016:RNA Nanotechnology and Therapeutics
401:The Φ29 pRNA is a highly versatile
903:10.1111/j.1432-1033.1977.tb11290.x
14:
1297:"40 Years with Bacteriophage ø29"
499:Φ29 is one of many phages with a
249:In nature, the Φ29 phage infects
1883:. IntechOpen. pp. 179–206.
954:Annals of Biomedical Engineering
890:European Journal of Biochemistry
633:to enhance therapeutic effects.
606:(TNBC) is an aggressive form of
488:of the genome are capped with a
47:
1907:Bacteriophage φ29 DNA packaging
1426:Journal of Biological Chemistry
1295:Salas, Margarita (2007-10-01).
455:was used to determine that the
2015:Advanced Drug Delivery Reviews
727:10.1128/MMBR.65.2.261-287.2001
1:
1916:10.1016/s0065-3527(02)58007-6
1723:10.1016/S1097-2765(00)80124-0
1535:10.1016/S1097-2765(00)80124-0
1487:10.1016/S0022-2836(05)80168-8
1301:Annual Review of Microbiology
787:10.1016/S0065-3527(08)60785-X
604:Triple-negative breast cancer
373:-powered motor that converts
1475:Journal of Molecular Biology
1253:10.1128/jb.89.3.782-790.1965
1123:Journal of Nanobiotechnology
587:for specific treatments for
769:Ackermann, Hans-W. (1998).
2261:
2027:10.1016/j.addr.2013.11.006
1136:10.1186/s12951-020-00758-4
775:Advances in Virus Research
361:(gp16), and the packaging
261:endospore-forming bacteria
1759:Annual Review of Virology
1379:Annual Review of Genetics
966:10.1007/s10439-009-9723-0
314:, viral replication, and
182:(bacteriophage Φ29) is a
42:
37:
28:
21:
1024:10.1201/9781003001560-21
439:cryo-electron microscopy
1970:10.1021/acsnano.6b05737
1664:10.1073/pnas.1016690108
1349:University of Minnesota
1241:Journal of Bacteriology
300:University of Minnesota
269:gastrointestinal tracts
1439:10.1074/jbc.M112061200
869:: CS1 maint: others (
711:"φ29 Family of Phages"
684:10.5281/zenodo.5111609
618:to resist therapeutic
554:large-scale production
529:
477:
468:Genome and replication
331:
174:
576:in tight spaces like
533:Nanoparticle assembly
527:
475:
329:
277:terrestrial organisms
224:prohead packaging RNA
172:
2170:Bacillus phage phi29
2131:Bacillus virus phi29
2101:Bacillus virus phi29
1834:10.3390/ijms23020635
1195:10.1099/mic.0.000922
1018:, pp. 141–157,
496:during replication.
449:anomalous scattering
433:basis rather than a
44:Virus classification
1655:2011PNAS..108.7357D
1587:2000Natur.408..745S
1432:(23): 20794–20803.
441:to determine that,
349:DNA packaging motor
184:double-stranded DNA
1345:"About | Virology"
653:Φ29 DNA polymerase
648:Bacteriophage pRNA
545:for nanomedicinal
530:
478:
332:
179:Bacillus virus Φ29
175:
160:Bacillus virus Φ29
23:Bacillus virus Φ29
2222:
2221:
2093:Taxon identifiers
1925:978-0-12-039858-4
1890:978-953-307-775-8
1649:(18): 7357–7362.
1581:(6813): 745–750.
1080:10.1002/wrna.1111
1033:978-1-003-00156-0
960:(10): 2064–2081.
848:978-1-904455-14-1
796:978-0-12-039851-5
490:covalently bonded
379:mechanical energy
304:molecular biology
267:, as well as the
263:that is found in
252:Bacillus subtilis
167:
166:
2252:
2215:
2214:
2202:
2201:
2189:
2188:
2187:
2161:
2160:
2148:
2147:
2135:
2134:
2133:
2120:
2119:
2118:
2088:
2081:
2080:
2078:
2077:
2063:
2057:
2056:
2046:
2006:
2000:
1999:
1989:
1964:(2): 1142–1164.
1949:
1943:
1942:
1941:
1940:
1901:
1895:
1894:
1874:
1865:
1864:
1854:
1836:
1812:
1801:
1800:
1790:
1750:
1744:
1743:
1725:
1701:
1695:
1694:
1684:
1666:
1634:
1625:
1624:
1614:
1595:10.1038/35047129
1565:
1556:
1555:
1537:
1513:
1507:
1506:
1466:
1460:
1459:
1441:
1417:
1411:
1410:
1370:
1364:
1363:
1361:
1360:
1351:. Archived from
1341:
1335:
1334:
1316:
1292:
1283:
1282:
1272:
1232:
1226:
1225:
1215:
1197:
1173:
1167:
1166:
1156:
1138:
1114:
1101:
1100:
1082:
1058:
1043:
1042:
1041:
1040:
1007:
996:
995:
985:
945:
924:
923:
905:
881:
875:
874:
868:
860:
833:
827:
826:
816:
766:
757:
756:
746:
706:
693:
692:
691:
690:
669:
52:
51:
33:
19:
16:Species of virus
2260:
2259:
2255:
2254:
2253:
2251:
2250:
2249:
2245:Bacillus phages
2235:Model organisms
2225:
2224:
2223:
2218:
2210:
2205:
2197:
2192:
2183:
2182:
2177:
2164:
2156:
2151:
2143:
2138:
2129:
2128:
2123:
2114:
2113:
2108:
2095:
2085:
2084:
2075:
2073:
2065:
2064:
2060:
2008:
2007:
2003:
1951:
1950:
1946:
1938:
1936:
1926:
1903:
1902:
1898:
1891:
1876:
1875:
1868:
1814:
1813:
1804:
1752:
1751:
1747:
1703:
1702:
1698:
1636:
1635:
1628:
1567:
1566:
1559:
1515:
1514:
1510:
1468:
1467:
1463:
1419:
1418:
1414:
1372:
1371:
1367:
1358:
1356:
1343:
1342:
1338:
1294:
1293:
1286:
1234:
1233:
1229:
1175:
1174:
1170:
1116:
1115:
1104:
1060:
1059:
1046:
1038:
1036:
1034:
1009:
1008:
999:
947:
946:
927:
883:
882:
878:
861:
849:
835:
834:
830:
797:
768:
767:
760:
708:
707:
696:
688:
686:
671:
670:
666:
661:
639:
620:pharmaceuticals
601:
562:
537:Versatility in
535:
522:
514:DNA replication
470:
453:crystallography
399:
375:chemical energy
351:
324:
289:
255:, a species of
163:
46:
17:
12:
11:
5:
2258:
2256:
2248:
2247:
2242:
2237:
2227:
2226:
2220:
2219:
2217:
2216:
2203:
2190:
2174:
2172:
2166:
2165:
2163:
2162:
2149:
2136:
2121:
2105:
2103:
2097:
2096:
2091:
2083:
2082:
2071:www.cancer.org
2058:
2001:
1944:
1924:
1896:
1889:
1866:
1802:
1765:(1): 351–378.
1745:
1716:(1): 149–155.
1710:Molecular Cell
1696:
1626:
1557:
1528:(1): 149–155.
1522:Molecular Cell
1508:
1481:(4): 559–566.
1461:
1412:
1365:
1336:
1284:
1247:(3): 782–790.
1227:
1188:(5): 425–427.
1168:
1102:
1073:(4): 469–481.
1044:
1032:
997:
925:
876:
847:
828:
795:
758:
721:(2): 261–287.
694:
663:
662:
660:
657:
656:
655:
650:
645:
638:
635:
600:
597:
561:
558:
534:
531:
521:
518:
501:DNA polymerase
494:DNA polymerase
469:
466:
398:
395:
383:ATP hydrolysis
350:
347:
340:capsid protein
323:
320:
296:microbiologist
288:
285:
236:nanotechnology
211:Salasmaviridae
165:
164:
157:
155:
151:
150:
143:
139:
138:
135:Salasmaviridae
131:
127:
126:
119:
115:
114:
111:Caudoviricetes
107:
103:
102:
95:
91:
90:
87:Heunggongvirae
83:
79:
78:
71:
64:
63:
58:
54:
53:
40:
39:
35:
34:
26:
25:
15:
13:
10:
9:
6:
4:
3:
2:
2257:
2246:
2243:
2241:
2238:
2236:
2233:
2232:
2230:
2213:
2208:
2204:
2200:
2195:
2191:
2186:
2180:
2176:
2175:
2173:
2171:
2167:
2159:
2154:
2150:
2146:
2141:
2137:
2132:
2126:
2122:
2117:
2111:
2107:
2106:
2104:
2102:
2098:
2094:
2089:
2072:
2068:
2062:
2059:
2054:
2050:
2045:
2040:
2036:
2032:
2028:
2024:
2020:
2016:
2012:
2005:
2002:
1997:
1993:
1988:
1983:
1979:
1975:
1971:
1967:
1963:
1959:
1955:
1948:
1945:
1935:
1931:
1927:
1921:
1917:
1913:
1909:
1908:
1900:
1897:
1892:
1886:
1882:
1881:
1873:
1871:
1867:
1862:
1858:
1853:
1848:
1844:
1840:
1835:
1830:
1826:
1822:
1818:
1811:
1809:
1807:
1803:
1798:
1794:
1789:
1784:
1780:
1776:
1772:
1768:
1764:
1760:
1756:
1749:
1746:
1741:
1737:
1733:
1729:
1724:
1719:
1715:
1711:
1707:
1700:
1697:
1692:
1688:
1683:
1678:
1674:
1670:
1665:
1660:
1656:
1652:
1648:
1644:
1640:
1633:
1631:
1627:
1622:
1618:
1613:
1608:
1604:
1600:
1596:
1592:
1588:
1584:
1580:
1576:
1572:
1564:
1562:
1558:
1553:
1549:
1545:
1541:
1536:
1531:
1527:
1523:
1519:
1512:
1509:
1504:
1500:
1496:
1492:
1488:
1484:
1480:
1476:
1472:
1465:
1462:
1457:
1453:
1449:
1445:
1440:
1435:
1431:
1427:
1423:
1416:
1413:
1408:
1404:
1400:
1396:
1392:
1388:
1384:
1380:
1376:
1369:
1366:
1355:on 2022-11-01
1354:
1350:
1346:
1340:
1337:
1332:
1328:
1324:
1320:
1315:
1310:
1306:
1302:
1298:
1291:
1289:
1285:
1280:
1276:
1271:
1266:
1262:
1258:
1254:
1250:
1246:
1242:
1238:
1231:
1228:
1223:
1219:
1214:
1209:
1205:
1201:
1196:
1191:
1187:
1183:
1179:
1172:
1169:
1164:
1160:
1155:
1150:
1146:
1142:
1137:
1132:
1128:
1124:
1120:
1113:
1111:
1109:
1107:
1103:
1098:
1094:
1090:
1086:
1081:
1076:
1072:
1068:
1064:
1057:
1055:
1053:
1051:
1049:
1045:
1035:
1029:
1025:
1021:
1017:
1013:
1006:
1004:
1002:
998:
993:
989:
984:
979:
975:
971:
967:
963:
959:
955:
951:
944:
942:
940:
938:
936:
934:
932:
930:
926:
921:
917:
913:
909:
904:
899:
895:
891:
887:
880:
877:
872:
866:
858:
854:
850:
844:
840:
839:
832:
829:
824:
820:
815:
810:
806:
802:
798:
792:
788:
784:
780:
776:
772:
765:
763:
759:
754:
750:
745:
740:
736:
732:
728:
724:
720:
716:
712:
705:
703:
701:
699:
695:
685:
681:
677:
676:
668:
665:
658:
654:
651:
649:
646:
644:
643:Bacteriophage
641:
640:
636:
634:
632:
628:
623:
621:
617:
613:
609:
608:breast cancer
605:
598:
596:
594:
590:
586:
581:
579:
578:blood vessels
575:
571:
567:
560:Drug delivery
559:
557:
555:
550:
548:
544:
543:nanoparticles
540:
532:
526:
519:
517:
515:
510:
506:
502:
497:
495:
491:
487:
483:
474:
467:
465:
462:
458:
454:
450:
446:
445:
440:
436:
432:
428:
424:
420:
416:
412:
408:
404:
396:
394:
392:
388:
384:
380:
376:
372:
368:
364:
360:
356:
348:
346:
343:
341:
337:
328:
321:
319:
317:
316:transcription
313:
312:DNA packaging
309:
308:morphogenesis
305:
301:
297:
294:
286:
284:
282:
278:
274:
270:
266:
262:
258:
257:gram-positive
254:
253:
247:
245:
241:
240:drug delivery
237:
233:
229:
225:
220:
218:
217:
212:
209:, and family
208:
207:
202:
201:
196:
193:
189:
188:bacteriophage
185:
181:
180:
171:
162:
161:
156:
153:
152:
149:
148:
144:
141:
140:
137:
136:
132:
129:
128:
125:
124:
120:
117:
116:
113:
112:
108:
105:
104:
101:
100:
96:
93:
92:
89:
88:
84:
81:
80:
77:
76:
75:Duplodnaviria
72:
69:
66:
65:
62:
59:
56:
55:
50:
45:
41:
36:
32:
27:
24:
20:
2169:
2100:
2074:. Retrieved
2070:
2061:
2018:
2014:
2004:
1961:
1957:
1947:
1937:, retrieved
1906:
1899:
1879:
1824:
1820:
1762:
1758:
1748:
1713:
1709:
1699:
1646:
1642:
1578:
1574:
1525:
1521:
1511:
1478:
1474:
1464:
1429:
1425:
1415:
1382:
1378:
1368:
1357:. Retrieved
1353:the original
1348:
1339:
1304:
1300:
1244:
1240:
1230:
1185:
1182:Microbiology
1181:
1171:
1126:
1122:
1070:
1066:
1037:, retrieved
1015:
957:
953:
896:(1): 39–55.
893:
889:
879:
837:
831:
778:
774:
718:
714:
687:, retrieved
674:
667:
624:
616:cancer cells
612:Chemotherapy
602:
592:
582:
563:
551:
547:therapeutics
536:
520:Applications
498:
479:
456:
442:
400:
352:
344:
333:
290:
281:human beings
279:, including
250:
248:
244:therapeutics
221:
214:
210:
206:Caudovirales
204:
198:
178:
177:
176:
159:
158:
146:
134:
123:Caudovirales
122:
110:
98:
86:
74:
67:
57:(unranked):
22:
2240:Podoviridae
2125:Wikispecies
1385:: 647–681.
1307:(1): 1–22.
781:: 135–201.
631:Doxorubicin
505:polymerases
461:flexibility
387:piconewtons
271:of various
232:replication
195:icosahedral
99:Uroviricota
2229:Categories
2076:2022-11-01
1939:2022-10-24
1827:(2): 635.
1359:2022-10-31
1039:2022-11-01
689:2021-07-17
659:References
435:microscopy
407:polymerize
306:, such as
200:Salasvirus
147:Salasvirus
2116:Q24808071
2035:0169-409X
2021:: 74–89.
1978:1936-0851
1843:1422-0067
1779:2327-056X
1732:1097-2765
1673:0027-8424
1603:1476-4687
1544:1097-2765
1495:0022-2836
1448:0021-9258
1399:0066-4197
1323:0066-4227
1261:0021-9193
1204:1350-0872
1145:1477-3155
1129:(1): 13.
974:1573-9686
912:0014-2956
865:cite book
805:0065-3527
735:1092-2172
585:multimers
566:ribozymes
423:pentamers
419:tetramers
405:that can
363:substrate
355:procapsid
322:Structure
291:In 1965,
154:Species:
82:Kingdom:
2199:11459168
2185:Q8084000
2179:Wikidata
2110:Wikidata
2053:24270010
1996:28045501
1958:ACS Nano
1934:12205781
1861:35054821
1797:26958920
1691:21471452
1621:11130079
1456:11886855
1407:18687036
1331:17441785
1279:14273661
1222:32391747
1163:33413427
1097:12631001
1089:22362726
992:19495981
857:86168751
753:11381102
637:See also
593:in vivo.
589:diseases
570:aptamers
451:(SIRAS)
427:hexamers
403:molecule
389:(pN) of
381:through
336:proteins
310:, viral
293:American
216:Bacillus
203:, order
186:(dsDNA)
130:Family:
94:Phylum:
2044:3955949
1987:5333189
1852:8775771
1788:4785836
1740:9702202
1682:3088594
1651:Bibcode
1612:4151180
1583:Bibcode
1552:9702202
1503:1700132
1213:7376258
1154:7792131
983:2855900
823:9891587
814:7173057
486:5’ ends
484:. Both
457:in vivo
444:in vivo
431:genetic
415:trimers
287:History
230:during
192:prolate
190:with a
142:Genus:
118:Order:
106:Class:
2212:816282
2051:
2041:
2033:
1994:
1984:
1976:
1932:
1922:
1887:
1859:
1849:
1841:
1795:
1785:
1777:
1738:
1730:
1689:
1679:
1671:
1619:
1609:
1601:
1575:Nature
1550:
1542:
1501:
1493:
1454:
1446:
1405:
1397:
1329:
1321:
1277:
1270:277537
1267:
1259:
1220:
1210:
1202:
1161:
1151:
1143:
1095:
1087:
1030:
990:
980:
972:
920:402269
918:
910:
855:
845:
821:
811:
803:
793:
751:
741:
733:
425:, and
411:dimers
367:energy
359:enzyme
273:marine
242:, and
228:genome
2207:WoRMS
2194:IRMNG
2158:10756
1093:S2CID
744:99027
627:siRNA
574:drugs
482:bases
409:into
391:force
68:Realm
61:Virus
2153:NCBI
2145:K922
2049:PMID
2031:ISSN
1992:PMID
1974:ISSN
1930:PMID
1920:ISBN
1885:ISBN
1857:PMID
1839:ISSN
1793:PMID
1775:ISSN
1736:PMID
1728:ISSN
1687:PMID
1669:ISSN
1617:PMID
1599:ISSN
1548:PMID
1540:ISSN
1499:PMID
1491:ISSN
1452:PMID
1444:ISSN
1403:PMID
1395:ISSN
1327:PMID
1319:ISSN
1275:PMID
1257:ISSN
1218:PMID
1200:ISSN
1159:PMID
1141:ISSN
1085:PMID
1028:ISBN
988:PMID
970:ISSN
916:PMID
908:ISSN
871:link
853:OCLC
843:ISBN
819:PMID
801:ISSN
791:ISBN
749:PMID
731:ISSN
568:and
397:pRNA
275:and
265:soil
2140:CoL
2039:PMC
2023:doi
1982:PMC
1966:doi
1912:doi
1847:PMC
1829:doi
1783:PMC
1767:doi
1718:doi
1677:PMC
1659:doi
1647:108
1607:PMC
1591:doi
1579:408
1530:doi
1483:doi
1479:215
1434:doi
1430:277
1387:doi
1309:doi
1265:PMC
1249:doi
1208:PMC
1190:doi
1186:166
1149:PMC
1131:doi
1075:doi
1020:doi
978:PMC
962:doi
898:doi
809:PMC
783:doi
739:PMC
723:doi
680:doi
539:RNA
377:to
371:ATP
2231::
2209::
2196::
2181::
2155::
2142::
2127::
2112::
2069:.
2047:.
2037:.
2029:.
2019:66
2017:.
2013:.
1990:.
1980:.
1972:.
1962:11
1960:.
1956:.
1928:,
1918:,
1869:^
1855:.
1845:.
1837:.
1825:23
1823:.
1819:.
1805:^
1791:.
1781:.
1773:.
1761:.
1757:.
1734:.
1726:.
1712:.
1708:.
1685:.
1675:.
1667:.
1657:.
1645:.
1641:.
1629:^
1615:.
1605:.
1597:.
1589:.
1577:.
1573:.
1560:^
1546:.
1538:.
1524:.
1520:.
1497:.
1489:.
1477:.
1473:.
1450:.
1442:.
1428:.
1424:.
1401:.
1393:.
1383:42
1381:.
1377:.
1347:.
1325:.
1317:.
1305:61
1303:.
1299:.
1287:^
1273:.
1263:.
1255:.
1245:89
1243:.
1239:.
1216:.
1206:.
1198:.
1184:.
1180:.
1157:.
1147:.
1139:.
1127:19
1125:.
1121:.
1105:^
1091:.
1083:.
1069:.
1065:.
1047:^
1026:,
1014:,
1000:^
986:.
976:.
968:.
958:37
956:.
952:.
928:^
914:.
906:.
894:73
892:.
888:.
867:}}
863:{{
851:.
817:.
807:.
799:.
789:.
779:51
777:.
773:.
761:^
747:.
737:.
729:.
719:65
717:.
713:.
697:^
678:,
622:.
580:.
509:3’
421:,
417:,
413:,
318:.
283:.
259:,
246:.
238:,
70::
2079:.
2055:.
2025::
1998:.
1968::
1914::
1893:.
1863:.
1831::
1799:.
1769::
1763:2
1742:.
1720::
1714:2
1693:.
1661::
1653::
1623:.
1593::
1585::
1554:.
1532::
1526:2
1505:.
1485::
1458:.
1436::
1409:.
1389::
1362:.
1333:.
1311::
1281:.
1251::
1224:.
1192::
1165:.
1133::
1099:.
1077::
1071:3
1022::
994:.
964::
922:.
900::
873:)
859:.
825:.
785::
755:.
725::
682::
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.