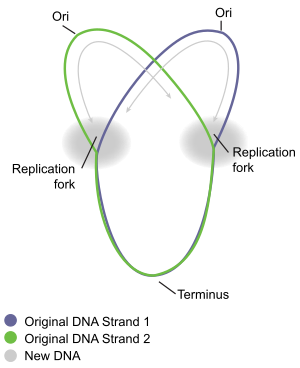
440:. It occurs in the terminus region, approximately opposite oriC on the chromosome (Fig 5). The terminus region contains several DNA replication terminator sites, or "Ter" sites. A special "replication terminator" protein must be bound at the Ter site for it to pause replication. Each Ter site has polarity of action, that is, it will arrest a replication fork approaching the Ter site from one direction, but will allow unimpeded fork movement through the Ter site from the other direction. The arrangement of the Ter sites forms two opposed groups that forces the two forks to meet each other within the region they span. This arrangement is called the "replication fork trap."
428:
95:
151:
142:, which moves with the helicase (together with other proteins) to synthesise a complementary copy of each strand. In this way, two identical copies of the original DNA are created. Eventually, the two replication forks moving around the circular chromosome meet in a specific zone of the chromosome, approximately opposite oriC, called the terminus region. The elongation enzymes then disassemble, and the two "daughter" chromosomes are resolved before cell division is completed.
31:
519:, Khodursky and Cozzarelli in 1997, it was found that topoisomerase IV is the only important decatenase of DNA replication intermediates in bacteria. When DNA gyrase alone was inhibited, most of the catenanes were unlinked. However, when Topoisomerase IV alone was inhibited, decatenation was almost completely blocked. This suggests that Topoisomerase IV is the primary protein for decatenation of interlinked chromosomes
512:
supercoils that form during DNA replication. Topoisomerase IV also relaxes positive supercoils, therefore, DNA Gyrase and topoisomerase IV play an almost identical role in removing the positive supercoils ahead of a translocating DNA polymerase, allowing DNA replication to continue unhindered by topological strain.
326:
and examining the developed film microscopically. This allowed the researchers to see where replication was taking place. The first conclusive observations of bidirectional replication was from studies of B. subtilis. Shortly after, the E. coli chromosome was also shown to replicate bidirectionally.
199:
that are important for its function include DnaA boxes, a 9-mer repeat with a highly conserved consensus sequence 5' – TTATCCACA – 3', that are recognized by the DnaA protein. DnaA protein plays a crucial role in the initiation of chromosomal DNA replication. Bound to ATP, and with the assistance of
511:
DNA gyrase also has topoisomerase type II activity; thus, with it being a homologue of topoisomerase IV (also having topoisomerase II activity) we expect similarity in the two proteins' functions. DNA gyrase's preliminary role is to introduce negative super coils into DNA, thereby relaxing positive
431:
Most circular bacterial chromosomes are replicated bidirectionally, starting at one point of origin and replicating in two directions away from the origin. This results in semiconservative replication, in which each new identical DNA molecule contains one template strand from the original molecule,
391:
are then added to this primer by a single DNA polymerase III dimer, in an integrated complex with DnaB helicase. Leading strand synthesis then proceeds continuously, while the DNA is concurrently unwound at the replication fork. In contrast, lagging strand synthesis is accomplished in short
Okazaki
924:
DelVecchio, VG; Kapatral, V; Redkar, RJ; Patra, G; Mujer, C; Los, T; Ivanova, N; Anderson, I; Bhattacharyya, A; Lykidis, A; Reznik, G; Jablonski, L; Larsen, N; D'Souza, M; Bernal, A; Mazur, M; Goltsman, E; Selkov, E; Elzer, PH; Hagius, S; O'Callaghan, D; Letesson, JJ; Haselkorn, R; Kyrpides, N;
126:) of the chromosome, in opposite directions away from the oriC, replicating the DNA to create two identical copies. This process is known as bidirectional replication. The entire assembly of molecules involved in DNA replication on each arm is called a
34:
A circular chromosome, showing DNA replication proceeding bidirectionally, with two replication forks generated at the "origin". Each half of the chromosome replicated by one replication fork is called a "replichore". (Graphic computer art by Daniel
82:. However, a circular chromosome has the disadvantage that after replication, the two progeny circular chromosomes can remain interlinked or tangled, and they must be extricated so that each cell inherits one complete copy of the chromosome during
681:
499:
plays in decatenation. To define the nomenclature, there are two types of topoisomerases: type I produces transient single-strand breaks in DNA and types II produces transient double-strand breaks. As a result, the type I enzyme removes
669:
This is based on an article by Imalda
Devaparanam and David Tribe made available under CC by SA licensing conditions from a university course activity at the Department of Microbiology and Immunology, University of Melbourne, 2007.
114:. Chromosome replication proceeds in three major stages: initiation, elongation and termination. The initiation stage starts with the ordered assembly of "initiator" proteins at the origin region of the chromosome, called
246:, from the DnaB-DnaC complex to the unwound region to form the pre-priming complex. After DnaB translocates to the apex of each replication fork, the helicase both unwinds the parental DNA and interacts momentarily with
717:
Hirota Y, Mordoh J and Jacob F (1970) On the process of cellular division in
Escherichia coli III. Thermosensitive mutants of Escherichia coli altered in the process of DNA initiation. J Mol Biol, 53, 369–387.
492:, DNA topoisomerase IV plays the major role in the separation of the catenated chromosomes, transiently breaking both DNA strands of one chromosome and allowing the other chromosome to pass through the break.
399:
When the synthesis of an
Okazaki fragment has been completed, replication halts and the core subunits of DNA Pol III dissociates from the β sliding clamp . The RNA primer is removed and replaced with DNA by
905:
Chris
Ullsperger and Nicholas R. Cozzarelli. Contrasting Enzymatic Activities of Topoisomerase IV and DNA Gyrase from Escherichia coli. Volume 271, Number 49, Issue of December 6, 1996, pp. 31549-31555
484:’ or topologically interlinked circles. The circles are not covalently but mechanically linked, because they are interwound and each is covalently closed. The catenated circles require the action of
1152:
914:
E L Zechiedrich, A B Khodursky, N R Cozzarelli. Topoisomerase IV, not gyrase, decatenates products of site-specific recombination in
Escherichia coli. Genes Dev. 1997 Oct 1;11 (19):2580-92 9334322
708:
C Weigel, A Schmidt, B Rückert, R Lurz, and W Messer. DnaA protein binding to individual DnaA boxes in the
Escherichia coli replication origin, oriC. EMBO J. 1997 November 3; 16(21): 6574–6583.
1084:
Lasek, Robert; Szuplewska, Magdalena; Mitura, Monika; Decewicz, Przemysław; Chmielowska, Cora; Pawłot, Aleksandra; Sentkowska, Dorota; Czarnecki, Jakub; Bartosik, Dariusz (25 October 2018).
685:
318:
As described above, bacterial chromosomal replication occurs in a bidirectional manner. This was first demonstrated by specifically labelling replicating bacterial chromosomes with
585:
has two circular chromosomes and one large plasmid, carrying genes not essential for survival but key to its biochemical behavior. The second chromosome has also been called a
415:
or strand break when cells are grown under normal laboratory conditions (without an exogenous DNA damaging treatment). The encountered DNA damages are ordinarily processed by
735:
Sekimizu K, Bramhill D and
Kornberg A (1987) ATP activates dnaA protein in initiating replication of plasmids bearing the origin of the E.coli chromosome. Cell, 50, 259–265
753:
Kowalski D, Eddy MJ. 1989. The DNA unwinding element: a novel, cis-acting component that facilitates opening of the
Escherichia coli replication origin. EMBO J. 8:4335–44
1145:
726:
Bramhill D, Kornberg A. 1988. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell 52:743–55
1022:
Si, YY; Xu, KH; Yu, XY; Wang, MF; Chen, XH (July 2019). "Complete genome sequence of
Paracoccus denitrificans ATCC 19367 and its denitrification characteristics".
118:. These assembly stages are regulated to ensure that chromosome replication occurs only once in each cell cycle. During the elongation phase of replication, the
677:
474:
of the Ter DNA-Tus protein complex (A) showing the nonblocking and the fork-blocking faces of Tus. (B) A cross-sectional view of the helicase-arresting surface.
807:
O'Donnell M., Jeruzalmi D., Kuriyan J. Clamp loader structure predicts the architecture of DNA polymerase III holoenzyme and RFC. Curr. Biol. 11 R935-R946 2001
880:
Kamada K, Horiuchi T, Ohsumi K, Shimamoto N, Morikawa K. 1996. Structure of a replication-terminator protein complexed with DNA. Nature, 17;383(6601):598–603.
1138:
593:
to the chromosome, are essential to life like the main chromosome, but has plasmid-type elements such as the origin of replication. Many other sequenced
762:
Carr KM, Kaguni JM. 2001. Stoichiometry of DnaA and DnaB protein in initiation at the Escherichia coli chromosomal origin. J. Biol. Chem. 276:44919–25
816:
Indiani C, O'Donnell M. Mechanism of the delta wrench in opening the beta sliding clamp. J Biol Chem. 2003 Oct 10;278(41):40272-81. Epub 2003 Jul 8.
392:
fragments. First, an RNA primer is synthesized by primase, and, like that in leading strand synthesis, DNA Pol III binds to the RNA primer and adds
871:
Duggin IG, Wake RG, Bell SD, Hill TM. 2008. The replication fork trap and termination of chromosome replication. Mol Microbiol. Dec;70(6):1323–33.
744:
Gotoh O, Tagashira Y. 1981. Locations of frequently opening regions on natural DNAs and their relation to functional loci. Biopolymers 20:1043–58
297:
base was incorporated uniformly into the bacterial chromosome. He then isolated the chromosomes by lysing the cells gently and placed them on an
798:
Prescott D.M., Kuempel P.L. 1972. Bidirectional replication of the chromosome in Escherichia coli. Proc Natl Acad Sci U S A. Oct;69(10):2842-5.
475:
449:
1086:"Genome Structure of the Opportunistic Pathogen Paracoccus yeei (Alphaproteobacteria) and Identification of Putative Virulence Factors"
771:
Tougu K, Marians KJ. 1996. The interaction between helicase and primase sets the replication fork clock. J. Biol. Chem. 271:21398–405
508:
and eukaryotes are the type I topoisomerase. The eukaryotic topo II, bacterial gyrase, and bacterial topo IV belong to the type II.
987:
Harrison, PW; Lower, RP; Kim, NK; Young, JP (April 2010). "Introducing the bacterial 'chromid': not a chromosome, not a plasmid".
412:
436:
Termination is the process of fusion of replication forks and disassembly of the replisomes to yield two separate and complete
1806:
1401:
78:. By contrast, most eukaryotes have linear DNA requiring elaborate mechanisms to maintain the stability of the telomeres and
699:
Jon M. Kaguni DnaA: Controlling the Initiation of Bacterial DNA Replication and More. Annu. Rev. Microbiol. 2006. 60:351–71
285:
of E. coli chromosomal replication in 1963, using an innovative method to visualize DNA replication. In his experiment, he
1942:
346:
305:
film for two months. This Experiment clearly demonstrates the theta replication model of circular bacterial chromosomes.
1590:
1510:
1266:
335:
chromosome from cells labeled for 19 min with thymine, followed by labeling for 2.5 min with thymine and thymidine.
647:
231:
is important as it alters the conformation of DNA to promote strand separation, and it appears that this region of
70:
Most prokaryote chromosomes contain a circular DNA molecule. This has the major advantage of having no free ends (
1811:
427:
94:
1658:
1559:
1476:
789:
Wake, R.G. 1972. Visualization of reinitiated chromosomes in Bacillus subtilis. J Mol Biol. Jul 28;68(3):501-9.
581:
541:
416:
278:
480:
Replication of the DNA separating the opposing replication forks leaves the completed chromosomes joined as ‘
1685:
1461:
257:
are needed to prevent the single strands of DNA from forming secondary structures and to prevent them from
1913:
1229:
889:
Kaplan DL, Bastia D. 2009. Mechanisms of polar arrest of a replication fork. Mol Microbiol. 72(2):279-85.
595:
223:(Dam), an enzyme that modifies the adenine base when this sequence is unmethylated or hemimethylated. The
192:
that will eventually lead to the establishment of two complete replisomes for bidirectional replication.
1980:
1975:
1673:
1646:
504:
from DNA one at a time, whereas the type II enzyme removes supercoils two at a time. The topo I of both
463:
220:
216:
165:
150:
1970:
1617:
1612:
1488:
938:
286:
1066:
1853:
1695:
1651:
1345:
561:
556:
393:
319:
298:
258:
209:
1903:
1047:
854:
181:
322:. The regions of DNA undergoing replication during the experiment were then visualized by using
1898:
1602:
1117:
1039:
1004:
966:
890:
846:
471:
358:
110:
60:
52:
188:
species. DnaA binding to the origin initiates the regulated recruitment of other enzymes and
102:
The circular bacteria chromosome replication is best understood in the well-studied bacteria
1634:
1528:
1161:
1107:
1097:
1031:
996:
956:
946:
838:
516:
401:
388:
370:
273:
When the replication fork moves around the circle, a structure shaped like the Greek letter
135:
104:
411:
A substantial proportion (10-15%) of the replication forks originating at oriC encounter a
1891:
1585:
1564:
1493:
1296:
657:
282:
254:
79:
56:
30:
627:
942:
1881:
1768:
1639:
1498:
1481:
1286:
1216:
1112:
1085:
374:
366:
331:
See Figure 4 of D. M. Prescott, and P. L. Kuempel (1972): A grain track produced by an
323:
313:
139:
1130:
961:
926:
1964:
1921:
1886:
1729:
1719:
1690:
1323:
1316:
842:
652:
642:
485:
350:
243:
83:
1051:
515:
DNA gyrase is not the sole enzyme responsible for decatenation. In an experiment by
265:
is needed to relieve the topological stress created by the action of DnaB helicase.
1908:
1751:
1711:
1678:
1450:
1436:
1334:
927:"The genome sequence of the facultative intracellular pathogen Brucella melitensis"
858:
454:
The Ter sites specifically interact with the replication terminator protein called
437:
362:
173:
131:
448:(A) Map showing the ori and the 10 Ter sites. (B) The consensus sequence of Ter.
336:
1932:
1798:
1746:
1741:
1254:
673:
611:-type origin, but the latter uses a P1 plasmid-type origin, making it a chromid.
590:
455:
224:
931:
Proceedings of the National Academy of Sciences of the United States of America
1840:
1835:
1788:
1781:
1776:
1761:
1756:
1597:
1532:
1281:
1223:
1000:
829:
Cox MM (1998). "A broadening view of recombinational DNA repair in bacteria".
780:
Cairns, J.P.: Cold Spring Harbor Symposia on Quantitative Biology 28:44, 1963.
505:
496:
405:
378:
294:
262:
204:-like proteins DnaA then unwinds an AT-rich region near the left boundary of
123:
1102:
17:
1845:
1830:
1817:
1569:
1357:
1276:
1271:
1261:
1249:
1188:
501:
365:
component . DNA Pol III uses one set of its core subunits to synthesize the
354:
290:
127:
64:
1121:
1043:
1008:
970:
951:
894:
1035:
850:
569:-stype origin of replication. In other cases, such as the closely related
1876:
1822:
1724:
1668:
1663:
1554:
1515:
1471:
1373:
1178:
632:
622:
535:
481:
239:
185:
71:
44:
432:
shown as the solid lines, and one new strand, shown as the dotted lines.
1858:
1736:
1520:
1505:
1208:
1183:
637:
586:
574:
521:
247:
228:
201:
189:
48:
122:
that were assembled at oriC during initiation proceed along each arm (
1949:
1927:
1624:
1547:
1542:
1307:
1198:
1193:
603:
547:
119:
573:, the smaller chromosome replicates like a plasmid and is clearly a
369:
continuously, while the other set of core subunits cycles from one
1431:
1423:
1241:
1203:
426:
302:
274:
93:
607:
have two circular chromosomes. The larger one uses a traditional
408:, which then ligates these fragments to form the lagging strand.
289:
the chromosome by growing his cultures in a medium containing 3H-
1937:
1629:
1170:
419:
repair enzymes to allow continued replication fork progression.
382:
377:. Leading strand synthesis begins with the synthesis of a short
177:
115:
1134:
138:. The two unwound single strands of DNA serve as templates for
75:
682:
Creative Commons Attribution-ShareAlike 3.0 Unported License
444:
See locations and sequences of the replication termini of
253:
In order for DNA replication to continue, single stranded
59:, in the form of a molecule of circular DNA, unlike the
309:
See Autoradiograph of intact replicating chromosome of
134:
that unwinds the two strands of DNA, creating a moving
208:, which carries three 13-mer motifs, and opens up the
559:
have two circular molecules. In some cases, such as
1869:
1797:
1710:
1578:
1460:
1449:
1422:
1394:
1344:
1333:
1306:
1295:
1240:
1169:
98:
Bidirectional replication in a circular chromosome.
381:at the replication origin by the enzyme Primase (
349:is a 900 kD complex, possessing an essentially a
824:
822:
678:Replication of a circular bacterial chromosome
1146:
495:There has been some confusion about the role
8:
672:This article incorporates material from the
466:of DnaB in an orientation-dependent manner.
212:for entrance of other replication proteins.
488:to separate the circles (decatenation). In
1457:
1341:
1303:
1153:
1139:
1131:
982:
980:
1111:
1101:
960:
950:
130:. At the forefront of the replisome is a
1067:"Paracoccus denitrificans - microbewiki"
525:, with DNA gyrase playing a minor role.
149:
29:
692:
565:, both appear chromosome-like with an
533:Several groups of bacteria, including
215:This region also contains four “GATC”
7:
551:have multiple circular chromosomes.
404:and the remaining nick is sealed by
235:C has a natural tendency to unwind.
238:DnaA then recruits the replicative
462:. The Tus-Ter complex impedes the
25:
219:sequences that are recognized by
1024:Canadian Journal of Microbiology
843:10.1046/j.1365-2443.1998.00175.x
1065:Larsen, Rachel; Pogliano, Kit.
680:", which is licensed under the
1807:Last universal common ancestor
1402:Defective interfering particle
925:Overbeek, R (8 January 2002).
301:(EM) grid which he exposed to
1:
1943:Clonally transmissible cancer
1379:Satellite-like nucleic acids
529:Multiple circular chromosomes
357:unit has a catalytic core, a
347:DNA polymerase III holoenzyme
195:DNA sequence elements within
589:, in that they have similar
176:that are recognised by the
1997:
1499:Class II or DNA transposon
1494:Class I or retrotransposon
648:Rolling circle replication
373:to the next on the looped
1812:Earliest known life forms
1686:Repeated sequences in DNA
1090:Frontiers in Microbiology
1001:10.1016/j.tim.2009.12.010
180:protein, which is highly
1659:Endogenous viral element
1477:Horizontal gene transfer
1103:10.3389/fmicb.2018.02553
582:Paracoccus denitrificans
542:Paracoccus denitrificans
1356:dsDNA satellite virus (
1914:Helper dependent virus
1230:Biological dark matter
1071:microbewiki.kenyon.edu
989:Trends in Microbiology
952:10.1073/pnas.221575398
658:Theta type replication
464:DNA unwinding activity
433:
158:
99:
36:
1674:Endogenous retrovirus
1647:Origin of replication
1363:ssDNA satellite virus
1353:ssRNA satellite virus
1036:10.1139/cjm-2019-0037
430:
287:radioactively labeled
221:DNA adenine methylase
217:DNA unwinding element
166:origin of replication
153:
97:
33:
1618:Secondary chromosome
1613:Extrachromosomal DNA
1489:Transposable element
394:deoxyribonucleotides
320:radioactive isotopes
1854:Model lipid bilayer
1696:Interspersed repeat
943:2002PNAS...99..443D
599:also have chromids.
562:Brucella melitensis
557:Alphaproteobacteria
299:electron micrograph
210:double-stranded DNA
43:is a chromosome in
41:circular chromosome
1164:organic structures
684:but not under the
434:
184:amongst different
159:
157:motifs in bacteria
100:
37:
27:Type of chromosome
1958:
1957:
1899:Non-cellular life
1706:
1705:
1445:
1444:
1418:
1417:
1372:ssRNA satellite (
472:crystal structure
281:demonstrated the
111:Bacillus subtilis
80:replicate the DNA
61:linear chromosome
16:(Redirected from
1988:
1635:Gene duplication
1458:
1454:self-replication
1342:
1304:
1162:Self-replicating
1155:
1148:
1141:
1132:
1126:
1125:
1115:
1105:
1081:
1075:
1074:
1062:
1056:
1055:
1019:
1013:
1012:
984:
975:
974:
964:
954:
921:
915:
912:
906:
903:
897:
887:
881:
878:
872:
869:
863:
862:
826:
817:
814:
808:
805:
799:
796:
790:
787:
781:
778:
772:
769:
763:
760:
754:
751:
745:
742:
736:
733:
727:
724:
718:
715:
709:
706:
700:
697:
402:DNA polymerase I
389:Deoxynucleotides
371:Okazaki fragment
353:structure. Each
255:binding proteins
136:replication fork
105:Escherichia coli
21:
1996:
1995:
1991:
1990:
1989:
1987:
1986:
1985:
1961:
1960:
1959:
1954:
1904:Synthetic virus
1892:Artificial cell
1865:
1793:
1702:
1591:RNA replication
1586:DNA replication
1574:
1565:Group II intron
1463:
1453:
1441:
1432:Mammalian prion
1414:
1390:
1369:dsRNA satellite
1366:ssDNA satellite
1336:
1329:
1298:
1291:
1236:
1165:
1159:
1129:
1083:
1082:
1078:
1064:
1063:
1059:
1021:
1020:
1016:
986:
985:
978:
923:
922:
918:
913:
909:
904:
900:
888:
884:
879:
875:
870:
866:
828:
827:
820:
815:
811:
806:
802:
797:
793:
788:
784:
779:
775:
770:
766:
761:
757:
752:
748:
743:
739:
734:
730:
725:
721:
716:
712:
707:
703:
698:
694:
667:
662:
618:
531:
425:
417:recombinational
361:subunit, and a
324:autoradiography
283:theta structure
271:
261:. In addition,
148:
92:
28:
23:
22:
15:
12:
11:
5:
1994:
1992:
1984:
1983:
1978:
1973:
1963:
1962:
1956:
1955:
1953:
1952:
1947:
1946:
1945:
1940:
1930:
1924:
1918:
1917:
1916:
1911:
1901:
1896:
1895:
1894:
1889:
1879:
1873:
1871:
1867:
1866:
1864:
1863:
1862:
1861:
1856:
1848:
1843:
1838:
1833:
1827:
1826:
1825:
1814:
1809:
1803:
1801:
1795:
1794:
1792:
1791:
1786:
1785:
1784:
1779:
1771:
1769:Kappa organism
1766:
1765:
1764:
1759:
1754:
1749:
1744:
1734:
1733:
1732:
1727:
1716:
1714:
1708:
1707:
1704:
1703:
1701:
1700:
1699:
1698:
1693:
1683:
1682:
1681:
1676:
1671:
1666:
1656:
1655:
1654:
1644:
1643:
1642:
1640:Non-coding DNA
1637:
1632:
1622:
1621:
1620:
1615:
1610:
1605:
1595:
1594:
1593:
1582:
1580:
1576:
1575:
1573:
1572:
1567:
1562:
1560:Group I intron
1557:
1552:
1551:
1550:
1540:
1539:
1538:
1535:
1526:
1523:
1518:
1513:
1503:
1502:
1501:
1496:
1486:
1485:
1484:
1482:Genomic island
1479:
1468:
1466:
1462:Mobile genetic
1455:
1447:
1446:
1443:
1442:
1440:
1439:
1434:
1428:
1426:
1420:
1419:
1416:
1415:
1413:
1412:
1411:
1410:
1407:
1398:
1396:
1392:
1391:
1389:
1388:
1387:
1386:
1383:
1377:
1370:
1367:
1364:
1361:
1354:
1350:
1348:
1339:
1331:
1330:
1328:
1327:
1320:
1312:
1310:
1301:
1293:
1292:
1290:
1289:
1287:dsDNA-RT virus
1284:
1282:ssRNA-RT virus
1279:
1277:(−)ssRNA virus
1274:
1272:(+)ssRNA virus
1269:
1264:
1259:
1258:
1257:
1246:
1244:
1238:
1237:
1235:
1234:
1233:
1232:
1227:
1217:Incertae sedis
1213:
1212:
1211:
1206:
1201:
1196:
1186:
1181:
1175:
1173:
1167:
1166:
1160:
1158:
1157:
1150:
1143:
1135:
1128:
1127:
1076:
1057:
1030:(7): 486–495.
1014:
976:
916:
907:
898:
882:
873:
864:
818:
809:
800:
791:
782:
773:
764:
755:
746:
737:
728:
719:
710:
701:
691:
666:
663:
661:
660:
655:
650:
645:
640:
635:
630:
625:
619:
617:
614:
613:
612:
600:
578:
530:
527:
486:topoisomerases
478:
477:
452:
451:
424:
421:
375:lagging strand
367:leading strand
340:
339:
316:
315:
270:
267:
147:
144:
140:DNA polymerase
91:
88:
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
1993:
1982:
1979:
1977:
1974:
1972:
1969:
1968:
1966:
1951:
1948:
1944:
1941:
1939:
1936:
1935:
1934:
1931:
1929:
1925:
1923:
1922:Nanobacterium
1919:
1915:
1912:
1910:
1907:
1906:
1905:
1902:
1900:
1897:
1893:
1890:
1888:
1887:Cell division
1885:
1884:
1883:
1880:
1878:
1875:
1874:
1872:
1868:
1860:
1857:
1855:
1852:
1851:
1849:
1847:
1844:
1842:
1839:
1837:
1834:
1832:
1828:
1824:
1821:
1820:
1819:
1815:
1813:
1810:
1808:
1805:
1804:
1802:
1800:
1796:
1790:
1787:
1783:
1780:
1778:
1775:
1774:
1772:
1770:
1767:
1763:
1760:
1758:
1755:
1753:
1750:
1748:
1745:
1743:
1740:
1739:
1738:
1735:
1731:
1730:Hydrogenosome
1728:
1726:
1723:
1722:
1721:
1720:Mitochondrion
1718:
1717:
1715:
1713:
1712:Endosymbiosis
1709:
1697:
1694:
1692:
1691:Tandem repeat
1689:
1688:
1687:
1684:
1680:
1677:
1675:
1672:
1670:
1667:
1665:
1662:
1661:
1660:
1657:
1653:
1650:
1649:
1648:
1645:
1641:
1638:
1636:
1633:
1631:
1628:
1627:
1626:
1623:
1619:
1616:
1614:
1611:
1609:
1606:
1604:
1601:
1600:
1599:
1596:
1592:
1589:
1588:
1587:
1584:
1583:
1581:
1579:Other aspects
1577:
1571:
1568:
1566:
1563:
1561:
1558:
1556:
1553:
1549:
1546:
1545:
1544:
1541:
1536:
1534:
1530:
1527:
1524:
1522:
1519:
1517:
1514:
1512:
1509:
1508:
1507:
1504:
1500:
1497:
1495:
1492:
1491:
1490:
1487:
1483:
1480:
1478:
1475:
1474:
1473:
1470:
1469:
1467:
1465:
1459:
1456:
1452:
1448:
1438:
1435:
1433:
1430:
1429:
1427:
1425:
1421:
1408:
1405:
1404:
1403:
1400:
1399:
1397:
1393:
1384:
1381:
1380:
1378:
1375:
1371:
1368:
1365:
1362:
1359:
1355:
1352:
1351:
1349:
1347:
1343:
1340:
1338:
1332:
1326:
1325:
1324:Avsunviroidae
1321:
1319:
1318:
1317:Pospiviroidae
1314:
1313:
1311:
1309:
1305:
1302:
1300:
1294:
1288:
1285:
1283:
1280:
1278:
1275:
1273:
1270:
1268:
1265:
1263:
1260:
1256:
1253:
1252:
1251:
1248:
1247:
1245:
1243:
1239:
1231:
1228:
1226:
1225:
1221:
1220:
1219:
1218:
1214:
1210:
1207:
1205:
1202:
1200:
1197:
1195:
1192:
1191:
1190:
1187:
1185:
1182:
1180:
1177:
1176:
1174:
1172:
1171:Cellular life
1168:
1163:
1156:
1151:
1149:
1144:
1142:
1137:
1136:
1133:
1123:
1119:
1114:
1109:
1104:
1099:
1095:
1091:
1087:
1080:
1077:
1072:
1068:
1061:
1058:
1053:
1049:
1045:
1041:
1037:
1033:
1029:
1025:
1018:
1015:
1010:
1006:
1002:
998:
994:
990:
983:
981:
977:
972:
968:
963:
958:
953:
948:
944:
940:
936:
932:
928:
920:
917:
911:
908:
902:
899:
896:
892:
886:
883:
877:
874:
868:
865:
860:
856:
852:
848:
844:
840:
836:
832:
825:
823:
819:
813:
810:
804:
801:
795:
792:
786:
783:
777:
774:
768:
765:
759:
756:
750:
747:
741:
738:
732:
729:
723:
720:
714:
711:
705:
702:
696:
693:
690:
689:
687:
683:
679:
675:
664:
659:
656:
654:
653:Topoisomerase
651:
649:
646:
644:
643:Ribbon theory
641:
639:
636:
634:
631:
629:
626:
624:
621:
620:
615:
610:
606:
605:
601:
598:
597:
592:
588:
584:
583:
579:
576:
572:
568:
564:
563:
558:
554:
553:
552:
550:
549:
544:
543:
538:
537:
528:
526:
524:
523:
518:
513:
509:
507:
503:
498:
493:
491:
487:
483:
476:
473:
469:
468:
467:
465:
461:
457:
450:
447:
443:
442:
441:
439:
438:DNA molecules
429:
422:
420:
418:
414:
409:
407:
403:
397:
395:
390:
386:
384:
380:
376:
372:
368:
364:
360:
356:
352:
348:
345:
337:
334:
330:
329:
328:
325:
321:
314:
312:
308:
307:
306:
304:
300:
296:
292:
288:
284:
280:
277:Ө is formed.
276:
268:
266:
264:
260:
256:
251:
249:
245:
241:
236:
234:
230:
226:
222:
218:
213:
211:
207:
203:
198:
193:
191:
187:
183:
179:
175:
174:DNA sequences
171:
167:
164:
156:
152:
145:
143:
141:
137:
133:
129:
125:
121:
117:
113:
112:
107:
106:
96:
89:
87:
85:
84:cell division
81:
77:
73:
68:
66:
62:
58:
54:
50:
46:
42:
32:
19:
18:Bacterial DNA
1981:Cell anatomy
1976:Bacteriology
1909:Viral vector
1752:Gerontoplast
1679:Transpoviron
1607:
1451:Nucleic acid
1437:Fungal prion
1335:Helper-virus
1322:
1315:
1222:
1215:
1093:
1089:
1079:
1070:
1060:
1027:
1023:
1017:
995:(4): 141–8.
992:
988:
937:(1): 443–8.
934:
930:
919:
910:
901:
885:
876:
867:
837:(2): 65–78.
834:
830:
812:
803:
794:
785:
776:
767:
758:
749:
740:
731:
722:
713:
704:
695:
671:
668:
628:Möbius strip
608:
602:
594:
580:
571:Ochrobactrum
570:
566:
560:
546:
540:
534:
532:
520:
514:
510:
494:
489:
479:
459:
453:
445:
435:
410:
398:
387:
363:processivity
359:dimerization
343:
341:
332:
317:
310:
272:
259:re-annealing
252:
237:
232:
214:
205:
196:
194:
172:consists of
169:
162:
160:
154:
132:DNA helicase
109:
103:
101:
69:
57:chloroplasts
53:mitochondria
40:
38:
1971:Chromosomes
1933:Cancer cell
1799:Abiogenesis
1747:Chromoplast
1742:Chloroplast
1525:Degradative
1267:dsRNA virus
1262:ssDNA virus
1255:Giant virus
1250:dsDNA virus
831:Genes Cells
674:Citizendium
591:codon usage
517:Zechiedrich
506:prokaryotes
423:Termination
279:John Cairns
225:methylation
90:Replication
1965:Categories
1841:Proteinoid
1836:Coacervate
1789:Nitroplast
1782:Trophosome
1777:Bacteriome
1762:Apicoplast
1757:Leucoplast
1598:Chromosome
1516:Resistance
1224:Parakaryon
665:References
596:Paracoccus
502:supercoils
497:DNA gyrase
413:DNA damage
406:DNA ligase
385:protein).
379:RNA primer
295:nucleoside
269:Elongation
263:DNA gyrase
200:bacterial
146:Initiation
124:replichore
65:eukaryotes
1850:Research
1831:Protocell
1570:Retrozyme
1529:Virulence
1511:Fertility
1358:Virophage
1346:Satellite
1337:dependent
1189:Eukaryota
676:article "
482:catenanes
355:monomeric
291:thymidine
186:bacterial
182:conserved
168:, called
128:replisome
74:) to the
72:telomeres
1877:Organism
1870:See also
1846:Sulphobe
1823:Ribozyme
1818:RNA life
1725:Mitosome
1669:Prophage
1664:Provirus
1652:Replicon
1608:Circular
1555:Phagemid
1472:Mobilome
1464:elements
1374:Virusoid
1297:Subviral
1209:Protista
1194:Animalia
1179:Bacteria
1122:30410477
1096:: 2553.
1052:85445608
1044:30897350
1009:20080407
971:11756688
895:19298368
633:Nucleoid
623:Catenane
616:See also
536:Brucella
446:E. coli.
240:helicase
229:adenines
190:proteins
63:of most
45:bacteria
1859:Jeewanu
1773:Organs
1737:Plastid
1537:Cryptic
1506:Plasmid
1204:Plantae
1184:Archaea
1113:6209633
939:Bibcode
859:2723712
851:9605402
638:Plasmid
587:chromid
575:chromid
522:in vivo
490:E. coli
460:E. coli
351:dimeric
344:E. coli
333:E. coli
248:primase
202:histone
163:E. coli
120:enzymes
49:archaea
1950:Virome
1928:Nanobe
1625:Genome
1603:Linear
1548:Fosmid
1543:Cosmid
1308:Viroid
1299:agents
1120:
1110:
1050:
1042:
1007:
969:
962:117579
959:
893:
857:
849:
604:Vibrio
548:Vibrio
545:, and
311:E.coli
293:. The
55:, and
1424:Prion
1395:Other
1242:Virus
1199:Fungi
1048:S2CID
855:S2CID
555:Many
303:X-ray
275:theta
35:Yuen)
1938:HeLa
1882:Cell
1630:Gene
1118:PMID
1040:PMID
1005:PMID
967:PMID
891:PMID
847:PMID
686:GFDL
609:oriC
567:oriC
470:The
383:DnaG
342:The
244:DnaB
206:oriC
197:oriC
178:DnaA
170:oriC
161:The
155:oriC
116:oriC
108:and
1521:Col
1409:DNA
1406:RNA
1385:DNA
1382:RNA
1108:PMC
1098:doi
1032:doi
997:doi
957:PMC
947:doi
839:doi
458:in
456:Tus
233:ori
227:of
76:DNA
1967::
1533:Ti
1116:.
1106:.
1092:.
1088:.
1069:.
1046:.
1038:.
1028:65
1026:.
1003:.
993:18
991:.
979:^
965:.
955:.
945:.
935:99
933:.
929:.
853:.
845:.
833:.
821:^
539:,
396:.
250:.
242:,
86:.
67:.
51:,
47:,
39:A
1926:?
1920:?
1829:†
1816:?
1531:/
1376:)
1360:)
1154:e
1147:t
1140:v
1124:.
1100::
1094:9
1073:.
1054:.
1034::
1011:.
999::
973:.
949::
941::
861:.
841::
835:3
688:.
577:.
338:.
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.