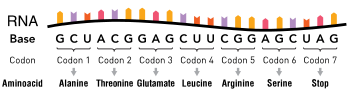448:. Beta-hemoglobin is created from the genetic information on the HBB, or "hemoglobin, beta" gene found on chromosome 11p15.5. A single point mutation in this polypeptide chain, which is 147 amino acids long, results in the disease known as Sickle Cell Anemia. Sickle-cell anemia is an autosomal recessive disorder that affects 1 in 500 African Americans, and is one of the most common blood disorders in the United States. The single replacement of the sixth amino acid in the beta-globin, glutamic acid, with valine results in deformed red blood cells. These sickle-shaped cells cannot carry nearly as much oxygen as normal red blood cells and they get caught more easily in the capillaries, cutting off blood supply to vital organs. The single nucleotide change in the beta-globin means that even the smallest of exertions on the part of the carrier results in severe pain and even heart attack. Below is a chart depicting the first thirteen amino acids in the normal and abnormal
367:
in the function, protein localization, stability of the protein or protein complex. Many methods have been proposed to predict the effects of missense mutations on proteins. Machine learning algorithms train their models to distinguish known disease-associated from neutral mutations whereas other methods do not explicitly train their models but almost all methods exploit the evolutionary conservation assuming that changes at conserved positions tend to be more deleterious. While majority of methods provide a binary classification of effects of mutations into damaging and benign, a new level of annotation is needed to offer an explanation of why and how these mutations damage proteins.
217:), resulting in abnormal extension of a protein's carboxyl terminus. Start-gain creates an AUG start codon upstream of the original start site. If the new AUG is near the original start site, in-frame within the processed transcript and downstream to a ribosomal binding site, it can be used to initiate translation. The likely effect is additional amino acids added to the amino terminus of the original protein. Frame-shift mutations are also possible in start-gain mutations, but typically do not affect translation of the original protein. Start-loss is a point mutation in a transcript's AUG start codon, resulting in the reduction or elimination of protein production.
39:
359:, it can prohibit mitosis from occurring due to the lack of a complete chromosome. Problems can also arise during the processes of transcription and replication of DNA. These all prohibit the cell from reproduction and thus lead to the death of the cell. Long-term effects can be a permanent changing of a chromosome, which can lead to a mutation. These mutations can be either beneficial or detrimental.
31:
343:. The theory explains the diversity and history of living organisms on Earth. In relation to point mutations, it states that beneficial mutations allow the organism to thrive and reproduce, thereby passing its positively affected mutated genes on to the next generation. On the other hand, harmful mutations cause the organism to die or be less likely to reproduce in a phenomenon known as
272:. A single nucleotide can change, but the new codon specifies the same amino acid, resulting in an unmutated protein. This type of change is called synonymous change since the old and new codon code for the same amino acid. This is possible because 64 codons specify only 20 amino acids. Different codons can lead to differential protein expression levels, however.
160:
847:, RIP mutations are found in single copy regions, adjacent to the repeated elements. These regions are either non-coding regions or genes encoding small secreted proteins including avirulence genes. The degree of RIP within these single copy regions was proportional to their proximity to repetitive elements.
223:
code for a different amino acid. A missense mutation changes a codon so that a different protein is created, a non-synonymous change. Conservative mutations result in an amino acid change. However, the properties of the amino acid remain the same (e.g., hydrophobic, hydrophilic, etc.). At times, a
145:
and higher-frequency light have ionizing capability, which in turn can affect DNA. Reactive oxygen molecules with free radicals, which are a byproduct of cellular metabolism, can also be very harmful to DNA. These reactants can lead to both single-stranded and double-stranded DNA breaks. Third, bonds
1019:
Hertwig studied sea urchins, and noticed that each egg contained one nucleus prior to fertilization and two nuclei after. This discovery proved that one spermatozoon could fertilize an egg, and therefore proved the process of meiosis. Hermann Fol continued
Hertwig's research by testing the effects
378:
If the mutation occurs in the region of the gene where transcriptional machinery binds to the protein, the mutation can affect the way in which transcription factors bind to the protein. The mechanisms of transcription bind to a protein through recognition of short nucleotide sequences. A mutation
1046:
acknowledged that the structure of DNA did indicate that there is some form of replicating process. However, there was not a lot of research done on this aspect of DNA until after Watson and Crick. People considered all possible methods of determining the replication process of DNA, but none were
850:
Rep and
Kistler have speculated that the presence of highly repetitive regions containing transposons, may promote mutation of resident effector genes. So the presence of effector genes within such regions is suggested to promote their adaptation and diversification when exposed to strong selection
370:
Moreover, if the mutation occurs in the region of the gene where transcriptional machinery binds to the protein, the mutation can affect the binding of the transcription factors because the short nucleotide sequences recognized by the transcription factors will be altered. Mutations in this region
366:
Other effects of point mutations, or single nucleotide polymorphisms in DNA, depend on the location of the mutation within the gene. For example, if the mutation occurs in the region of the gene responsible for coding, the amino acid sequence of the encoded protein may be altered, causing a change
1023:
Flemming began his research of cell division starting in 1868. The study of cells was an increasingly popular topic in this time period. By 1873, Schneider had already begun to describe the steps of cell division. Flemming furthered this description in 1874 and 1875 as he explained the steps in
374:
Point mutations can have several effects on the behavior and reproduction of a protein depending on where the mutation occurs in the amino acid sequence of the protein. If the mutation occurs in the region of the gene that is responsible for coding for the protein, the amino acid may be altered.
433:
The β-globin gene is found on the short arm of chromosome 11. The association of two wild-type α-globin subunits with two mutant β-globin subunits forms hemoglobin S (HbS). Under low-oxygen conditions (being at high altitude, for example), the absence of a polar amino acid at position six of the
711:
The HEXA gene makes part of an enzyme called beta-hexosaminidase A, which plays a critical role in the nervous system. This enzyme helps break down a fatty substance called GM2 ganglioside in nerve cells. Mutations in the HEXA gene disrupt the activity of beta-hexosaminidase A, preventing the
213:), which signals the end of translation. This interruption causes the protein to be abnormally shortened. The number of amino acids lost mediates the impact on the protein's functionality and whether it will function whatsoever. Stop-loss is a mutation in the original termination codon (
1024:
more detail. He also argued with
Schneider's findings that the nucleus separated into rod-like structures by suggesting that the nucleus actually separated into threads that in turn separated. Flemming concluded that cells replicate through cell division, to be more specific mitosis.
862:
whereby leakage of RIP was detected in single copy sequences at least 930 bp from the boundary of neighbouring duplicated sequences. To elucidate the mechanism of detection of repeated sequences leading to RIP may allow to understand how the flanking sequences may also be affected.
712:
breakdown of the fatty substances. As a result, the fatty substances accumulate to deadly levels in the brain and spinal cord. The buildup of GM2 ganglioside causes progressive damage to the nerve cells. This is the cause of the signs and symptoms of Tay-Sachs disease.
224:
change to one amino acid in the protein is not detrimental to the organism as a whole. Most proteins can withstand one or two point mutations before their function changes. Non-conservative mutations result in an amino acid change that has different properties than the
334:
based on the environment where the organism lives. An advantageous mutation can create an advantage for that organism and lead to the trait's being passed down from generation to generation, improving and benefiting the entire population. The scientific theory of
890:
mutations within repeats, however, the mechanism that detects the repeated sequences is unknown. RID is the only known protein essential for RIP. It is a DNA methyltransferease-like protein, that when mutated or knocked out results in loss of RIP. Deletion of the
375:
This slight change in the sequence of amino acids can cause a change in the function, activation of the protein meaning how it binds with a given enzyme, where the protein will be located within the cell, or the amount of free energy stored within the protein.
379:
in this region may alter these sequences and, thus, change the way the transcription factors bind to the protein. Mutations in this region can affect the efficiency of gene transcription, which controls both the levels of mRNA and overall protein levels.
82:
sequence of an organism's genome. Point mutations have a variety of effects on the downstream protein product—consequences that are moderately predictable based upon the specifics of the mutation. These consequences can range from no effect (e.g.
103:. DNA replication occurs when one double-stranded DNA molecule creates two single strands of DNA, each of which is a template for the creation of the complementary strand. A single point mutation can change the whole DNA sequence. Changing one
1047:
successful until
Meselson and Stahl. Meselson and Stahl introduced a heavy isotope into some DNA and traced its distribution. Through this experiment, Meselson and Stahl were able to prove that DNA reproduces semi-conservatively.
983:
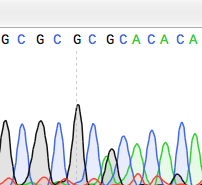
and go through its sexual cycle to activate the RIP machinery. Many different mutations within the duplicated gene are obtained from even a single fertilization event so that inactivated alleles, usually due to
1509:
Serra, E; Ars, E; Ravella, A; Sánchez, A; Puig, S; Rosenbaum, T; Estivill, X; Lázaro, C (2001). "Somatic NF1 mutational spectrum in benign neurofibromas: MRNA splice defects are common among point mutations".
350:
There are different short-term and long-term effects that can arise from mutations. Smaller ones would be a halting of the cell cycle at numerous points. This means that a codon coding for the amino acid
146:
in DNA eventually degrade, which creates another problem to keep the integrity of DNA to a high standard. There can also be replication errors that lead to substitution, insertion, or deletion mutations.
355:
may be changed to a stop codon, causing the proteins that should have been produced to be deformed and unable to complete their intended tasks. Because the mutations can affect the DNA and thus the
288:
or deletions of a single base pair (which has more of an adverse effect on the synthesized protein due to the nucleotides' still being read in triplets, but in different frames: a mutation called a
138:
or extreme heat, or chemical (molecules that misplace base pairs or disrupt the helical shape of DNA). Mutagens associated with cancers are often studied to learn about cancer and its prevention.
430:
is caused by a point mutation in the β-globin chain of hemoglobin, causing the hydrophilic amino acid glutamic acid to be replaced with the hydrophobic amino acid valine at the sixth position.
1326:
Li, Minghui; Goncearenco, Alexander; Panchenko, Anna R. (2017). "Annotating
Mutational Effects on Proteins and Protein Interactions: Designing Novel and Revisiting Existing Protocols".
258:
gene; this leads to an activation of the RAF protein which causes unlimited proliferative signalling in cancer cells. These are both examples of a non-conservative (missense) mutation.
836:
identity may also be subject to RIP. Though the exact mechanism of repeat recognition and mutagenesis are poorly understood, RIP results in repeated sequences undergoing multiple
440:
is a protein found in red blood cells, and is responsible for the transportation of oxygen through the body. There are two subunits that make up the hemoglobin protein:
434:β-globin chain promotes the non-covalent polymerisation (aggregation) of hemoglobin, which distorts red blood cells into a sickle shape and decreases their elasticity.
323:. If the original protein functions in cellular reproduction then this single point mutation can change the entire process of cellular reproduction for this organism.
59:
2898:
2178:
186:
with another pyrimidine. Transversions are replacement of a purine with a pyrimidine or vice versa. There is a systematic difference in mutation rates for
300:
Point mutations that occur in non-coding sequences are most often without consequences, although there are exceptions. If the mutated base pair is in the
1973:
Idnurm A, Howlett BJ (June 2003). "Analysis of loss of pathogenicity mutants reveals that repeat-induced point mutations can occur in the
Dothideomycete
1770:
755:
mutations. Genomic evidence indicates that RIP occurs or has occurred in a variety of fungi while experimental evidence indicates that RIP is active in
254:. The protein may also exhibit a "gain of function" or become activated, such is the case with the mutation changing a valine to glutamic acid in the
858:
suggested that leakage of RIP mutation might occur within a relatively short distance of a RIP-affected repeat. Indeed, this has been reported in
1199:
2666:
2594:
1343:
54:
when translated to protein. When one of these codons is changed by a point mutation, the corresponding amino acid of the protein is changed.
708:
is a genetic defect that is passed from parent to child. This genetic defect is located in the HEXA gene, which is found on chromosome 15.
2499:"A gene essential for de novo methylation and development in Ascobolus reveals a novel type of eukaryotic DNA methyltransferase structure"
2716:
920:
397:
174:
coined the terms "transitions" or "transversions" to categorize different types of point mutations. Transitions are replacement of a
1836:
Selker EU, Cambareri EB, Jensen BC, Haack KR (December 1987). "Rearrangement of duplicated DNA in specialized cells of
Neurospora".
371:
can affect rate of efficiency of gene transcription, which in turn can alter levels of mRNA and, thus, protein levels in general.
1602:
Mohyuddin, A; Neary, W. J.; Wallace, A; Wu, C. L.; Purcell, S; Reid, H; Ramsden, R. T.; Read, A; Black, G; Evans, D. G. (2002).
1385:
304:
sequence of a gene, then the expression of the gene may change. Also, if the mutation occurs in the splicing site of an
1020:
of injecting several spermatozoa into an egg, and found that the process did not work with more than one spermatozoon.
843:
The RIP mutations do not seem to be limited to repeated sequences. Indeed, for example, in the phytopathogenic fungus
58:
2012:
Cuomo CA, Güldener U, Xu JR, Trail F, Turgeon BG, Di Pietro A, Walton JD, Ma LJ, et al. (September 2007). "The
854:
As RIP mutation is traditionally observed to be restricted to repetitive regions and not single copy regions, Fudal
2908:
2875:
2214:
2067:
Coleman JJ, Rounsley SD, Rodriguez-Carres M, Kuo A, Wasmann CC, Grimwood J, Schmutz J, et al. (August 2009).
2825:
2328:"High frequency repeat-induced point mutation (RIP) is not associated with efficient recombination in Neurospora"
972:
2688:
1774:
1555:"Somatic NF1 mutation spectra in a family with neurofibromatosis type 1: Toward a theory of genetic modifiers"
1796:
Clutterbuck AJ (2011). "Genomic evidence of repeat-induced point mutation (RIP) in filamentous ascomycetes".
2870:
963:
Because RIP is so efficient at detecting and mutating repeats, fungal biologists often use it as a tool for
825:
265:
84:
2709:
2657:
Meselson, Stahl, and the replication of DNA : a history of "the most beautiful experiment in biology"
775:
951:
of the sequence (if there were any to begin with). In addition, many of the C-bearing nucleotides become
911:
705:
2830:
2794:
2757:
887:
752:
187:
163:
947:
in the coding sequence. This hypermutation of G-C to A-T in repetitive sequences eliminates functional
1203:
2739:
2451:
2392:
2190:
2025:
1604:"Molecular genetic analysis of the NF2 gene in young patients with unilateral vestibular schwannomas"
1463:
1284:
1233:
1119:
897:
787:
320:
285:
88:
2440:"A cytosine methyltransferase homologue is essential for sexual development in Aspergillus nidulans"
2327:
1401:"Messing up disorder: how do missense mutations in the tumor suppressor protein APC lead to cancer?"
396:. For instance, point mutations in Adenomatous Polyposis Coli promote tumorigenesis. A novel assay,
2804:
2744:
837:
445:
289:
229:
2283:"Repeat-induced point mutation (RIP) as an alternative mechanism of evolution toward virulence in
228:. The protein may lose its function, which can result in a disease in the organism. For example,
2635:
2528:
2049:
1910:
1861:
1584:
1535:
1308:
1257:
989:
985:
769:
763:
427:
301:
220:
202:
1747:
2903:
2789:
2784:
2702:
2662:
2627:
2590:
2563:
2520:
2479:
2420:
2357:
2308:
2263:
2206:
2159:
2104:
2041:
1994:
1955:
1902:
1853:
1813:
1684:
1633:
1576:
1527:
1491:
1432:
1367:
1349:
1339:
1300:
1249:
1147:
944:
940:
820:
793:
757:
721:
408:
344:
327:
206:
142:
131:
2582:
2860:
2809:
2619:
2555:
2510:
2469:
2459:
2410:
2400:
2347:
2339:
2298:
2253:
2198:
2149:
2139:
2094:
2084:
2033:
1986:
1945:
1928:
Ikeda K, Nakayashiki H, Kataoka T, Tamba H, Hashimoto Y, Tosa Y, Mayama S (September 2002).
1892:
1845:
1805:
1676:
1623:
1615:
1566:
1519:
1481:
1471:
1422:
1412:
1357:
1331:
1292:
1241:
1174:
1137:
1127:
1027:
1013:
1656:. National Center for Biotechnology Information (US). 29 September 1998 – via PubMed.
2799:
2684:
2377:"A cytosine methyltransferase homologue is essential for repeat-induced point mutation in
1452:"Determining biophysical protein stability in lysates by a fast proteolysis assay, FASTpp"
1039:
1035:
815:
798:
781:
416:
412:
400:, might help swift screening of specific stability defects in individual cancer patients.
261:
123:
100:
2559:
2497:
Malagnac F, Wendel B, Goyon C, Faugeron G, Zickler D, Rossignol JL, et al. (1997).
2455:
2396:
2194:
2029:
1467:
1288:
1237:
1218:
1123:
2655:
2546:
Selker EU (1990). "Premeiotic instability of repeated sequences in
Neurospora crassa".
2474:
2439:
2352:
2154:
2123:
2099:
2068:
1628:
1603:
1486:
1451:
1427:
1400:
1362:
1031:
340:
2515:
2498:
2415:
2376:
1990:
1178:
1142:
1107:
205:
include stop-gain and start-loss. Stop-gain is a mutation that results in a premature
38:
17:
2892:
2639:
2258:
2237:
1950:
1929:
1897:
1880:
1849:
1165:
Freese, Ernst (1959). "The
Specific Mutagenic Effect of Base Analogues on Phage T4".
1056:
1043:
1005:
666:
544:
537:
251:
2532:
2053:
1914:
1865:
1588:
1108:"The difference between spontaneous and base-analogue induced mutations of phage T4"
2752:
2128:
is affected by genomic environment and exposure to resistance genes in host plants"
2016:
genome reveals a link between localized polymorphism and pathogen specialization".
1879:
Graïa F, Lespinet O, Rimbault B, Dequard-Chablat M, Coppin E, Picard M (May 2001).
1539:
1312:
1261:
948:
808:
191:
171:
2343:
1275:
Hoeijmakers JH (May 2001). "Genome maintenance mechanisms for preventing cancer".
194:(Beta). Transition mutations are about ten times more common than transversions.
30:
2464:
2144:
2089:
1476:
1335:
1881:"Genome quality control: RIP (repeat-induced point mutation) comes to Podospora"
1680:
1080:
964:
952:
449:
255:
42:
Schematic of a single-stranded RNA molecule illustrating a series of three-base
1727:
1723:
1651:
2202:
1809:
1061:
936:
932:
833:
437:
331:
244:
233:
183:
112:
108:
51:
47:
1732:. Bethesda MD: National Center for Biotechnology Information. 1998. NBK22183.
1417:
1353:
319:
may change, thereby changing the entire protein. The new protein is called a
2610:
Paweletz N (January 2001). "Walther
Flemming: pioneer of mitosis research".
2037:
829:
691:
649:
639:
569:
527:
517:
356:
336:
330:
can lead to beneficial as well as harmful traits or diseases. This leads to
225:
2631:
2483:
2424:
2405:
2312:
2303:
2282:
2267:
2210:
2163:
2108:
2045:
1998:
1959:
1906:
1817:
1637:
1580:
1531:
1495:
1436:
1371:
1304:
1253:
1151:
2567:
2524:
2361:
1857:
1688:
1523:
1132:
159:
34:
Point mutations of a codon, classified by their impact on protein sequence
2865:
2855:
2850:
2725:
1619:
876:
741:
309:
119:
71:
1245:
1571:
1554:
1009:
1001:
884:
880:
872:
812:
749:
745:
737:
681:
654:
644:
559:
532:
522:
352:
316:
269:
232:
is caused by a single point mutation (a missense mutation) in the beta-
127:
74:
where a single nucleotide base is changed, inserted or deleted from a
2623:
2179:"The genomic organization of plant pathogenicity in Fusarium species"
1296:
980:
976:
686:
676:
671:
659:
634:
564:
554:
549:
512:
393:
360:
305:
247:
179:
175:
104:
27:
Replacement, insertion, or deletion of a single DNA or RNA nucleotide
939:
by invading and multiplying within the genome. RIP creates multiple
308:, then this may interfere with correct splicing of the transcribed
1702:
1553:
Wiest, V; Eisenbarth, I; Schmegner, C; Krone, W; Assum, G (2003).
1330:. Methods in Molecular Biology. Vol. 1550. pp. 235–260.
240:
135:
57:
43:
1934:: implications for its sexual cycle in the natural field context"
91:), with regard to protein production, composition, and function.
968:
236:
2698:
931:
RIP is believed to have evolved as a defense mechanism against
2073:: contribution of supernumerary chromosomes to gene expansion"
1667:
Hsia CC (January 1998). "Respiratory function of hemoglobin".
733:
441:
268:"). A silent mutation does not affect the functioning of the
79:
75:
1388:. Genetic Engineering & Biotechnology News. 18 June 2008.
141:
There are multiple ways for point mutations to occur. First,
832:
in length are vulnerable to RIP. Repeats with as low as 80%
392:
Point mutations in multiple tumor suppressor proteins cause
2694:
2122:
Van de Wouw AP, Cozijnsen AJ, Hane JK, et al. (2010).
2587:
The organic codes: an introduction to semantic biology
1217:
Davies H, Bignell GR, Cox C, et al. (June 2002).
1399:
Minde DP, Anvarian Z, Rüdiger SG, Maurice MM (2011).
905:, results in loss of fertility while deletion of the
62:
A to G point mutation detected with Sanger sequencing
2375:
Freitag M, Williams RL, Kothe GO, Selker EU (2002).
2281:
Fudal I, Ross S, Brun H, et al. (August 2009).
2843:
2818:
2775:
2768:
2732:
2326:Irelan JT, Hagemann AT, Selker EU (December 1994).
130:. Mutagens can be physical, such as radiation from
2654:
2438:Lee DW, Freitag M, Selker EU, Aramayo R (2008).
1194:
1192:
1190:
1188:
1831:
1829:
1827:
1012:was discovered several years later in 1882 by
363:is an example of how they can be detrimental.
2710:
2124:"Evolution of linked avirulence effectors in
8:
1219:"Mutations of the BRAF gene in human cancer"
315:By altering just one amino acid, the entire
1718:
1716:
919:, results in fertility defects and loss of
383:Specific diseases caused by point mutations
339:is greatly dependent on point mutations in
126:. The rate of mutation may be increased by
118:Point mutations may arise from spontaneous
2772:
2717:
2703:
2695:
2589:. Cambridge University Press. p. 13.
1741:
1739:
99:Point mutations usually take place during
2687:at the U.S. National Library of Medicine
2514:
2473:
2463:
2414:
2404:
2351:
2302:
2257:
2153:
2143:
2098:
2088:
1949:
1896:
1627:
1570:
1485:
1475:
1450:Minde DP, Maurice MM, Rüdiger SG (2012).
1426:
1416:
1361:
1141:
1131:
276:Single base pair insertions and deletions
2844:Mutation with respect to overall fitness
1930:"Repeat-induced point mutation (RIP) in
921:methylation induced premeiotically (MIP)
579:
457:
158:
37:
29:
1072:
807:RIP occurs during the sexual stage in
155:Transition/transversion categorization
2242:: the world of the end as we know it"
1386:"A Shortcut to Personalized Medicine"
1000:The cellular reproduction process of
797:, sequences mutated by RIP are often
7:
2238:"Telomeres in the rice blast fungus
581:Sequence for sickle-cell hemoglobin
411:is caused by point mutations in the
2899:Modification of genetic information
2560:10.1146/annurev.ge.24.120190.003051
1034:are credited with the discovery of
111:may change the amino acid that the
2769:Mutation with respect to structure
2653:Holmes, Frederic Lawrence (2001).
398:Fast parallel proteolysis (FASTpp)
25:
2177:Rep M, Kistler HC (August 2010).
967:. A second copy of a single-copy
955:, thus decreasing transcription.
811:after fertilization but prior to
264:code for the same amino acid (a "
166:(Alpha) and transversions (Beta).
2259:10.1111/j.1574-6968.2007.00812.x
1951:10.1046/j.1365-2958.2002.03101.x
1898:10.1046/j.1365-2958.2001.02367.x
988:, as well as alleles containing
1709:. National Library of Medicine.
459:Sequence for normal hemoglobin
87:) to deleterious effects (e.g.
1:
2516:10.1016/S0092-8674(00)80410-9
1991:10.1016/S1087-1845(02)00588-1
1179:10.1016/S0022-2836(59)80038-3
726:repeat-induced point mutation
716:Repeat-induced point mutation
2465:10.1371/journal.pone.0002531
2145:10.1371/journal.ppat.1001180
2090:10.1371/journal.pgen.1000618
1850:10.1016/0092-8674(87)90097-3
1477:10.1371/journal.pone.0046147
1336:10.1007/978-1-4939-6747-6_17
1112:Proc. Natl. Acad. Sci. U.S.A
1106:Freese, Ernst (April 1959).
243:into GUG, which encodes the
2583:"The problem of generation"
2581:Barbieri, Marcello (2003).
2344:10.1093/genetics/138.4.1093
2291:Mol. Plant Microbe Interact
1681:10.1056/NEJM199801223380407
1608:Journal of Medical Genetics
2925:
2826:Chromosomal translocations
2661:. Yale University Press.
2236:Farman ML (August 2007).
2203:10.1016/j.pbi.2010.04.004
1810:10.1016/j.fgb.2010.09.002
198:Functional categorization
2689:Medical Subject Headings
2612:Nat. Rev. Mol. Cell Biol
2385:Proc Natl Acad Sci U S A
1703:"HBB — Hemoglobin, Beta"
1418:10.1186/1476-4598-10-101
959:Use in molecular biology
50:codon corresponds to an
2866:Nearly neutral mutation
2038:10.1126/science.1143708
1707:Genetics Home Reference
979:. The fungus must then
266:synonymous substitution
2876:Nonsynonymous mutation
2831:Chromosomal inversions
2733:Mechanisms of mutation
2406:10.1073/pnas.132212899
2304:10.1094/MPMI-22-8-0932
2285:Leptosphaeria maculans
2183:Curr. Opin. Plant Biol
2126:Leptosphaeria maculans
1975:Leptosphaeria maculans
776:Leptosphaeria maculans
732:is a process by which
167:
143:ultraviolet (UV) light
63:
55:
35:
18:Base-pair substitution
2856:Advantageous mutation
2795:Conservative mutation
1771:"Causes of Tay-Sachs"
1724:"Anemia, Sickle Cell"
1524:10.1007/s004390100514
1133:10.1073/pnas.45.4.622
933:transposable elements
162:
61:
41:
33:
2851:Deleterious mutation
2819:Large-scale mutation
2246:FEMS Microbiol. Lett
2071:Nectria haematococca
2014:Fusarium graminearum
1620:10.1136/jmg.39.5.315
898:Aspergillus nidulans
838:transition mutations
788:Nectria haematococca
296:General consequences
284:is used to describe
239:that converts a GAG
182:or replacement of a
89:frameshift mutations
85:synonymous mutations
2871:Synonymous mutation
2805:Frameshift mutation
2456:2008PLoSO...3.2531L
2397:2002PNAS...99.8802F
2195:2010COPB...13..420R
2030:2007Sci...317.1400C
1468:2012PLoSO...746147M
1289:2001Natur.411..366H
1246:10.1038/nature00766
1238:2002Natur.417..949D
1124:1959PNAS...45..622F
912:Ascobolus immersens
582:
460:
452:polypeptide chain.
290:frameshift mutation
280:Sometimes the term
230:sickle-cell disease
2240:Magnaporthe oryzae
1932:Magnaporthe grisea
1748:"Genetic Mutation"
1572:10.1002/humu.10272
1087:. 22 November 2016
1085:Biology Dictionary
1004:was discovered by
990:missense mutations
986:nonsense mutations
945:nonsense mutations
770:Magnaporthe grisea
764:Podospora anserina
580:
458:
428:Sickle-cell anemia
423:Sickle-cell anemia
328:germline mutations
221:Missense mutations
203:Nonsense mutations
178:base with another
168:
122:that occur during
64:
56:
36:
2909:Molecular biology
2884:
2883:
2839:
2838:
2790:Missense mutation
2785:Nonsense mutation
2668:978-0-300-08540-2
2596:978-0-521-53100-9
2379:Neurospora crassa
1979:Fungal Genet Biol
1798:Fungal Genet Biol
1746:Clancy S (2008).
1729:Genes and Disease
1653:Genes and Disease
1345:978-1-4939-6745-2
1206:on 11 April 2005.
1200:"Genetics Primer"
992:can be obtained.
935:, which resemble
821:Neurospora crassa
794:Neurospora crassa
758:Neurospora crassa
722:molecular biology
706:Tay–Sachs disease
700:Tay–Sachs disease
697:
696:
575:
574:
409:Neurofibromatosis
404:Neurofibromatosis
345:natural selection
211:a stop was gained
207:termination codon
16:(Redirected from
2916:
2861:Neutral mutation
2810:Dynamic mutation
2773:
2719:
2712:
2705:
2696:
2673:
2672:
2660:
2650:
2644:
2643:
2624:10.1038/35048077
2607:
2601:
2600:
2578:
2572:
2571:
2543:
2537:
2536:
2518:
2494:
2488:
2487:
2477:
2467:
2435:
2429:
2428:
2418:
2408:
2372:
2366:
2365:
2355:
2323:
2317:
2316:
2306:
2278:
2272:
2271:
2261:
2233:
2227:
2226:
2224:
2222:
2213:. Archived from
2174:
2168:
2167:
2157:
2147:
2138:(11): e1001180.
2119:
2113:
2112:
2102:
2092:
2064:
2058:
2057:
2024:(5843): 1400–2.
2009:
2003:
2002:
1970:
1964:
1963:
1953:
1944:(5): 1355–1364.
1925:
1919:
1918:
1900:
1876:
1870:
1869:
1833:
1822:
1821:
1793:
1787:
1786:
1784:
1782:
1777:on 6 August 2020
1773:. Archived from
1766:
1760:
1759:
1752:Nature Education
1743:
1734:
1733:
1720:
1711:
1710:
1699:
1693:
1692:
1664:
1658:
1657:
1648:
1642:
1641:
1631:
1599:
1593:
1592:
1574:
1550:
1544:
1543:
1506:
1500:
1499:
1489:
1479:
1447:
1441:
1440:
1430:
1420:
1396:
1390:
1389:
1382:
1376:
1375:
1365:
1323:
1317:
1316:
1297:10.1038/35077232
1283:(6835): 366–74.
1272:
1266:
1265:
1232:(6892): 949–54.
1223:
1214:
1208:
1207:
1202:. Archived from
1196:
1183:
1182:
1162:
1156:
1155:
1145:
1135:
1103:
1097:
1096:
1094:
1092:
1081:"Point Mutation"
1077:
1028:Matthew Meselson
1014:Walther Flemming
828:of at least 400
826:repeat sequences
662:
607:
583:
540:
485:
461:
262:Silent mutations
21:
2924:
2923:
2919:
2918:
2917:
2915:
2914:
2913:
2889:
2888:
2885:
2880:
2835:
2814:
2800:Silent mutation
2764:
2728:
2723:
2681:
2676:
2669:
2652:
2651:
2647:
2609:
2608:
2604:
2597:
2580:
2579:
2575:
2545:
2544:
2540:
2496:
2495:
2491:
2437:
2436:
2432:
2374:
2373:
2369:
2338:(4): 1093–103.
2325:
2324:
2320:
2280:
2279:
2275:
2235:
2234:
2230:
2220:
2218:
2176:
2175:
2171:
2121:
2120:
2116:
2083:(8): e1000618.
2069:"The genome of
2066:
2065:
2061:
2011:
2010:
2006:
1972:
1971:
1967:
1927:
1926:
1922:
1878:
1877:
1873:
1835:
1834:
1825:
1795:
1794:
1790:
1780:
1778:
1768:
1767:
1763:
1745:
1744:
1737:
1722:
1721:
1714:
1701:
1700:
1696:
1669:N. Engl. J. Med
1666:
1665:
1661:
1650:
1649:
1645:
1601:
1600:
1596:
1552:
1551:
1547:
1508:
1507:
1503:
1449:
1448:
1444:
1398:
1397:
1393:
1384:
1383:
1379:
1346:
1325:
1324:
1320:
1274:
1273:
1269:
1221:
1216:
1215:
1211:
1198:
1197:
1186:
1164:
1163:
1159:
1105:
1104:
1100:
1090:
1088:
1079:
1078:
1074:
1070:
1053:
1036:DNA replication
998:
961:
929:
869:
816:DNA replication
782:Gibberella zeae
718:
702:
660:
605:
577:
538:
483:
455:
425:
417:Neurofibromin 2
413:Neurofibromin 1
406:
390:
385:
321:protein variant
298:
278:
215:a stop was lost
200:
157:
152:
124:DNA replication
101:DNA replication
97:
28:
23:
22:
15:
12:
11:
5:
2922:
2920:
2912:
2911:
2906:
2901:
2891:
2890:
2882:
2881:
2879:
2878:
2873:
2868:
2863:
2858:
2853:
2847:
2845:
2841:
2840:
2837:
2836:
2834:
2833:
2828:
2822:
2820:
2816:
2815:
2813:
2812:
2807:
2802:
2797:
2792:
2787:
2781:
2779:
2777:Point mutation
2770:
2766:
2765:
2763:
2762:
2761:
2760:
2755:
2747:
2742:
2736:
2734:
2730:
2729:
2724:
2722:
2721:
2714:
2707:
2699:
2693:
2692:
2685:Point+Mutation
2680:
2679:External links
2677:
2675:
2674:
2667:
2645:
2602:
2595:
2573:
2548:Annu Rev Genet
2538:
2489:
2430:
2391:(13): 8802–7.
2367:
2318:
2273:
2228:
2217:on 2 June 2020
2169:
2114:
2059:
2004:
1965:
1920:
1891:(3): 586–595.
1871:
1844:(5): 741–752.
1823:
1788:
1761:
1735:
1712:
1694:
1659:
1643:
1594:
1559:Human Mutation
1545:
1512:Human Genetics
1501:
1462:(10): e46147.
1442:
1391:
1377:
1344:
1318:
1267:
1209:
1184:
1157:
1098:
1071:
1069:
1066:
1065:
1064:
1059:
1052:
1049:
1032:Franklin Stahl
997:
994:
960:
957:
928:
925:
868:
865:
809:haploid nuclei
717:
714:
701:
698:
695:
694:
689:
684:
679:
674:
669:
664:
657:
652:
647:
642:
637:
632:
628:
627:
624:
621:
618:
615:
612:
609:
602:
599:
596:
593:
590:
587:
573:
572:
567:
562:
557:
552:
547:
542:
535:
530:
525:
520:
515:
510:
506:
505:
502:
499:
496:
493:
490:
487:
480:
477:
474:
471:
468:
465:
424:
421:
405:
402:
389:
386:
384:
381:
297:
294:
282:point mutation
277:
274:
199:
196:
156:
153:
151:
150:Categorization
148:
96:
93:
68:point mutation
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
2921:
2910:
2907:
2905:
2902:
2900:
2897:
2896:
2894:
2887:
2877:
2874:
2872:
2869:
2867:
2864:
2862:
2859:
2857:
2854:
2852:
2849:
2848:
2846:
2842:
2832:
2829:
2827:
2824:
2823:
2821:
2817:
2811:
2808:
2806:
2803:
2801:
2798:
2796:
2793:
2791:
2788:
2786:
2783:
2782:
2780:
2778:
2774:
2771:
2767:
2759:
2756:
2754:
2751:
2750:
2749:Substitution
2748:
2746:
2743:
2741:
2738:
2737:
2735:
2731:
2727:
2720:
2715:
2713:
2708:
2706:
2701:
2700:
2697:
2690:
2686:
2683:
2682:
2678:
2670:
2664:
2659:
2658:
2649:
2646:
2641:
2637:
2633:
2629:
2625:
2621:
2617:
2613:
2606:
2603:
2598:
2592:
2588:
2584:
2577:
2574:
2569:
2565:
2561:
2557:
2553:
2549:
2542:
2539:
2534:
2530:
2526:
2522:
2517:
2512:
2509:(2): 281–90.
2508:
2504:
2500:
2493:
2490:
2485:
2481:
2476:
2471:
2466:
2461:
2457:
2453:
2449:
2445:
2441:
2434:
2431:
2426:
2422:
2417:
2412:
2407:
2402:
2398:
2394:
2390:
2386:
2382:
2380:
2371:
2368:
2363:
2359:
2354:
2349:
2345:
2341:
2337:
2333:
2329:
2322:
2319:
2314:
2310:
2305:
2300:
2297:(8): 932–41.
2296:
2292:
2288:
2286:
2277:
2274:
2269:
2265:
2260:
2255:
2252:(2): 125–32.
2251:
2247:
2243:
2241:
2232:
2229:
2216:
2212:
2208:
2204:
2200:
2196:
2192:
2188:
2184:
2180:
2173:
2170:
2165:
2161:
2156:
2151:
2146:
2141:
2137:
2133:
2129:
2127:
2118:
2115:
2110:
2106:
2101:
2096:
2091:
2086:
2082:
2078:
2074:
2072:
2063:
2060:
2055:
2051:
2047:
2043:
2039:
2035:
2031:
2027:
2023:
2019:
2015:
2008:
2005:
2000:
1996:
1992:
1988:
1984:
1980:
1976:
1969:
1966:
1961:
1957:
1952:
1947:
1943:
1939:
1938:Mol Microbiol
1935:
1933:
1924:
1921:
1916:
1912:
1908:
1904:
1899:
1894:
1890:
1886:
1885:Mol Microbiol
1882:
1875:
1872:
1867:
1863:
1859:
1855:
1851:
1847:
1843:
1839:
1832:
1830:
1828:
1824:
1819:
1815:
1811:
1807:
1804:(3): 306–26.
1803:
1799:
1792:
1789:
1776:
1772:
1765:
1762:
1757:
1753:
1749:
1742:
1740:
1736:
1731:
1730:
1725:
1719:
1717:
1713:
1708:
1704:
1698:
1695:
1690:
1686:
1682:
1678:
1675:(4): 239–47.
1674:
1670:
1663:
1660:
1655:
1654:
1647:
1644:
1639:
1635:
1630:
1625:
1621:
1617:
1614:(5): 315–22.
1613:
1609:
1605:
1598:
1595:
1590:
1586:
1582:
1578:
1573:
1568:
1564:
1560:
1556:
1549:
1546:
1541:
1537:
1533:
1529:
1525:
1521:
1518:(5): 416–29.
1517:
1513:
1505:
1502:
1497:
1493:
1488:
1483:
1478:
1473:
1469:
1465:
1461:
1457:
1453:
1446:
1443:
1438:
1434:
1429:
1424:
1419:
1414:
1410:
1406:
1402:
1395:
1392:
1387:
1381:
1378:
1373:
1369:
1364:
1359:
1355:
1351:
1347:
1341:
1337:
1333:
1329:
1322:
1319:
1314:
1310:
1306:
1302:
1298:
1294:
1290:
1286:
1282:
1278:
1271:
1268:
1263:
1259:
1255:
1251:
1247:
1243:
1239:
1235:
1231:
1227:
1220:
1213:
1210:
1205:
1201:
1195:
1193:
1191:
1189:
1185:
1180:
1176:
1173:(2): 87–105.
1172:
1168:
1161:
1158:
1153:
1149:
1144:
1139:
1134:
1129:
1125:
1121:
1118:(4): 622–33.
1117:
1113:
1109:
1102:
1099:
1086:
1082:
1076:
1073:
1067:
1063:
1060:
1058:
1057:Missense mRNA
1055:
1054:
1050:
1048:
1045:
1041:
1037:
1033:
1029:
1025:
1021:
1017:
1015:
1011:
1007:
1006:Oscar Hertwig
1003:
995:
993:
991:
987:
982:
978:
974:
970:
966:
958:
956:
954:
950:
949:gene products
946:
942:
938:
934:
926:
924:
922:
918:
914:
913:
908:
904:
900:
899:
894:
889:
886:
882:
878:
874:
866:
864:
861:
857:
852:
848:
846:
841:
839:
835:
831:
827:
823:
822:
817:
814:
810:
805:
803:
800:
796:
795:
790:
789:
784:
783:
778:
777:
772:
771:
766:
765:
760:
759:
754:
751:
747:
743:
739:
735:
731:
727:
723:
715:
713:
709:
707:
704:The cause of
699:
693:
690:
688:
685:
683:
680:
678:
675:
673:
670:
668:
665:
663:
658:
656:
653:
651:
648:
646:
643:
641:
638:
636:
633:
630:
629:
625:
622:
619:
616:
613:
610:
603:
600:
597:
594:
591:
588:
585:
584:
578:
571:
568:
566:
563:
561:
558:
556:
553:
551:
548:
546:
543:
541:
536:
534:
531:
529:
526:
524:
521:
519:
516:
514:
511:
508:
507:
503:
500:
497:
494:
491:
488:
481:
478:
475:
472:
469:
466:
463:
462:
456:
453:
451:
447:
446:alpha-globins
443:
439:
435:
431:
429:
422:
420:
418:
414:
410:
403:
401:
399:
395:
387:
382:
380:
376:
372:
368:
364:
362:
358:
354:
348:
346:
342:
338:
333:
329:
324:
322:
318:
313:
311:
307:
303:
295:
293:
291:
287:
283:
275:
273:
271:
267:
263:
259:
257:
253:
252:glutamic acid
249:
246:
242:
238:
235:
231:
227:
222:
218:
216:
212:
208:
204:
197:
195:
193:
192:transversions
189:
185:
181:
177:
173:
165:
161:
154:
149:
147:
144:
139:
137:
133:
129:
125:
121:
116:
114:
110:
106:
102:
94:
92:
90:
86:
81:
77:
73:
70:is a genetic
69:
60:
53:
49:
46:. Each three-
45:
40:
32:
19:
2886:
2776:
2753:Transversion
2656:
2648:
2615:
2611:
2605:
2586:
2576:
2551:
2547:
2541:
2506:
2502:
2492:
2450:(6): e2531.
2447:
2443:
2433:
2388:
2384:
2378:
2370:
2335:
2331:
2321:
2294:
2290:
2284:
2276:
2249:
2245:
2239:
2231:
2219:. Retrieved
2215:the original
2189:(4): 420–6.
2186:
2182:
2172:
2135:
2131:
2125:
2117:
2080:
2076:
2070:
2062:
2021:
2017:
2013:
2007:
1985:(1): 31–37.
1982:
1978:
1974:
1968:
1941:
1937:
1931:
1923:
1888:
1884:
1874:
1841:
1837:
1801:
1797:
1791:
1779:. Retrieved
1775:the original
1764:
1755:
1751:
1728:
1706:
1697:
1672:
1668:
1662:
1652:
1646:
1611:
1607:
1597:
1565:(6): 423–7.
1562:
1558:
1548:
1515:
1511:
1504:
1459:
1455:
1445:
1408:
1404:
1394:
1380:
1327:
1321:
1280:
1276:
1270:
1229:
1225:
1212:
1204:the original
1170:
1167:J. Mol. Biol
1166:
1160:
1115:
1111:
1101:
1089:. Retrieved
1084:
1075:
1026:
1022:
1018:
999:
962:
930:
927:Consequences
916:
910:
906:
902:
896:
892:
870:
859:
855:
853:
849:
844:
842:
819:
806:
801:
792:
786:
780:
774:
768:
762:
756:
736:accumulates
729:
725:
719:
710:
703:
576:
454:
442:beta-globins
436:
432:
426:
407:
391:
377:
373:
369:
365:
349:
325:
314:
299:
281:
279:
260:
250:rather than
219:
214:
210:
201:
190:(Alpha) and
172:Ernst Freese
169:
140:
117:
98:
67:
65:
2618:(1): 72–5.
2554:: 579–613.
2221:29 December
2132:PLOS Pathog
1781:28 December
1405:Mol. Cancer
973:transformed
965:mutagenesis
909:homolog in
895:homolog in
871:RIP causes
845:L. maculans
450:sickle cell
332:adaptations
188:transitions
164:Transitions
113:nucleotides
2893:Categories
2758:Transition
2077:PLOS Genet
1328:Proteomics
1068:References
1062:PAM matrix
1008:in 1876.
953:methylated
888:transition
851:pressure.
834:nucleotide
830:base pairs
799:methylated
753:transition
438:Hemoglobin
286:insertions
245:amino acid
234:hemoglobin
184:pyrimidine
115:code for.
109:pyrimidine
52:amino acid
48:nucleotide
2740:Insertion
2640:205011982
1758:(1): 187.
1354:1940-6029
975:into the
971:is first
937:parasites
867:Mechanism
860:N. crassa
357:chromatin
337:evolution
226:wild type
120:mutations
2904:Mutation
2745:Deletion
2726:Mutation
2632:11413469
2533:14143830
2484:18575630
2444:PLOS ONE
2425:12072568
2332:Genetics
2313:19589069
2268:17610516
2211:20471307
2164:21079787
2109:19714214
2054:11080216
2046:17823352
1999:12742061
1960:12207702
1915:25096512
1907:11359565
1866:23036409
1818:20854921
1769:eMedTV.
1638:12011146
1589:22140210
1581:14635100
1532:11409870
1496:23056252
1456:PLOS ONE
1437:21859464
1372:28188534
1305:11357144
1254:12068308
1152:16590424
1051:See also
941:missense
310:pre-mRNA
302:promoter
170:In 1959
128:mutagens
72:mutation
2568:2150906
2525:9346245
2475:2432034
2452:Bibcode
2393:Bibcode
2362:7896093
2353:1206250
2191:Bibcode
2155:2973834
2100:2725324
2026:Bibcode
2018:Science
1858:2960455
1689:9435331
1629:1735110
1540:2136834
1487:3463568
1464:Bibcode
1428:3170638
1411:: 101.
1363:5388446
1313:4337913
1285:Bibcode
1262:3071547
1234:Bibcode
1120:Bibcode
1010:Mitosis
1002:meiosis
996:History
813:meiotic
802:de novo
353:glycine
317:peptide
270:protein
132:UV rays
2691:(MeSH)
2665:
2638:
2630:
2593:
2566:
2531:
2523:
2482:
2472:
2423:
2416:124379
2413:
2360:
2350:
2311:
2266:
2209:
2162:
2152:
2107:
2097:
2052:
2044:
1997:
1958:
1913:
1905:
1864:
1856:
1816:
1687:
1636:
1626:
1587:
1579:
1538:
1530:
1494:
1484:
1435:
1425:
1370:
1360:
1352:
1342:
1311:
1303:
1277:Nature
1260:
1252:
1226:Nature
1150:
1143:222607
1140:
1091:17 May
1040:Watson
977:genome
856:et al.
419:gene.
394:cancer
388:Cancer
361:Cancer
326:Point
306:intron
248:valine
180:purine
176:purine
136:X-rays
105:purine
95:Causes
44:codons
2636:S2CID
2529:S2CID
2050:S2CID
1911:S2CID
1862:S2CID
1585:S2CID
1536:S2CID
1309:S2CID
1258:S2CID
1222:(PDF)
1044:Crick
917:masc1
818:. In
791:. In
631:START
509:START
341:cells
241:codon
2663:ISBN
2628:PMID
2591:ISBN
2564:PMID
2521:PMID
2503:Cell
2480:PMID
2421:PMID
2358:PMID
2309:PMID
2264:PMID
2223:2018
2207:PMID
2160:PMID
2105:PMID
2042:PMID
1995:PMID
1956:PMID
1903:PMID
1854:PMID
1838:Cell
1814:PMID
1783:2011
1685:PMID
1634:PMID
1577:PMID
1528:PMID
1492:PMID
1433:PMID
1368:PMID
1350:ISSN
1340:ISBN
1301:PMID
1250:PMID
1148:PMID
1093:2019
1042:and
1030:and
981:mate
969:gene
943:and
903:dmtA
785:and
626:ACU
504:ACU
444:and
256:BRAF
237:gene
2620:doi
2556:doi
2511:doi
2470:PMC
2460:doi
2411:PMC
2401:doi
2348:PMC
2340:doi
2336:138
2299:doi
2254:doi
2250:273
2199:doi
2150:PMC
2140:doi
2095:PMC
2085:doi
2034:doi
2022:317
1987:doi
1977:".
1946:doi
1893:doi
1846:doi
1806:doi
1677:doi
1673:338
1624:PMC
1616:doi
1567:doi
1520:doi
1516:108
1482:PMC
1472:doi
1423:PMC
1413:doi
1358:PMC
1332:doi
1293:doi
1281:411
1242:doi
1230:417
1175:doi
1138:PMC
1128:doi
1038:.
907:rid
893:rid
879:to
744:to
734:DNA
730:RIP
728:or
720:In
692:Thr
687:Val
682:Ala
677:Ser
672:Lys
667:Glu
661:Val
655:Pro
650:Thr
645:Leu
640:His
635:Val
623:GUU
620:GCC
617:UCU
614:AAG
611:GAG
601:CCU
598:ACU
595:CUG
592:CAC
589:GUG
586:AUG
570:Thr
565:Val
560:Ala
555:Ser
550:Lys
545:Glu
539:Glu
533:Pro
528:Thr
523:Leu
518:His
513:Val
501:GUU
498:GCC
495:UCU
492:AAG
489:GAG
479:CCU
476:ACU
473:CUG
470:CAC
467:GUG
464:AUG
415:or
292:).
107:or
80:RNA
78:or
76:DNA
2895::
2634:.
2626:.
2614:.
2585:.
2562:.
2552:24
2550:.
2527:.
2519:.
2507:91
2505:.
2501:.
2478:.
2468:.
2458:.
2446:.
2442:.
2419:.
2409:.
2399:.
2389:99
2387:.
2383:.
2356:.
2346:.
2334:.
2330:.
2307:.
2295:22
2293:.
2289:.
2262:.
2248:.
2244:.
2205:.
2197:.
2187:13
2185:.
2181:.
2158:.
2148:.
2134:.
2130:.
2103:.
2093:.
2079:.
2075:.
2048:.
2040:.
2032:.
2020:.
1993:.
1983:39
1981:.
1954:.
1942:45
1940:.
1936:.
1909:.
1901:.
1889:40
1887:.
1883:.
1860:.
1852:.
1842:51
1840:.
1826:^
1812:.
1802:48
1800:.
1754:.
1750:.
1738:^
1726:.
1715:^
1705:.
1683:.
1671:.
1632:.
1622:.
1612:39
1610:.
1606:.
1583:.
1575:.
1563:22
1561:.
1557:.
1534:.
1526:.
1514:.
1490:.
1480:.
1470:.
1458:.
1454:.
1431:.
1421:.
1409:10
1407:.
1403:.
1366:.
1356:.
1348:.
1338:.
1307:.
1299:.
1291:.
1279:.
1256:.
1248:.
1240:.
1228:.
1224:.
1187:^
1169:.
1146:.
1136:.
1126:.
1116:45
1114:.
1110:.
1083:.
1016:.
923:.
915:,
901:,
840:.
824:,
804:.
779:,
773:,
767:,
761:,
724:,
347:.
312:.
134:,
66:A
2718:e
2711:t
2704:v
2671:.
2642:.
2622::
2616:2
2599:.
2570:.
2558::
2535:.
2513::
2486:.
2462::
2454::
2448:3
2427:.
2403::
2395::
2381:"
2364:.
2342::
2315:.
2301::
2287:"
2270:.
2256::
2225:.
2201::
2193::
2166:.
2142::
2136:6
2111:.
2087::
2081:5
2056:.
2036::
2028::
2001:.
1989::
1962:.
1948::
1917:.
1895::
1868:.
1848::
1820:.
1808::
1785:.
1756:1
1691:.
1679::
1640:.
1618::
1591:.
1569::
1542:.
1522::
1498:.
1474::
1466::
1460:7
1439:.
1415::
1374:.
1334::
1315:.
1295::
1287::
1264:.
1244::
1236::
1181:.
1177::
1171:1
1154:.
1130::
1122::
1095:.
885:T
883::
881:A
877:C
875::
873:G
750:T
748::
746:A
742:C
740::
738:G
608:G
606:U
604:G
486:G
484:A
482:G
209:(
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.