293:
166:
600:
35:
625:-tryptophan is first turned into aminophenylpyrrole (APP) and then by subsequent steps to aminopyrrolnitrin and pyrrolnitrin. While these steps have not been described in more detail, prnB is able to produce APP, presumably from tryptophan as starting material. APP seems to be an unwanted side product. The gene coding for prnB also starts with the unusual GTG
443:
763:"The Ternary Complex of PrnB (the Second Enzyme in the Pyrrolnitrin Biosynthesis Pathway), Tryptophan, and Cyanide Yields New Mechanistic Insights into the Indolamine Dioxygenase Superfamily"
412:
456:
655:
Kirner, Sabine; Hammer, Philip E.; Hill, D. Steven; Altmann, Annett; Fischer, Ilona; Weislo, Laura J.; Lanahan, Mike; van Pée, Karl-Heinz; Ligon, James M. (April 1998).
858:
De
Laurentis, Walter; Khim, Leang; Anderson, J. L. Ross; Adam, Ariane; Phillips, Robert S.; Chapman, Stephen K.; van Pee, Karl-Heinz; Naismith, James H. (2007-10-01).
103:
342:
948:
Jespers, A.B.K.; Davidse, L.C.; Dewaard, M.A. (1993). "Biochemical
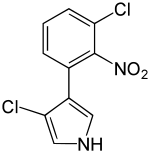
Effects of the Phenylpyrrole Fungicide Fenpiclonil in Fusarium sulphureum (Schlecht)".
814:"Production of the antifungal compounds phenazine and pyrrolnitrin from Pseudomonas chlororaphis O6 is differentially regulated by glucose"
999:
1004:
571:
307:
932:
463:
250:
271:
588:
prnC – chlorination of monodechloroaminopyrrolnitrin to form aminopyrrolnitrin (APRN), requiring NAD for its activity
860:"The Second Enzyme in Pyrrolnitrin Biosynthetic Pathway Is Related to the Heme-Dependent Dioxygenase Superfamily"
510:
173:
211:
555:
161:
495:
559:
487:
217:
989:
47:
288:
69:
517:
509:
as secondary metabolite. It is believed that the antifungal properties come from inhibition of
994:
965:
928:
897:
879:
835:
794:
743:
694:
676:
629:, further lowering the amount of prnB expressed and thus lowering the amount of present APP.
957:
920:
887:
871:
825:
784:
774:
733:
725:
684:
668:
365:
599:
259:
143:
578:
79:
812:
Park, J. Y.; Oh, S. A.; Anderson, A. J.; Neiswender, J.; Kim, J. -C.; Kim, Y. C. (2011).
551:
The products of these genes are similar in size and catalyze four subsequent reactions:
292:
165:
123:
892:
859:
789:
762:
615:
434:
738:
713:
689:
656:
983:
915:
Pillonel, Ch; Knauf-beiter, G.; Steinemann, A. (2003). "Fungicides, Phenylpyrroles".
830:
813:
483:
154:
672:
657:"Functions Encoded by Pyrrolnitrin Biosynthetic Genes from Pseudomonas fluorescens"
423:
418:
729:
239:
626:
592:
521:
501:
924:
506:
490:
396:
134:
969:
883:
680:
779:
17:
961:
901:
839:
798:
614:
Neither of the chlorinating enzymes, prnA nor prnC, show homology to known
747:
698:
316:
InChI=1S/C10H6Cl2N2O2/c11-8-3-1-2-6(10(8)14(15)16)7-4-13-5-9(7)12/h1-5,13H
34:
326:
InChI=1/C10H6Cl2N2O2/c11-8-3-1-2-6(10(8)14(15)16)7-4-13-5-9(7)12/h1-5,13H
226:
174:
875:
545:
433:
Except where otherwise noted, data are given for materials in their
200:
598:
114:
102:
92:
537:
191:
276:
585:-tryptophan to form monodechloroaminopyrrolnitrin (MAD)
451:
607:
Except for prnA, these enzymes are unable to act on
761:Zhu, X.; Van Pee, K. -H.; Naismith, J. H. (2010).
621:An alternative pathway was also suggested, where
238:
78:
714:"Systemic antifungal activity of pyrrolnitrin"
536:, biosynthesis of pyrrolnitrin requires four
8:
291:
164:
142:
26:
891:
829:
788:
778:
737:
688:
258:
524:are chemically related to pyrrolnitrin.
712:Gordee, R. S.; Matthews, T. R. (1969).
638:
347:
312:
287:
216:
155:
950:Pesticide Biochemistry and Physiology
319:Key: QJBZDBLBQWFTPZ-UHFFFAOYSA-N
122:
52:3-Chloro-4-(3-chloro-2-nitrophenyl)-1
7:
853:
851:
849:
650:
648:
646:
644:
642:
422:
595:to form nitro group of pyrrolnitrin
329:Key: QJBZDBLBQWFTPZ-UHFFFAOYAG
229:
199:
505:species produce pyrrolnitrin from
350:C1=CC(=C(C(=C1)Cl)(=O))C2=CNC=C2Cl
25:
831:10.1111/j.1472-765X.2011.03036.x
441:
33:
818:Letters in Applied Microbiology
767:Journal of Biological Chemistry
673:10.1128/JB.180.7.1939-1943.1998
570:-tryptophan (7-CLT), requiring
437:(at 25 °C , 100 kPa).
577:prnB – ring rearrangement and
1:
917:Encyclopedia of Agrochemicals
730:10.1128/AEM.17.5.690-694.1969
1021:
1000:Chlorobenzene derivatives
925:10.1002/047126363X.agr106
603:Pyrrolnitrin biosynthesis
516:The synthetic fungicides
511:electron transport system
431:
405:
358:
338:
303:
62:
46:
41:
32:
1005:Nitrobenzene derivatives
566:-tryptophan to 7-chloro-
780:10.1074/jbc.M110.120485
661:Journal of Bacteriology
544:arranged into a single
534:Pseudomonas fluorescens
401:257.07284
962:10.1006/pest.1993.1014
604:
496:Pseudomonas pyrrocinia
602:
718:Applied Microbiology
618:nor to one another.
591:prnD – oxidation of
48:Preferred IUPAC name
870:(43): 12393–12404.
773:(27): 21126–21133.
29:
605:
464:Infobox references
27:
876:10.1021/bi7012189
624:
610:
584:
569:
565:
486:chemical used an
472:Chemical compound
470:
469:
272:CompTox Dashboard
104:Interactive image
16:(Redirected from
1012:
974:
973:
945:
939:
938:
912:
906:
905:
895:
855:
844:
843:
833:
809:
803:
802:
792:
782:
758:
752:
751:
741:
709:
703:
702:
692:
667:(7): 1939–1943.
652:
622:
608:
582:
574:for its activity
567:
563:
454:
448:
445:
444:
426:
366:Chemical formula
296:
295:
280:
278:
262:
242:
231:
220:
203:
176:
168:
157:
146:
126:
106:
82:
37:
30:
21:
1020:
1019:
1015:
1014:
1013:
1011:
1010:
1009:
980:
979:
978:
977:
947:
946:
942:
935:
914:
913:
909:
857:
856:
847:
811:
810:
806:
760:
759:
755:
711:
710:
706:
654:
653:
640:
635:
616:haloperoxidases
579:decarboxylation
530:
473:
466:
461:
460:
459: ?)
450:
446:
442:
438:
415:
390:
386:
382:
378:
374:
368:
354:
351:
346:
345:
334:
331:
330:
327:
321:
320:
317:
311:
310:
299:
281:
274:
265:
245:
232:
206:
186:
149:
129:
109:
96:
85:
72:
58:
57:
23:
22:
15:
12:
11:
5:
1018:
1016:
1008:
1007:
1002:
997:
992:
982:
981:
976:
975:
956:(2): 116–129.
940:
933:
907:
845:
824:(5): 532–537.
804:
753:
724:(5): 690–694.
704:
637:
636:
634:
631:
597:
596:
589:
586:
575:
529:
526:
471:
468:
467:
462:
440:
439:
435:standard state
432:
429:
428:
416:
411:
408:
407:
403:
402:
399:
393:
392:
388:
384:
380:
376:
372:
369:
364:
361:
360:
356:
355:
353:
352:
349:
341:
340:
339:
336:
335:
333:
332:
328:
325:
324:
322:
318:
315:
314:
306:
305:
304:
301:
300:
298:
297:
284:
282:
270:
267:
266:
264:
263:
255:
253:
247:
246:
244:
243:
235:
233:
225:
222:
221:
214:
208:
207:
205:
204:
196:
194:
188:
187:
185:
184:
180:
178:
170:
169:
159:
151:
150:
148:
147:
139:
137:
131:
130:
128:
127:
119:
117:
111:
110:
108:
107:
99:
97:
90:
87:
86:
84:
83:
75:
73:
68:
65:
64:
60:
59:
51:
50:
44:
43:
39:
38:
24:
14:
13:
10:
9:
6:
4:
3:
2:
1017:
1006:
1003:
1001:
998:
996:
993:
991:
988:
987:
985:
971:
967:
963:
959:
955:
951:
944:
941:
936:
930:
926:
922:
918:
911:
908:
903:
899:
894:
889:
885:
881:
877:
873:
869:
865:
861:
854:
852:
850:
846:
841:
837:
832:
827:
823:
819:
815:
808:
805:
800:
796:
791:
786:
781:
776:
772:
768:
764:
757:
754:
749:
745:
740:
735:
731:
727:
723:
719:
715:
708:
705:
700:
696:
691:
686:
682:
678:
674:
670:
666:
662:
658:
651:
649:
647:
645:
643:
639:
632:
630:
628:
619:
617:
612:
611:-tryptophan.
601:
594:
590:
587:
580:
576:
573:
561:
557:
554:
553:
552:
550:
547:
543:
539:
535:
527:
525:
523:
519:
514:
512:
508:
504:
503:
498:
497:
492:
489:
485:
484:phenylpyrrole
481:
477:
465:
458:
453:
436:
430:
425:
420:
417:
414:
410:
409:
406:Pharmacology
404:
400:
398:
395:
394:
370:
367:
363:
362:
357:
348:
344:
337:
323:
313:
309:
302:
294:
290:
289:DTXSID9046867
286:
285:
283:
273:
269:
268:
261:
257:
256:
254:
252:
249:
248:
241:
237:
236:
234:
228:
224:
223:
219:
215:
213:
210:
209:
202:
198:
197:
195:
193:
190:
189:
182:
181:
179:
177:
172:
171:
167:
163:
160:
158:
156:ECHA InfoCard
153:
152:
145:
141:
140:
138:
136:
133:
132:
125:
121:
120:
118:
116:
113:
112:
105:
101:
100:
98:
94:
89:
88:
81:
77:
76:
74:
71:
67:
66:
61:
55:
49:
45:
40:
36:
31:
28:Pyrrolnitrin
19:
953:
949:
943:
916:
910:
867:
864:Biochemistry
863:
821:
817:
807:
770:
766:
756:
721:
717:
707:
664:
660:
620:
613:
606:
581:of 7-chloro-
560:chlorination
548:
541:
533:
531:
528:Biosynthesis
515:
500:
494:
479:
476:Pyrrolnitrin
475:
474:
63:Identifiers
53:
18:C10H6Cl2N2O2
990:Antifungals
627:start codon
593:amino group
522:fludioxonil
518:fenpiclonil
502:Pseudomonas
359:Properties
162:100.012.557
124:CHEBI:32079
984:Categories
934:047126363X
633:References
507:tryptophan
499:and other
491:antibiotic
488:antifungal
397:Molar mass
260:N0P24B6EDQ
135:ChemSpider
91:3D model (
70:CAS Number
970:0048-3575
884:0006-2960
681:0021-9193
183:213-812-7
175:EC Number
80:1018-71-9
995:Pyrroles
902:17924666
840:21362001
799:20421301
542:prnABCD,
540:, named
427:)
413:ATC code
56:-pyrrole
893:3326534
790:2898318
748:5785951
699:9537395
482:) is a
457:what is
455: (
421: (
419:D01AA07
391:
227:PubChem
218:D011764
968:
931:
900:
890:
882:
838:
797:
787:
746:
739:377781
736:
697:
690:107110
687:
679:
546:operon
452:verify
449:
343:SMILES
201:D01094
42:Names
538:genes
308:InChI
240:13916
144:13314
115:ChEBI
93:JSmol
966:ISSN
929:ISBN
898:PMID
880:ISSN
836:PMID
795:PMID
744:PMID
695:PMID
677:ISSN
556:prnA
520:and
251:UNII
212:MeSH
192:KEGG
958:doi
921:doi
888:PMC
872:doi
826:doi
785:PMC
775:doi
771:285
734:PMC
726:doi
685:PMC
669:doi
665:180
572:NAD
562:of
532:In
480:PRN
424:WHO
277:EPA
230:CID
986::
964:.
954:45
952:.
927:.
919:.
896:.
886:.
878:.
868:46
866:.
862:.
848:^
834:.
822:52
820:.
816:.
793:.
783:.
769:.
765:.
742:.
732:.
722:17
720:.
716:.
693:.
683:.
675:.
663:.
659:.
641:^
558:–
513:.
493:.
379:Cl
373:10
972:.
960::
937:.
923::
904:.
874::
842:.
828::
801:.
777::
750:.
728::
701:.
671::
623:L
609:D
583:L
568:L
564:L
549:.
478:(
447:N
389:2
387:O
385:2
383:N
381:2
377:6
375:H
371:C
279:)
275:(
95:)
54:H
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.