563:) manner forming a disk-like compact structure. By doing so, they form clefts that constitute the canonical ligand binding regions. In principle, the number of canonical binding sites matches the number of CBS domains within the molecule and are traditionally numbered according to the CBS domain that contains each of the conserved aspartate residues that potentially interact with the ribose of the nucleotides. However, not all of these cavities might necessarily bind nucleotides or be functional. Recently, a non-canonical site for AMP has also been described in protein MJ1225 from
594:, or s-adenosylmethionine, but they may also bind metallic ions such as Mg. Upon binding these different ligands the CBS domains regulate the activity of associated enzymatic domains. The molecular mechanisms underlying this regulation are just starting to be elucidated. At the moment, two different type of mechanisms have been proposed. The first one claims that the nucleotide portion of the ligand induces essentially no change in the protein structure, the
631:
571:
26:
1786:
Tuominen H, Salminen A, Oksanen E, Jämsen J, Heikkilä O, Lehtiö L, Magretova NN, Goldman A, Baykov AA, Lahti R (May 2010). "Crystal structures of the CBS and DRTGG domains of the regulatory region of
Clostridiumperfringens pyrophosphatase complexed with the inhibitor, AMP, and activator, diadenosine
1618:
Lucas M, Encinar JA, Arribas EA, Oyenarte I, García IG, Kortazar D, Fernández JA, Mato JM, Martínez-Chantar ML, Martínez-Cruz LA (February 2010). "Binding of S-methyl-5'-thioadenosine and S-adenosyl-L-methionine to protein MJ0100 triggers an open-to-closed conformational change in its CBS motif
627:, membrane transporters or DNA-binding domains. However, proteins that contain only CBS domains are also often found, particularly in prokaryotes. These standalone CBS domain proteins might form complexes upon binding to other proteins such as kinases to which they interact with and regulate.
598:
at the binding site being the most significant property of adenosine nucleotide binding. This "static" response would be involved in processes in which regulation by energy charge would be advantageous. On the contrary, the second type of mechanism (denoted as "dynamic") involves dramatic
1918:
Tuominen H, Salminen A, Oksanen E, et al. (May 2010). "Crystal
Structures of the CBS and DRTGG Domains of the Regulatory Region of Clostridium perfringens Pyrophosphatase Complexed with the Inhibitor, AMP, and Activator, Diadenosine Tetraphosphate".
1825:
Xiao B, Heath R, Saiu P, Leiper FC, Leone P, Jing C, Walker PA, Haire L, Eccleston JF, Davis CT, Martin SR, Carling D, Gamblin SJ (September 2007). "Structural basis for AMP binding to mammalian AMP-activated protein kinase".
1577:
Gómez-García I, Oyenarte I, Martínez-Cruz LA (May 2010). "The
Crystal Structure of Protein MJ1225 from Methanocaldococcus jannaschii Shows Strong Conservation of Key Structural Features Seen in the Eukaryal gamma-AMPK".
528:
on one side. CBS domains are always found in pairs in protein sequences and each pair of these domains tightly associate in a pseudo dimeric arrangement through their β-sheets forming a so-called CBS-pair or
1264:
Zhang R, Evans G, Rotella FJ, Westbrook EM, Beno D, Huberman E, Joachimiak A, Collart FR (April 1999). "Characteristics and crystal structure of bacterial inosine-5'-monophosphate dehydrogenase".
161:
1087:
Day P, Sharff A, Parra L, et al. (May 2007). "Structure of a CBS-domain pair from the regulatory gamma1 subunit of human AMPK in complex with AMP and ZMP".
488:(AMPK). CBS domains regulate the activity of associated enzymatic and transporter domains in response to binding molecules with adenosyl groups such as
1350:
Meyer S, Savaresi S, Forster IC, Dutzler R (January 2007). "Nucleotide recognition by the cytoplasmic domain of the human chloride transporter ClC-5".
1044:
Meyer S, Savaresi S, Forster IC, Dutzler R (January 2007). "Nucleotide recognition by the cytoplasmic domain of the human chloride transporter ClC-5".
73:
599:
conformational changes in the protein structure upon ligand binding and has been reported for the cytosolic domain of the Mg transporter MgtE from
1871:"Purification, crystallization and preliminary X-ray diffraction analysis of the CBS-domain pair from the Methanococcus jannaschii protein MJ0100"
660:
1396:
Amodeo GA, Rudolph MJ, Tong L (September 2007). "Crystal structure of the heterotrimer core of
Saccharomyces cerevisiae AMPK homologue SNF1".
97:
30:
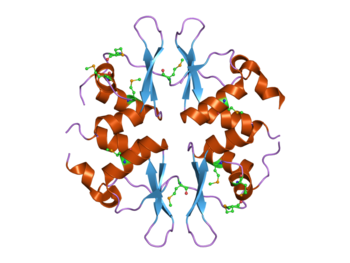
Structure of the yeast SNF4 protein that contains four CBS domains. This protein is part of the AMP-activated protein kinase (AMPK) complex.
477:
956:"Removal of gating in voltage-dependent ClC-2 chloride channel by point mutations affecting the pore and C-terminus CBS-2 domain"
2038:"Albers-Schönberg disease (autosomal dominant osteopetrosis, type II) results from mutations in the ClCN7 chloride channel gene"
682:
genes, and mutations in the CBS domains of several of these have been identified as the cause of genetic diseases. Mutations in
1956:"Mutations in the regulatory domain of cystathionine beta synthase can functionally suppress patient-derived mutations in cis"
886:
Ponting CP (March 1997). "CBS domains in CIC chloride channels implicated in myotonia and nephrolithiasis (kidney stones)".
181:
485:
809:
Bateman A (January 1997). "The structure of a domain common to archaebacteria and the homocystinuria disease protein".
656:
644:
574:
473:
577:
of CBS domains showing secondary structures above. Yellow arrows represent beta strands and red boxes alpha helices.
1005:"The structure of the cytoplasmic domain of the chloride channel ClC-Ka reveals a conserved interaction interface"
117:
643:
Mutations in some human CBS domain-containing proteins leads to genetic diseases. For example, mutations in the
2082:
1739:"CBS domains form energy-sensing modules whose binding of adenosine ligands is disrupted by disease mutations"
169:
476:, one of the proteins it is found in. CBS domains are also found in a wide variety of other proteins such as
611:
595:
587:
489:
848:
1273:
591:
493:
165:
699:
668:
601:
497:
110:
1278:
122:
78:
1311:"Structure of the Bateman2 domain of yeast Snf4: dimeric association and relevance for AMP binding"
1124:"Structure of the Bateman2 domain of yeast Snf4: dimeric association and relevance for AMP binding"
770:"Structure of the Bateman2 domain of yeast Snf4: dimeric association and relevance for AMP binding"
509:
468:
found in a range of proteins in all species from bacteria to humans. It was first identified as a
1851:
1477:
1421:
1375:
1069:
517:
469:
2059:
2018:
1977:
1936:
1900:
1843:
1804:
1768:
1719:
1692:
1636:
1595:
1559:
1518:
1469:
1413:
1367:
1332:
1291:
1246:
1197:
1145:
1104:
1061:
1026:
1004:
985:
936:
914:
895:
868:
826:
791:
758:
711:
623:
CBS domains are often found in proteins that contain other domains. These domains are usually
188:
156:
2049:
2008:
1967:
1928:
1890:
1882:
1835:
1796:
1758:
1750:
1682:
1672:
1628:
1587:
1549:
1508:
1459:
1448:"Crystal structures of the adenylate sensor from fission yeast AMP-activated protein kinase"
1405:
1359:
1322:
1283:
1236:
1228:
1187:
1179:
1135:
1096:
1053:
1016:
975:
967:
926:
860:
818:
781:
735:
679:
481:
148:
630:
513:
655:). Mutations in the gamma subunit of the AMPK enzyme have been shown to lead to familial
1895:
1870:
1687:
1660:
980:
955:
648:
465:
1763:
1738:
1241:
1216:
1192:
1167:
822:
2076:
1309:
Rudolph MJ, Amodeo GA, Iram SH, Hong SP, Pirino G, Carlson M, Tong L (January 2007).
768:
Rudolph MJ, Amodeo GA, Iram SH, Hong SP, Pirino G, Carlson M, Tong L (January 2007).
723:
102:
1661:"Mg2+-sensing mechanism of Mg2+ transporter MgtE probed by molecular dynamics study"
1481:
1379:
1073:
570:
54:
1855:
1425:
144:
971:
66:
586:
It has been shown that CBS domains bind to adenosyl groups in molecules such as
525:
954:
Yusef YR, Zúñiga L, Catalán M, Niemeyer MI, Cid LP, Sepúlveda FV (April 2006).
864:
1932:
1886:
1800:
1659:
Ishitani R, Sugita Y, Dohmae N, Furuya N, Hattori M, Nureki O (October 2008).
1632:
1591:
1554:
1537:
1513:
1496:
1327:
1310:
1140:
1123:
1100:
1021:
931:
786:
769:
667:). Mutations in the CBS domains of the IMPDH enzyme lead to the eye condition
521:
2054:
2037:
1677:
1464:
1447:
2063:
2022:
1981:
1972:
1955:
1940:
1904:
1847:
1808:
1772:
1723:
1696:
1640:
1599:
1563:
1522:
1473:
1417:
1371:
1336:
1295:
1250:
1201:
1149:
1108:
1065:
1030:
989:
940:
915:"Crystal structure of the cytoplasmic domain of the chloride channel ClC-0"
872:
795:
899:
830:
106:
25:
687:
533:. These CBS domain pairs can associate in a head-to-head (i.e. PDB codes
61:
1997:"Myotonia caused by mutations in the muscle chloride channel gene CLCN1"
1839:
1409:
2013:
1996:
85:
1287:
762:
559:
553:
547:
541:
535:
446:
440:
434:
428:
422:
416:
410:
404:
398:
392:
386:
380:
374:
368:
362:
356:
350:
344:
338:
332:
326:
320:
314:
308:
302:
296:
290:
284:
278:
272:
266:
260:
254:
248:
242:
236:
230:
224:
218:
212:
206:
200:
194:
1754:
1363:
1232:
1183:
1057:
731:
624:
176:
90:
1217:"Bateman domains and adenosine derivatives form a binding contract"
1168:"Bateman domains and adenosine derivatives form a binding contract"
849:"CBS domains: structure, function, and pathology in human proteins"
719:
707:
695:
683:
629:
569:
1869:
Lucas M, Kortazar D, Astigarraga E, et al. (October 2008).
739:
727:
715:
703:
691:
672:
664:
652:
138:
49:
2036:
Cleiren E, Bénichou O, Van Hul E, et al. (December 2001).
647:
protein lead to an inherited disorder of the metabolism called
1497:"Structural insight into AMPK regulation: ADP comes into play"
1954:
Shan X, Dunbrack RL, Christopher SA, Kruger WD (March 2001).
1122:
Rudolph MJ, Amodeo GA, Iram SH, et al. (January 2007).
1737:
Scott JW, Hawley SA, Green KA, et al. (January 2004).
508:
The CBS domain is composed of a beta-alpha-beta-beta-alpha
634:
Example protein domains found associated with CBS domains
187:
175:
155:
137:
132:
116:
96:
84:
72:
60:
48:
40:
35:
18:
1538:"AMPK structure and regulation from three angles"
1654:
1652:
1650:
1441:
1439:
1437:
1435:
1161:
1159:
567:, though its functional role is still unknown.
1536:Kemp BE, Oakhill JS, Scott JW (October 2007).
1391:
1389:
842:
840:
1613:
1611:
1609:
8:
1820:
1818:
1495:Jin X, Townley R, Shapiro L (October 2007).
520:that contains a three-stranded antiparallel
605:, the unknown function protein MJ0100 from
551:) or a head-to-tail (i.e. PDB codes
129:
2053:
2012:
1971:
1894:
1762:
1686:
1676:
1553:
1512:
1463:
1326:
1277:
1240:
1191:
1139:
1020:
979:
930:
785:
847:Ignoul S, Eggermont J (December 2005).
750:
1710:Hattori M, Nureki O (March 2008). "".
678:Humans have a number of voltage-gated
15:
7:
913:Meyer S, Dutzler R (February 2006).
1446:Townley R, Shapiro L (March 2007).
1003:Markovic S, Dutzler R (June 2007).
478:inosine monophosphate dehydrogenase
14:
1875:Acta Crystallographica Section F
24:
700:idiopathic generalised epilepsy
482:voltage gated chloride channels
472:region in 1997 and named after
661:Wolff–Parkinson–White syndrome
1:
853:Am. J. Physiol., Cell Physiol
823:10.1016/S0968-0004(96)30046-7
609:and the regulatory region of
133:Available protein structures:
1665:Proc. Natl. Acad. Sci. U.S.A
972:10.1113/jphysiol.2005.102392
639:Mutations leading to disease
486:AMP-activated protein kinase
657:hypertrophic cardiomyopathy
645:cystathionine beta synthase
575:Multiple sequence alignment
474:cystathionine beta synthase
2099:
1712:Tanpakushitsu Kakusan Koso
865:10.1152/ajpcell.00282.2005
460:In molecular biology, the
1933:10.1016/j.jmb.2010.03.019
1887:10.1107/S1744309108027930
1801:10.1016/j.jmb.2010.03.019
1633:10.1016/j.jmb.2009.12.012
1592:10.1016/j.jmb.2010.03.045
1555:10.1016/j.str.2007.09.006
1514:10.1016/j.str.2007.07.017
1328:10.1016/j.str.2006.11.014
1141:10.1016/j.str.2006.11.014
1101:10.1107/S0907444907009110
1022:10.1016/j.str.2007.04.013
932:10.1016/j.str.2005.10.008
787:10.1016/j.str.2006.11.014
128:
23:
1215:Kemp BE (January 2004).
1166:Kemp BE (January 2004).
1678:10.1073/pnas.0802991105
1465:10.1126/science.1137503
612:Clostridium perfringens
596:electrostatic potential
2055:10.1093/hmg/10.25.2861
1995:Pusch M (April 2002).
1352:Nat. Struct. Mol. Biol
1046:Nat. Struct. Mol. Biol
635:
578:
633:
573:
1973:10.1093/hmg/10.6.635
730:), and mutations in
669:retinitis pigmentosa
602:Thermus thermophilus
498:s-adenosylmethionine
1840:10.1038/nature06161
1410:10.1038/nature06127
1089:Acta Crystallogr. D
811:Trends Biochem. Sci
510:secondary structure
2014:10.1002/humu.10063
636:
619:Associated domains
579:
518:tertiary structure
470:conserved sequence
1881:(Pt 10): 936–41.
1834:(7161): 496–500.
1787:tetraphosphate".
1288:10.1021/bi982858v
615:pyrophosphatase.
458:
457:
454:
453:
182:structure summary
2090:
2068:
2067:
2057:
2033:
2027:
2026:
2016:
1992:
1986:
1985:
1975:
1951:
1945:
1944:
1915:
1909:
1908:
1898:
1866:
1860:
1859:
1822:
1813:
1812:
1783:
1777:
1776:
1766:
1755:10.1172/JCI19874
1734:
1728:
1727:
1707:
1701:
1700:
1690:
1680:
1656:
1645:
1644:
1615:
1604:
1603:
1574:
1568:
1567:
1557:
1533:
1527:
1526:
1516:
1492:
1486:
1485:
1467:
1458:(5819): 1726–9.
1443:
1430:
1429:
1393:
1384:
1383:
1364:10.1038/nsmb1188
1347:
1341:
1340:
1330:
1306:
1300:
1299:
1281:
1272:(15): 4691–700.
1261:
1255:
1254:
1244:
1233:10.1172/JCI20846
1212:
1206:
1205:
1195:
1184:10.1172/JCI20846
1163:
1154:
1153:
1143:
1119:
1113:
1112:
1095:(Pt 5): 587–96.
1084:
1078:
1077:
1058:10.1038/nsmb1188
1041:
1035:
1034:
1024:
1000:
994:
993:
983:
966:(Pt 1): 173–81.
951:
945:
944:
934:
910:
904:
903:
883:
877:
876:
844:
835:
834:
806:
800:
799:
789:
765:
755:
736:Bartter syndrome
718:), mutations in
706:), mutations in
694:), mutations in
680:chloride channel
562:
556:
550:
544:
538:
516:into a globular
512:pattern that is
449:
443:
437:
431:
425:
419:
413:
407:
401:
395:
389:
383:
377:
371:
365:
359:
353:
347:
341:
335:
329:
323:
317:
311:
305:
299:
293:
287:
281:
275:
269:
263:
257:
251:
245:
239:
233:
227:
221:
215:
209:
203:
197:
130:
28:
16:
2098:
2097:
2093:
2092:
2091:
2089:
2088:
2087:
2083:Protein domains
2073:
2072:
2071:
2042:Hum. Mol. Genet
2035:
2034:
2030:
1994:
1993:
1989:
1960:Hum. Mol. Genet
1953:
1952:
1948:
1917:
1916:
1912:
1868:
1867:
1863:
1824:
1823:
1816:
1785:
1784:
1780:
1743:J. Clin. Invest
1736:
1735:
1731:
1714:(in Japanese).
1709:
1708:
1704:
1671:(40): 15393–8.
1658:
1657:
1648:
1617:
1616:
1607:
1576:
1575:
1571:
1535:
1534:
1530:
1507:(10): 1285–95.
1494:
1493:
1489:
1445:
1444:
1433:
1404:(7161): 492–5.
1395:
1394:
1387:
1349:
1348:
1344:
1308:
1307:
1303:
1279:10.1.1.488.2542
1263:
1262:
1258:
1221:J. Clin. Invest
1214:
1213:
1209:
1172:J. Clin. Invest
1165:
1164:
1157:
1121:
1120:
1116:
1086:
1085:
1081:
1043:
1042:
1038:
1002:
1001:
997:
953:
952:
948:
912:
911:
907:
885:
884:
880:
859:(6): C1369–78.
846:
845:
838:
808:
807:
803:
767:
757:
756:
752:
748:
641:
621:
584:
558:
552:
546:
540:
534:
506:
445:
439:
433:
427:
421:
415:
409:
403:
397:
391:
385:
379:
373:
367:
361:
355:
349:
343:
337:
331:
325:
319:
313:
307:
301:
295:
289:
283:
277:
271:
265:
259:
253:
247:
241:
235:
229:
223:
217:
211:
205:
199:
193:
31:
12:
11:
5:
2096:
2094:
2086:
2085:
2075:
2074:
2070:
2069:
2048:(25): 2861–7.
2028:
1987:
1946:
1927:(3): 400–413.
1910:
1861:
1814:
1778:
1729:
1702:
1646:
1605:
1569:
1548:(10): 1161–3.
1528:
1487:
1431:
1385:
1342:
1301:
1256:
1207:
1155:
1114:
1079:
1036:
995:
946:
925:(2): 299–307.
905:
878:
836:
801:
749:
747:
744:
712:Dent's disease
649:homocystinuria
640:
637:
620:
617:
583:
582:Ligand binding
580:
531:Bateman domain
505:
502:
466:protein domain
456:
455:
452:
451:
191:
185:
184:
179:
173:
172:
159:
153:
152:
142:
135:
134:
126:
125:
120:
114:
113:
100:
94:
93:
88:
82:
81:
76:
70:
69:
64:
58:
57:
52:
46:
45:
42:
38:
37:
33:
32:
29:
21:
20:
13:
10:
9:
6:
4:
3:
2:
2095:
2084:
2081:
2080:
2078:
2065:
2061:
2056:
2051:
2047:
2043:
2039:
2032:
2029:
2024:
2020:
2015:
2010:
2007:(4): 423–34.
2006:
2002:
1998:
1991:
1988:
1983:
1979:
1974:
1969:
1966:(6): 635–43.
1965:
1961:
1957:
1950:
1947:
1942:
1938:
1934:
1930:
1926:
1922:
1914:
1911:
1906:
1902:
1897:
1892:
1888:
1884:
1880:
1876:
1872:
1865:
1862:
1857:
1853:
1849:
1845:
1841:
1837:
1833:
1829:
1821:
1819:
1815:
1810:
1806:
1802:
1798:
1795:(3): 400–13.
1794:
1790:
1782:
1779:
1774:
1770:
1765:
1760:
1756:
1752:
1749:(2): 274–84.
1748:
1744:
1740:
1733:
1730:
1725:
1721:
1717:
1713:
1706:
1703:
1698:
1694:
1689:
1684:
1679:
1674:
1670:
1666:
1662:
1655:
1653:
1651:
1647:
1642:
1638:
1634:
1630:
1627:(3): 800–20.
1626:
1622:
1614:
1612:
1610:
1606:
1601:
1597:
1593:
1589:
1585:
1581:
1573:
1570:
1565:
1561:
1556:
1551:
1547:
1543:
1539:
1532:
1529:
1524:
1520:
1515:
1510:
1506:
1502:
1498:
1491:
1488:
1483:
1479:
1475:
1471:
1466:
1461:
1457:
1453:
1449:
1442:
1440:
1438:
1436:
1432:
1427:
1423:
1419:
1415:
1411:
1407:
1403:
1399:
1392:
1390:
1386:
1381:
1377:
1373:
1369:
1365:
1361:
1357:
1353:
1346:
1343:
1338:
1334:
1329:
1324:
1320:
1316:
1312:
1305:
1302:
1297:
1293:
1289:
1285:
1280:
1275:
1271:
1267:
1260:
1257:
1252:
1248:
1243:
1238:
1234:
1230:
1226:
1222:
1218:
1211:
1208:
1203:
1199:
1194:
1189:
1185:
1181:
1177:
1173:
1169:
1162:
1160:
1156:
1151:
1147:
1142:
1137:
1133:
1129:
1125:
1118:
1115:
1110:
1106:
1102:
1098:
1094:
1090:
1083:
1080:
1075:
1071:
1067:
1063:
1059:
1055:
1051:
1047:
1040:
1037:
1032:
1028:
1023:
1018:
1015:(6): 715–25.
1014:
1010:
1006:
999:
996:
991:
987:
982:
977:
973:
969:
965:
961:
957:
950:
947:
942:
938:
933:
928:
924:
920:
916:
909:
906:
901:
897:
893:
889:
882:
879:
874:
870:
866:
862:
858:
854:
850:
843:
841:
837:
832:
828:
824:
820:
816:
812:
805:
802:
797:
793:
788:
783:
779:
775:
771:
764:
760:
754:
751:
745:
743:
741:
737:
733:
729:
725:
724:osteopetrosis
721:
717:
713:
709:
705:
701:
697:
693:
689:
685:
681:
676:
674:
670:
666:
662:
658:
654:
650:
646:
638:
632:
628:
626:
618:
616:
614:
613:
608:
607:M. jannaschii
604:
603:
597:
593:
589:
581:
576:
572:
568:
566:
565:M. jannaschii
561:
555:
549:
543:
537:
532:
527:
523:
519:
515:
511:
503:
501:
499:
495:
491:
487:
483:
479:
475:
471:
467:
463:
448:
442:
436:
430:
424:
418:
412:
406:
400:
394:
388:
382:
376:
370:
364:
358:
352:
346:
340:
334:
328:
322:
316:
310:
304:
298:
292:
286:
280:
274:
268:
262:
256:
250:
244:
238:
232:
226:
220:
214:
208:
202:
196:
192:
190:
186:
183:
180:
178:
174:
171:
167:
163:
160:
158:
154:
150:
146:
143:
140:
136:
131:
127:
124:
121:
119:
115:
112:
108:
104:
101:
99:
95:
92:
89:
87:
83:
80:
77:
75:
71:
68:
65:
63:
59:
56:
53:
51:
47:
43:
39:
34:
27:
22:
17:
2045:
2041:
2031:
2004:
2000:
1990:
1963:
1959:
1949:
1924:
1920:
1913:
1878:
1874:
1864:
1831:
1827:
1792:
1789:J. Mol. Biol
1788:
1781:
1746:
1742:
1732:
1718:(3): 242–8.
1715:
1711:
1705:
1668:
1664:
1624:
1621:J. Mol. Biol
1620:
1586:(1): 53–70.
1583:
1579:
1572:
1545:
1541:
1531:
1504:
1500:
1490:
1455:
1451:
1401:
1397:
1355:
1351:
1345:
1321:(1): 65–74.
1318:
1314:
1304:
1269:
1266:Biochemistry
1265:
1259:
1227:(2): 182–4.
1224:
1220:
1210:
1178:(2): 182–4.
1175:
1171:
1134:(1): 65–74.
1131:
1127:
1117:
1092:
1088:
1082:
1049:
1045:
1039:
1012:
1008:
998:
963:
959:
949:
922:
918:
908:
894:(3): 160–3.
891:
887:
881:
856:
852:
814:
810:
804:
780:(1): 65–74.
777:
773:
753:
734:can lead to
722:can lead to
710:can lead to
698:can lead to
677:
642:
622:
610:
606:
600:
585:
564:
530:
507:
461:
459:
1358:(1): 60–7.
1052:(1): 60–7.
888:J. Mol. Med
817:(1): 12–3.
36:Identifiers
2001:Hum. Mutat
1921:J Mol Biol
1580:J Mol Biol
960:J. Physiol
746:References
462:CBS domain
145:structures
19:CBS domain
1542:Structure
1501:Structure
1315:Structure
1274:CiteSeerX
1128:Structure
1009:Structure
919:Structure
774:Structure
766:;
625:enzymatic
557:,
545:,
539:,
526:α-helices
524:with two
504:Structure
444:,
438:,
432:,
426:,
420:,
414:,
408:,
402:,
396:,
390:,
384:,
378:,
372:,
366:,
360:,
354:,
348:,
342:,
336:,
330:,
324:,
318:,
312:,
306:,
300:,
294:,
288:,
282:,
276:,
270:,
264:,
258:,
252:,
246:,
240:,
234:,
228:,
222:,
216:,
210:,
204:,
198:,
67:IPR000644
2077:Category
2064:11741829
2023:11933197
1982:11230183
1941:20303981
1905:18931440
1848:17851531
1809:20303981
1773:14722619
1724:18326297
1697:18832160
1641:20026078
1600:20382158
1564:17937905
1523:17937917
1482:38983201
1474:17289942
1418:17851534
1380:20733119
1372:17195847
1337:17223533
1296:10200156
1251:14722609
1202:14722609
1150:17223533
1109:17452784
1074:20733119
1066:17195847
1031:17562318
990:16469788
941:16472749
873:16275737
796:17223533
688:myotonia
686:lead to
162:RCSB PDB
62:InterPro
1896:2564890
1856:4345919
1688:2563093
1619:pair".
1452:Science
1426:4342092
981:1779660
900:9106071
831:9020585
738:(OMIM:
726:(OMIM:
714:(OMIM:
702:(OMIM:
690:(OMIM:
671:(OMIM:
663:(OMIM:
651:(OMIM:
522:β-sheet
450:
123:cd02205
91:PS51371
86:PROSITE
55:PF00571
2062:
2021:
1980:
1939:
1903:
1893:
1854:
1846:
1828:Nature
1807:
1771:
1764:311435
1761:
1722:
1695:
1685:
1639:
1598:
1562:
1521:
1480:
1472:
1424:
1416:
1398:Nature
1378:
1370:
1335:
1294:
1276:
1249:
1242:311445
1239:
1200:
1193:311445
1190:
1148:
1107:
1072:
1064:
1029:
988:
978:
939:
898:
871:
829:
794:
740:241200
732:CLCNKB
728:259700
716:300009
704:600699
692:160800
673:180105
665:600858
653:236200
514:folded
177:PDBsum
151:
141:
111:SUPFAM
41:Symbol
1852:S2CID
1478:S2CID
1422:S2CID
1376:S2CID
1070:S2CID
720:CLCN7
708:CLCN5
696:CLCN2
684:CLCN1
659:with
496:, or
464:is a
107:SCOPe
98:SCOP2
74:SMART
2060:PMID
2019:PMID
1978:PMID
1937:PMID
1901:PMID
1844:PMID
1805:PMID
1769:PMID
1720:PMID
1693:PMID
1637:PMID
1596:PMID
1560:PMID
1519:PMID
1470:PMID
1414:PMID
1368:PMID
1333:PMID
1292:PMID
1247:PMID
1198:PMID
1146:PMID
1105:PMID
1062:PMID
1027:PMID
986:PMID
937:PMID
896:PMID
869:PMID
827:PMID
792:PMID
763:2nye
590:and
560:1PBJ
554:1O50
548:2OOX
542:1PVM
536:3KPC
492:and
484:and
447:3ddj
441:2yzq
435:2yzi
429:2yvx
423:2v9j
417:2v92
411:2v8q
405:2rih
399:2rif
393:2rc3
387:2qh1
381:2oux
375:2oox
369:2o16
363:2nye
357:2nyc
351:2j9l
345:2ef7
339:2d4z
333:2cu0
327:1zfj
321:1yav
315:1y5h
309:1xkf
303:1vrd
297:1vr9
291:1pvn
285:1pvm
279:1pbj
273:1o50
267:1nfb
261:1nf7
255:1mew
249:1mei
243:1meh
237:1me9
231:1me8
225:1me7
219:1lrt
213:1jr1
207:1jcn
201:1b3o
195:1ak5
170:PDBj
166:PDBe
149:ECOD
139:Pfam
103:1zfj
50:Pfam
2050:doi
2009:doi
1968:doi
1929:doi
1925:398
1891:PMC
1883:doi
1836:doi
1832:449
1797:doi
1793:398
1759:PMC
1751:doi
1747:113
1683:PMC
1673:doi
1669:105
1629:doi
1625:396
1588:doi
1584:399
1550:doi
1509:doi
1460:doi
1456:315
1406:doi
1402:449
1360:doi
1323:doi
1284:doi
1237:PMC
1229:doi
1225:113
1188:PMC
1180:doi
1176:113
1136:doi
1097:doi
1054:doi
1017:doi
976:PMC
968:doi
964:572
927:doi
861:doi
857:289
819:doi
782:doi
759:PDB
742:).
675:).
592:ATP
588:AMP
494:ATP
490:AMP
189:PDB
157:PDB
118:CDD
79:CBS
44:CBS
2079::
2058:.
2046:10
2044:.
2040:.
2017:.
2005:19
2003:.
1999:.
1976:.
1964:10
1962:.
1958:.
1935:.
1923:.
1899:.
1889:.
1879:64
1877:.
1873:.
1850:.
1842:.
1830:.
1817:^
1803:.
1791:.
1767:.
1757:.
1745:.
1741:.
1716:53
1691:.
1681:.
1667:.
1663:.
1649:^
1635:.
1623:.
1608:^
1594:.
1582:.
1558:.
1546:15
1544:.
1540:.
1517:.
1505:15
1503:.
1499:.
1476:.
1468:.
1454:.
1450:.
1434:^
1420:.
1412:.
1400:.
1388:^
1374:.
1366:.
1356:14
1354:.
1331:.
1319:15
1317:.
1313:.
1290:.
1282:.
1270:38
1268:.
1245:.
1235:.
1223:.
1219:.
1196:.
1186:.
1174:.
1170:.
1158:^
1144:.
1132:15
1130:.
1126:.
1103:.
1093:63
1091:.
1068:.
1060:.
1050:14
1048:.
1025:.
1013:15
1011:.
1007:.
984:.
974:.
962:.
958:.
935:.
923:14
921:.
917:.
892:75
890:.
867:.
855:.
851:.
839:^
825:.
815:22
813:.
790:.
778:15
776:.
772:.
761::
500:.
480:,
168:;
164:;
147:/
109:/
105:/
2066:.
2052::
2025:.
2011::
1984:.
1970::
1943:.
1931::
1907:.
1885::
1858:.
1838::
1811:.
1799::
1775:.
1753::
1726:.
1699:.
1675::
1643:.
1631::
1602:.
1590::
1566:.
1552::
1525:.
1511::
1484:.
1462::
1428:.
1408::
1382:.
1362::
1339:.
1325::
1298:.
1286::
1253:.
1231::
1204:.
1182::
1152:.
1138::
1111:.
1099::
1076:.
1056::
1033:.
1019::
992:.
970::
943:.
929::
902:.
875:.
863::
833:.
821::
798:.
784::
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.