104:, identifying RNA binding sites that contained the expected Nova-binding motifs. Sequencing of the cDNA library identified many positions close to alternative exons, several of which were found to require Nova1/2 for their brains-specific splicing patterns. In 2008, CLIP was combined with high-throughput sequencing (termed "HITS-CLIP") to generate genome-wide protein-RNA interaction maps for Nova; since then a number of other RNA-binding proteins have been studied with CLIP, including PTBP1, RbFox2 (where it was referred to as "CLIP-seq"), SFRS1, Argonaute, hnRNP C, the Fragile-X mental retardation protein FMRP, Ptbp2 (in the mouse brain), Mbnl2, the nElavl proteins (the neuron-specific Hu proteins), and has been applied to RNA binding proteins from all kingdoms of life, including prokaryotes. CLIP analysis of the RNA-binding protein
193:(photoactivatable ribonucleoside–enhanced cross-linking and immunoprecipitation) is also used for identifying the binding sites of cellular RNA-binding proteins (RBPs) and microRNA-containing ribonucleoprotein complexes (miRNPs). The method relies on the incorporation of photoreactive ribonucleoside analogs, such as 4-thiouridine (4-SU) and 6-thioguanosine (6-SG) into nascent RNA transcripts by living cells. Irradiation of the cells by UV light of 365 nm induces efficient cross-linking of photoreactive nucleoside-labeled cellular RNAs to interacting RBPs. Immunoprecipitation of the RBP of interest is followed by the isolation of the cross-linked and co-immunoprecipitated RNA. The isolated RNA is converted into a cDNA library and deep sequenced using
254:
the targeted RBP. After ligation of a 3’ RNA adapter, immunoprecipitated material (as well as a paired input sample) are run on denaturing protein gels and transferred to nitrocellulose membranes. A region from the protein size to 75 kDa above is cut from the membrane and treated with
Proteinase K to release RNA. After cleanup, RNA is then reverse transcribed to ssDNA, when a second adapter is ligated. By ligating the second adapter to cDNAs, eCLIP can identify truncated cDNAs, similar to iCLIP, and thereby study RNA-protein interaction sites with high resolution. PCR amplification is then used to obtain sufficient material for high-throughput sequencing. eCLIP can also be used to identify miRNA targets and profile RNA modifications such as m6A.
265:
the amount of input material and omitting several purification steps. Additionally, it permits a radiolabel-free visualization of immunoprecipitated RNA by using a highly sensitive biotin-based labeling technique. Along with a bioinformatical platform this method is designed to provide deep insights into RNA–protein interactomes in biomedical science, where the amount of starting material is often limited (i.e. in case of precious clinical samples). Additional iCLIP variants have also been developed that retain the individual nucleotide resolution but differ in one or more steps from the original iCLIP method. These include iCLIP2, irCLIP, iiCLIP, and iCLIP1.5, a few to name.
251:
publication, eCLIP was reported to increase such efficiency by >1000-fold, which not only decreases wasted sequencing of PCR duplicate molecules, but also dramatically decreases experimental failures during the CLIP procedure. Additionally, the amplification in eCLIP is now comparable to RNA-seq, enabling rigorous quantitative normalization against paired input controls (to remove background at ribosomal and other highly abundant RNAs) as well as quantitative comparison across peaks and samples, enabling the ability to detect allele-specific binding or differential RNA binding between conditions.
238:) along with experimental barcodes to the primer used for reverse transcription, thereby barcoding unique cDNAs to minimise any errors or quantitative biases of PCR, and thus improving the quantification of binding events. Enabling amplification of truncated cDNAs led to identification of the sites of RNA-protein interactions at high resolution by analysing the starting position of truncated cDNAs, as well as their precise quantification using UMIs with software called "
242:". These innovations of iCLIP were adopted by later variants of CLIP such as eCLIP and irCLIP. Another modification of iCLIP, miCLIP, identifies methylated RNA sites with use of mutant enzyme or modification-specific antibody. The quantitative nature of iCLIP enabled comparison across samples at the level of full RNAs, or to study competitive binding of multiple RNA-binding proteins or subtle changes in binding of a mutant protein at the level of binding peaks.
59:
300:
223:(individual nucleotide–resolution crosslinking and immunoprecipitation) is a variant of CLIP that enabled amplification of truncated cDNAs, which are produced when reverse transcription stops prematurely at the cross-link site. Other approaches to identify protein-RNA crosslink sites include mutational analysis of read-through cDNAs, such as nucleotide transitions in
143:
181:
287:
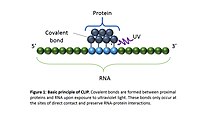
to preserve RNA-protein interactions, but these also generate protein-protein cross-links. By employing UV crosslinking that is specific to direct protein-RNA contacts, CLIP avoid protein-protein cross-links and ensures high specificity, while also obtaining positional information on the sites of protein-RNA interactions.
211:
71:
isolated via immunoprecipitation. In order to allow for priming of reverse transcription, RNA adapters are ligated to the 3' ends, and RNA fragments are labelled to enable the analysis of the RNA-protein complexes after they have been separated from free RNA using gel electrophoresis and membrane transfer.
1724:
Ke, Shengdong; Alemu, Endalkachew A.; Mertens, Claudia; Gantman, Emily Conn; Fak, John J.; Mele, Aldo; Haripal, Bhagwattie; Zucker-Scharff, Ilana; Moore, Michael J.; Park, Christopher Y.; Vågbø, Cathrine
Broberg; Kusśnierczyk, Anna; Klungland, Arne; Darnell, James E.; Darnell, Robert B. (2015-10-01).
286:
have been demonstrated to be dependent on the reaction conditions of the experiment, such as protein concentrations and ionic conditions, and reassociation of RNA-binding proteins following cell lysis could lead to detection of artificial interactions. Formaldehyde crosslinking methods have been used
281:
RNA-binding proteins are frequently components of multi-protein complexes, and RNAs from various genes are present in cells at a range of abundance, therefore it is common that RNAs bound to co-purified proteins or non-specifically sticking to beads may be isolated when immunoprecipitating a specific
253:
As in other CLIP methods, eCLIP relies on RBP-RNA interactions covalently linked using UV crosslinking of live cells. Cells are then lysed, and RNA is fragmented using limited RNase treatment. A specific RBP (and its bound RNA) is then immunoprecipitated using an antibody that specifically recognizes
201:
However, PAR-CLIP is limited mainly to cultured cells, and nucleoside cytotoxicity is a concern; it has been reported that 4-SU inhibits ribosomal RNA synthesis, induces a nucleolar stress response, and reduces cell proliferation. PAR-CLIP has been employed to determine the transcriptome-wide binding
2157:
Charizanis, K; Lee, KY; Batra, R; Goodwin, M; Zhang, C; Yuan, Y; Shiue, L; Cline, M; Scotti, MM; Xia, G; Kumar, A; Ashizawa, T; Clark, HB; Kimura, T; Takahashi, MP; Fujimura, H; Jinnai, K; Yoshikawa, H; Gomes–Pereira, M; Gourdon, G; Sakai, N; Nishino, S; Foster, TC; Ares, M; Darnell, RB; Swanson, MS
320:
are comparable to CLIP in identifying miRNA targets, raising questions as to its utility relative to existing predictions. Because CLIP methods rely on immunoprecipitation, crosslinked RNA could in some cases affect antibody-epitope interactions. Finally, significant differences have been observed.
70:
cross-linking of RNA-protein complexes using ultraviolet light (UV). Upon UV exposure, covalent bonds are formed between proteins and nucleic acids that are in close proximity (on the order of
Angstroms apart). The cross-linked cells are then lysed, RNA is fragmented, and the protein of interest is
290:
Since UV crosslinking creates a covalent bond, the crosslinked RNA fragments retain a short peptide after
Proteinase K digestion, which can be exploited to identify the crosslink site. Reverse transcription most often truncates at the crosslink sites, creating truncated cDNAs that are exploited by
264:
sCLIP (simple CLIP) is a technique that requires lower amounts of input RNA and omits radio-labeling of the immunoprecipitated RNA. The method is based on linear amplification of the immunoprecipitated RNA and thereby improves the complexity of the sequencing-library despite significantly reducing
307:
All CLIP library generation protocols require moderate quantities of cells or tissue (50–100 mg), require numerous enzymatic steps, and customised computational analyses. Certain steps are difficult to optimize and frequently have low efficiencies. For example, overdigestion with RNase can
308:
decrease the number of identified binding sites and thus needs to be optimised. Crosslinking efficiency also varies between proteins, and nucleotide bias of crosslinking has been reported, for example by comparing cross-linking sites and motifs enriched when protein-RNA complexes are studied
250:
eCLIP (enhanced CrossLinking and ImmunoPrecipitation followed by high-throughput sequencing) is also used to map RBP binding sites on RNAs transcriptome-wide. eCLIP was designed to improve upon iCLIP by increasing the efficiency in converting purified RNA fragments into cDNA library. At its
49:
on a transcriptome-wide scale, thereby increasing our understanding of post-transcriptional regulatory networks. CLIP can be used either with antibodies against endogenous proteins, or with common peptide tags (including FLAG, V5, HA, and others) or affinity purification, which enables the
167:
to identify binding sites of RNA-binding proteins. HITS-CLIP also introduced the addition of dinucleotide barcodes to primers, providing the ability to sequence and then deconvolute multiple experiments simultaneously. With analysis of cross-linking induced mutation sites (CIMS) at
1166:
Hafner, Markus; Landthaler, Markus; Burger, Lukas; Khorshid, Mohsen; Hausser, Jean; Berninger, Philipp; Rothballer, Andrea; Ascano, Manuel; Jungkamp, Anna-Carina; Munschauer, Mathias; Ulrich, Alexander; Wardle, Greg S.; Dewell, Scott; Zavolan, Mihaela; Tuschl, Thomas (2010-04-02).
1646:
Van
Nostrand, Eric L.; Freese, Peter; Pratt, Gabriel A.; Wang, Xiaofeng; Wei, Xintao; Xiao, Rui; Blue, Steven M.; Chen, Jia-Yu; Cody, Neal A. L.; Dominguez, Daniel; Olson, Sara; Sundararaman, Balaji; Zhan, Lijun; Bazile, Cassandra; Bouvrette, Louis Philip Benoit (July 2020).
1461:
Tollervey, James R.; Curk, Tomaž; Rogelj, Boris; Briese, Michael; Cereda, Matteo; Kayikci, Melis; König, Julian; Hortobágyi, Tibor; Nishimura, Agnes L.; Zupunski, Vera; Patani, Rickie; Chandran, Siddharthan; Rot, Gregor; Zupan, Blaž; Shaw, Christopher E. (April 2011).
1576:
Hallegger, Martina; Chakrabarti, Anob M.; Lee, Flora C. Y.; Lee, Bo Lim; Amalietti, Aram G.; Odeh, Hana M.; Copley, Katie E.; Rubien, Jack D.; Portz, Bede; Kuret, Klara; Huppertz, Ina; Rau, Frédérique; Patani, Rickie; Fawzi, Nicolas L.; Shorter, James (2021-09-02).
75:
digestion is then performed in order to remove protein from the crosslinked RNA, which leaves a few amino acids at the crosslink site. This often leads to truncation of cDNAs at the crosslinked nucleotide, which is exploited in variants such as
80:
to increase the resolution of the method. cDNA is then synthesized via RT-PCR followed by high-throughput sequencing followed by mapping the reads back to the transcriptome and other computational analyses to study the interaction sites.
197:
technology. Cross-linking the 4-SU and 6-SG analogs results in thymidine to cytidine, and guanosine to adenosine transitions respectively. As a result, PAR-CLIP can identify binding site locations with high accuracy.
1343:
Hussain, Shobbir; Sajini, Abdulrahim A.; Blanco, Sandra; Dietmann, Sabine; Lombard, Patrick; Sugimoto, Yoichiro; Paramor, Maike; Gleeson, Joseph G.; Odom, Duncan T.; Ule, Jernej; Frye, Michaela (2013-07-25).
2453:
Hafner, M; Landthaler, M; Burger, L; Khorshid, M; Hausser, J; Berninger, P; Rothballer, A; Ascano, M; Jungkamp, AC; Munschauer, M; Ulrich, A; Wardle, GS; Dewell, S; Zavolan, M; Tuschl, T (2010b).
2410:
Hafner, M; Landthaler, M; Burger, L; Khorshid, M; Hausser, J; Berninger, P; Rothballer, A; Ascano, M; Jungkamp, AC; Munschauer, M; Ulrich, A; Wardle, GS; Dewell, S; Zavolan, M; Tuschl, T (2010).
1519:
Zarnack, Kathi; König, Julian; Tajnik, Mojca; Martincorena, Iñigo; Eustermann, Sebastian; Stévant, Isabelle; Reyes, Alejandro; Anders, Simon; Luscombe, Nicholas M.; Ule, Jernej (2013-01-31).
342:
2578:
Ke, S; Alemu, EA; Mertens, C; Gantman, EC; Fak, JJ; Mele, A; Haripal, B; Zucker-Scharff, I; Moore, MJ; Park, CY; Vågbø, CB; Kusnierczyk, A; Klungland, A; Darnell, JE; Darnell, RB (2015).
2373:
Fecko, CJ; Munson, KM; Saunders, A; Sun, G; Begley, TP; Lis, JT; Webb, WW (2007). "Comparison of femtosecond laser and continuous wave UV sources for protein-nucleic acid crosslinking".
333:
studies enrichment of full RNAs after immunoprecipitation of specific protein followed by microarray analysis, but without using cross-linking, it does not identify RNA binding sites
1871:
Knörlein, Anna; Sarnowski, Chris P.; de Vries, Tebbe; Stoltz, Moritz; Götze, Michael; Aebersold, Ruedi; Allain, Frédéric H.-T.; Leitner, Alexander; Hall, Jonathan (2022-05-17).
268:
As a modification of CLIP, methylated RNA sites were identified with the use of mutant enzyme or modification-specific antibody with the methods termed miCLIP or m6A-CLIP.
946:
Hale, Caryn R.; Sawicka, Kirsty; Mora, Kevin; Fak, John J.; Kang, Jin Joo; Cutrim, Paula; Cialowicz, Katarzyna; Carroll, Thomas S.; Darnell, Robert B. (2021-12-23).
2535:
Ince-Dunn, G; Okano, HJ; Jensen, KB; Park, WY; Zhong, R; Ule, J; Mele, A; Fak, JJ; Yang, CW; Zhang, C; Yoo, J; Herre, M; Okano, H; Noebels, JL; Darnell, RB (2012).
3447:"sCLIP—an integrated platform to study RNA–protein interactomes in biomedical research: identification of CSTF2tau in alternative processing of small nuclear RNAs"
2252:
Darnell, JC; Van
Driesche, SJ; Zhang, C; Hung, KY; Mele, A; Fraser, CE; Stone, EF; Chen, C; Fak, JJ; Chi, SW; Licatalosi, DD; Richter, JD; Darnell, RB (2011).
202:
sites of several known RBPs and microRNA-containing ribonucleoprotein complexes at high resolution. This includes the miRNA targeting AGO and TNRC6 proteins.
2707:
König, J; McGlincy, NJ; Ule, J (2012). "Analysis of
Protein-RNA Interactions with Single-Nucleotide Resolution Using iCLIP and Next-Generation Sequencing".
739:
König, Julian; Zarnack, Kathi; Rot, Gregor; Curk, Tomaz; Kayikci, Melis; Zupan, Blaz; Turner, Daniel J.; Luscombe, Nicholas M.; Ule, Jernej (July 2010).
321:
Therefore, raw CLIP results require further computational analyses to thoroughly investigate RNA-protein binding site interactions within the cell.
561:
Hafner, Markus; Katsantoni, Maria; Köster, Tino; Marks, James; Mukherjee, Joyita; Staiger, Dorothee; Ule, Jernej; Zavolan, Mihaela (2021-03-04).
2951:"Mapping Argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis"
3228:"Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping"
2812:
Licatalosi, DD; Mele, A; Fak, JJ; Ule, J; Kayikci, M; Chi, SW; Clark, TA; Schweitzer, AC; Blume, JE; Wang, X; Darnell, JC; Darnell, RB (2008).
466:: dCLIP is a Perl program for discovering differential binding regions in two comparative CLIP-Seq (HITS-CLIP, PAR-CLIP or iCLIP) experiments.
316:, though methods are being developed to minimise such bias for enriched motif discovery. Computationally predicted miRNA targets derived from
2908:"Evidence for reassociation of RNA-binding proteins after cell lysis: Implications for the interpretation of immunoprecipitation analyses"
3152:
Ule, J; Jensen, K; Ruggiu, M; Mele, A; Ule, A; Darnell, RB (Nov 14, 2003). "CLIP identified Nova-regulated RNA networks in the brain".
2724:
1404:
Linder, Bastian; Grozhik, Anya V.; Olarerin-George, Anthony O.; Meydan, Cem; Mason, Christopher E.; Jaffrey, Samie R. (August 2015).
3197:
Ule, J; Jensen, K; Mele, A; Darnell, RB (2005). "CLIP: a method for identifying protein-RNA interactions sites in living cells".
2623:"m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover"
363:: a database for exploring miRNA-lncRNA, miRNA-mRNA, miRNA-sncRNA, miRNA-circRNA, protein-lncRNA, protein-RNA interactions and
2537:"Neuronal Elav-like (Hu) proteins regulate RNA splicing and abundance to control glutamate levels and neuronal excitability"
3543:
283:
2621:
Ke, S; Pandya-Jones, A; Saito, Y; Fak, JJ; Vågbø, CB; Geula, S; Hanna, JH; Black, DL; Darnell, JE; Darnell, RB (2017).
235:
2494:"Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo"
2779:"Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs"
2114:
Burger, K; Mühl, B; Kellner, M; Rohrmoser, M; Gruber-Eber, A; Windhager, L; Friedel, CC; Dölken, L; Eick, D (2013).
1346:"NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs"
194:
164:
3271:"starBase: a database for exploring microRNA–mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data"
2865:"Ptbp2 represses adult-specific splicing to regulate the generation of neuronal precursors in the embryonic brain"
2160:"Muscleblind-like 2-mediated alternative splicing in the developing brain and dysregulation in myotonic dystrophy"
1080:
Wolf, Joshua J.; Dowell, Robin D.; Mahony, Shaun; Rabani, Michal; Gifford, David K.; Fink, Gerald R. (June 2010).
3226:
Xue, Y; Zhou, Y; Wu, T; Zhu, T; Ju, X; Kwon, YS; Zhang, C; Yeo, G; Black, DL; Sun, H; Fu, XD; Zhang, Y (2009).
1521:"Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements"
113:
3123:
Uhl, M; Houwaart, T; Corrado, G; Wright, PR; Backofen, R (2017). "Computational analysis of CLIP-seq data".
291:
iCLIP, while read-through cDNAs often contain mutations at the crosslink site (see HITS-CLIP and PAR-CLIP).
1082:"Feed-forward regulation of a cell fate determinant by an RNA-binding protein generates asymmetry in yeast"
227:, or rare errors introduced by reverse transcriptase when it reads through the crosslink sites in standard
1950:"Positional motif analysis reveals the extent of specificity of protein-RNA interactions observed by CLIP"
948:"FMRP regulates mRNAs encoding distinct functions in the cell body and dendrites of CA1 pyramidal neurons"
360:
121:
3314:"An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells"
1948:
Kuret, Klara; Amalietti, Aram Gustav; Jones, D. Marc; Capitanchik, Charlotte; Ule, Jernej (2022-09-09).
2492:
Holmqvist, E; Wright, PR; Li, L; Bischler, T; Barquist, L; Reinhardt, R; Backofen, R; Vogel, J (2016).
487:
Ule, Jernej; Jensen, Kirk B.; Ruggiu, Matteo; Mele, Aldo; Ule, Aljaz; Darnell, Robert B. (2003-11-14).
2664:
König, J; Zarnack, K; Rot, G; Curk, T; Kayikci, M; Zupan, B; Turner, DJ; Luscombe, NM; Ule, J (2010).
1794:
Chakrabarti, Anob M.; Haberman, Nejc; Praznik, Arne; Luscombe, Nicholas M.; Ule, Jernej (2018-07-20).
256:
eCLIP datasets have been produced for over 150 RBPs with validated commercially available antibodies.
3492:"Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP)"
3161:
2825:
2214:
1884:
1873:"Nucleotide-amino acid π-stacking interactions initiate photo cross-linking in RNA-protein complexes"
1660:
500:
126:
160:
90:
38:
3037:"Analysis of CLIP and iCLIP methods for nucleotide-resolution studies of protein-RNA interactions"
2073:"Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis"
3185:
2398:
2340:"CLIP (Cross-Linking and Immunoprecipitation) Identification of RNAs Bound by a Specific Protein"
1841:
592:
532:
2992:
Sanford, JR; Wang, X; Mort, M; Fanduyn, N; Cooper, DN; Mooney, SD; Edenberg, HJ; Liu, Y (2009).
2580:"A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation"
2412:"Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP"
1727:"A majority of m6A residues are in the last exons, allowing the potential for 3' UTR regulation"
1169:"Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP"
2666:"iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution"
741:"iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution"
3521:
3476:
3433:
3386:
3357:"Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data"
3343:
3300:
3257:
3214:
3177:
3140:
3111:
3068:
3023:
2980:
2937:
2894:
2851:
2765:
2720:
2695:
2652:
2609:
2566:
2523:
2480:
2455:"PAR-CliP - A Method to Identify Transcriptome-wide the Binding Sites of RNA Binding Proteins"
2441:
2390:
2361:
2326:
2283:
2240:
2189:
2145:
2102:
2059:
1989:
1971:
1918:
1900:
1833:
1815:
1764:
1746:
1694:
1676:
1616:
1598:
1558:
1540:
1501:
1483:
1443:
1425:
1383:
1365:
1322:
1293:"Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data"
1206:
1188:
1119:
1101:
987:
969:
916:
898:
778:
760:
701:
693:
649:
631:
584:
524:
516:
27:
1811:
112:-mRNA and protein-RNA interaction maps in the mouse brain and subsequently in budding yeast (
3511:
3503:
3466:
3458:
3423:
3415:
3398:
Zisoulis, DG; Lovci, MT; Wilbert, ML; Hutt, KR; Liang, TY; Pasquinelli, AE; Yeo, GW (2010).
3376:
3368:
3333:
3325:
3290:
3282:
3247:
3239:
3206:
3169:
3132:
3101:
3093:
3058:
3048:
3013:
3005:
2970:
2962:
2927:
2919:
2884:
2876:
2841:
2833:
2798:
2790:
2755:
2747:
2712:
2685:
2677:
2642:
2634:
2599:
2591:
2556:
2548:
2513:
2505:
2470:
2462:
2431:
2423:
2382:
2351:
2316:
2308:
2273:
2265:
2230:
2222:
2179:
2171:
2135:
2127:
2092:
2084:
2049:
2039:
1979:
1961:
1908:
1892:
1823:
1807:
1754:
1738:
1684:
1668:
1606:
1590:
1548:
1532:
1491:
1475:
1433:
1417:
1373:
1357:
1312:
1304:
1196:
1180:
1109:
1093:
977:
959:
906:
890:
768:
752:
683:
639:
623:
574:
508:
169:
94:
3165:
2863:
Licatalosi, DD; Yano, M; Fak, JJ; Mele, A; Grabinski, SE; Zhang, C; Darnell, RB (2012).
2829:
2777:
Leung, AK; Young, AG; Bhutkar, A; Zheng, GX; Bosson, AD; Nielsen, CB; Sharp, PA (2011).
2218:
1888:
1664:
504:
58:
50:
possibility of profiling model organisms or RBPs otherwise lacking suitable antibodies.
3516:
3491:
3471:
3446:
3428:
3399:
3381:
3356:
3338:
3313:
3295:
3270:
3252:
3227:
3106:
3081:
3063:
3036:
3018:
2993:
2975:
2950:
2932:
2907:
2889:
2864:
2846:
2813:
2803:
2778:
2760:
2735:
2690:
2665:
2647:
2622:
2604:
2579:
2561:
2536:
2518:
2493:
2475:
2436:
2411:
2321:
2296:
2278:
2253:
2235:
2202:
2184:
2159:
2140:
2115:
2097:
2072:
2054:
2027:
1984:
1949:
1913:
1872:
1828:
1795:
1759:
1726:
1689:
1648:
1611:
1578:
1553:
1520:
1496:
1463:
1438:
1405:
1378:
1345:
1317:
1292:
1201:
1168:
1114:
1081:
982:
947:
911:
878:
773:
740:
644:
611:
282:
protein. The data specificity obtained using early immunoprecipitation methods such as
2994:"Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts"
2071:
Bohnsack, MT; Martin, R; Granneman, S; Ruprecht, M; Schleiff, E; Tollervey, D (2009).
3537:
3035:
Sugimoto, Y; König, J; Hussain, S; Zupan, B; Curk, T; Frye, M; Ule, J (Aug 3, 2012).
2386:
2254:"FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism"
1464:"Characterizing the RNA targets and position-dependent splicing regulation by TDP-43"
596:
3189:
2402:
1845:
536:
172:, crosslink sites can be differentiated from other sources of sequence variation.
3548:
1406:"Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome"
72:
1579:"TDP-43 condensation properties specify its RNA-binding and regulatory repertoire"
3243:
2552:
2175:
2088:
1796:"Data Science Issues in Studying Protein–RNA Interactions with CLIP Technologies"
1361:
688:
671:
89:
CLIP was originally undertaken to study interactions between the neuron-specific
3210:
3136:
2454:
1097:
627:
101:
2716:
2427:
2269:
1966:
1896:
1594:
1536:
1184:
579:
562:
2116:"4-thiouridine inhibits rRNA synthesis and causes a nucleolar stress response"
1672:
317:
299:
156:
34:
3053:
2814:"HITS-CLIP yields genome-wide insights into brain alternative RNA processing"
1975:
1904:
1819:
1750:
1680:
1602:
1544:
1487:
1429:
1369:
1192:
1105:
973:
902:
764:
697:
635:
588:
520:
3173:
2949:
Moore, JJ; Zhang, C; Gantman, EC; Mele, A; Darnell, JC; Darnell, RB (2014).
2509:
512:
418:
377:
228:
151:
105:
3525:
3480:
3437:
3390:
3347:
3304:
3261:
3218:
3181:
3144:
3115:
3072:
3027:
2984:
2966:
2941:
2898:
2880:
2855:
2794:
2769:
2699:
2656:
2638:
2613:
2595:
2570:
2527:
2484:
2445:
2394:
2365:
2356:
2339:
2330:
2287:
2244:
2193:
2149:
2106:
2063:
1993:
1922:
1837:
1768:
1742:
1698:
1620:
1562:
1505:
1447:
1387:
1326:
1210:
1123:
991:
920:
782:
705:
653:
528:
488:
3286:
3009:
3462:
3097:
450:
424:
336:
330:
224:
189:
109:
2923:
2837:
2226:
2044:
1649:"A large-scale binding and functional map of human RNA-binding proteins"
1226:
1224:
1222:
1220:
964:
3507:
1421:
672:"Advances in CLIP Technologies for Studies of Protein-RNA Interactions"
46:
3419:
3329:
2681:
2297:"HITS-CLIP: panoramic views of protein-RNA regulation in living cells"
2131:
1247:
1245:
1243:
1241:
1239:
894:
756:
142:
3372:
2736:"Protein-RNA interactions: new genomic technologies and perspectives"
1308:
877:
Chi, Sung Wook; Hannon, Gregory J.; Darnell, Robert B. (2012-02-12).
437:
2751:
2312:
1479:
860:
858:
180:
3312:
Yeo, GW; Coufal, NG; Liang, TY; Peng, GE; Fu, XD; Gage, FH (2009).
2005:
2003:
460:: a pipeline to analyze short RNA reads from HITS-CLIP experiments.
239:
3400:"Comprehensive discovery of endogenous Argonaute binding sites in
794:
792:
463:
430:
383:
364:
298:
219:
209:
179:
141:
97:
77:
57:
2466:
1711:
845:
843:
234:
iCLIP also added a random sequence (unique molecular identifier,
231:
methods, termed
Crosslink induced mutation site (CIMS) analysis.
210:
2028:"Predicting effective microRNA target sites in mammalian mRNAs"
1935:
401:
610:
Ule, Jernej; Hwang, Hun-Way; Darnell, Robert B. (2018-08-01).
457:
42:
3269:
Yang, JH; Li, JH; Shao, P; Zhou, H; Chen, YQ; Qu, LH (2011).
1063:
1061:
3445:
Kargapolova, Y; Levin, M; Lackner, K; Danckwardt, S (2017).
3082:"Experimental strategies for microRNA target identification"
2203:"Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps"
933:
612:"The Future of Cross-Linking and Immunoprecipitation (CLIP)"
393:
1262:
1260:
31:
1278:
489:"CLIP identifies Nova-regulated RNA networks in the brain"
339:, method for finding interactions with DNA rather than RNA
1633:
1230:
1040:
1016:
1136:
1028:
1251:
1148:
834:
108:
led to identification of microRNA targets by decoding
1052:
1004:
864:
717:
715:
2009:
1858:
879:"An alternative mode of microRNA target recognition"
798:
2734:König, J; Zarnack, K; Luscombe, NM; Ule, J (2012).
2026:Agarwal, V; Bell, GW; Nam, J-W; Bartel, DP (2015).
849:
822:
1291:Zhang, Chaolin; Darnell, Robert B (1 June 2011).
434:data and PICTAR microRNA target site predictions.
2201:Chi, SW; Zang, JB; Mele, A; Darnell, RB (2009).
3490:Van Nostrand, E; Pratt, G; Shishkin, A (2016).
349:method for finding a consensus binding sequence
3080:Thomson, DW; Bracken, CP; Goodall, GJ (2011).
1067:
8:
1266:
810:
670:Lee, Flora C. Y.; Ule, Jernej (2018-02-01).
440:: A computational approach for identifying
1781:
616:Cold Spring Harbor Perspectives in Biology
3515:
3470:
3427:
3380:
3337:
3294:
3251:
3105:
3062:
3052:
3017:
2974:
2931:
2888:
2845:
2802:
2759:
2689:
2646:
2603:
2560:
2517:
2474:
2435:
2355:
2320:
2277:
2234:
2183:
2139:
2096:
2053:
2043:
1983:
1965:
1912:
1827:
1758:
1688:
1610:
1552:
1495:
1437:
1377:
1316:
1200:
1113:
981:
963:
910:
883:Nature Structural & Molecular Biology
772:
745:Nature Structural & Molecular Biology
687:
643:
578:
1812:10.1146/annurev-biodatasci-080917-013525
1800:Annual Review of Biomedical Data Science
1634:Van Nostrand, Pratt & Shishkin 2016
721:
476:
1399:
1397:
1338:
1336:
1161:
1159:
1157:
20:Cross-linking and immunoprecipitation
7:
2709:Tag-Based Next Generation Sequencing
734:
732:
730:
665:
663:
556:
554:
552:
550:
548:
546:
482:
480:
1053:Thomson, Bracken & Goodall 2011
14:
2459:Journal of Visualized Experiments
396:, PicTar, RNA22, miRanda and PITA
2387:10.1111/j.1751-1097.2007.00179.x
563:"CLIP and complementary methods"
16:Method used in molecular biology
2375:Photochemistry and Photobiology
3355:Zhang, C; Darnell, RB (2011).
567:Nature Reviews Methods Primers
1:
3281:(Database issue): D202–D209.
3244:10.1016/j.molcel.2009.12.003
2906:Mili, S; Steitz, JA (2004).
2553:10.1016/j.neuron.2012.07.009
2344:Cold Spring Harbor Protocols
2176:10.1016/j.neuron.2012.05.029
2089:10.1016/j.molcel.2009.09.039
1362:10.1016/j.celrep.2013.06.029
689:10.1016/j.molcel.2018.01.005
442:microRNA-target interactions
3211:10.1016/j.ymeth.2005.07.018
3137:10.1016/j.ymeth.2017.02.006
1098:10.1534/genetics.110.113944
628:10.1101/cshperspect.a032243
404:: a database for exploring
3565:
2717:10.1002/9783527644582.ch10
2428:10.1016/j.cell.2010.03.009
2270:10.1016/j.cell.2011.06.013
1967:10.1186/s13059-022-02755-2
1897:10.1038/s41467-022-30284-w
1595:10.1016/j.cell.2021.07.018
1537:10.1016/j.cell.2012.12.023
1185:10.1016/j.cell.2010.03.009
580:10.1038/s43586-021-00018-1
272:Advantages and limitations
195:high-throughput sequencing
165:high-throughput sequencing
129:and tissue culture cells.
2301:Wiley Interdiscip Rev RNA
1673:10.1038/s41586-020-2077-3
3054:10.1186/gb-2012-13-8-r67
1267:Zhang & Darnell 2011
114:Saccharomyces cerevisiae
85:History and applications
3174:10.1126/science.1090095
2740:Nature Reviews Genetics
2627:Genes & Development
2584:Genes & Development
2510:10.15252/embj.201593360
1731:Genes & Development
1712:Kargapolova et al. 2017
513:10.1126/science.1090095
62:Basic Principle of CLIP
3402:Caenorhabditis elegans
3086:Nucleic Acids Research
2967:10.1038/nprot.2014.012
2881:10.1101/gad.191338.112
2795:10.1038/nsmb0911-1084a
2639:10.1101/gad.301036.117
2596:10.1101/gad.269415.115
2357:10.1101/pdb.prot072132
1782:Mili & Steitz 2004
1743:10.1101/gad.269415.115
1017:Charizanis et al. 2012
1005:Licatalosi et al. 2012
799:Licatalosi et al. 2008
444:using high-throughput
398:microRNA target sites.
304:
215:
185:
170:high sequencing depths
147:
122:Caenorhabditis elegans
63:
26:) is a method used in
3010:10.1101/gr.082503.108
1877:Nature Communications
1589:(18): 4680–4696.e22.
1041:Holmqvist et al. 2016
1029:Ince-Dunn et al. 2012
402:BIMSB doRiNA database
302:
213:
183:
145:
66:CLIP begins with the
61:
41:in order to identify
3361:Nature Biotechnology
2338:Darnell, RB (2012).
2295:Darnell, RB (2010).
1936:Bohnsack et al. 2009
1297:Nature Biotechnology
1231:Sugimoto et al. 2012
1137:Zisoulis et al. 2010
312:in living cells and
127:embryonic stem cells
3544:Genetics techniques
3408:Nat Struct Mol Biol
3318:Nat Struct Mol Biol
3287:10.1093/nar/gkq1056
3166:2003Sci...302.1212U
3160:(5648): 1212–1215.
2924:10.1261/rna.7151404
2838:10.1038/nature07488
2830:2008Natur.456..464L
2783:Nat Struct Mol Biol
2670:Nat Struct Mol Biol
2227:10.1038/nature08170
2219:2009Natur.460..479C
2045:10.7554/eLife.05005
2010:Agarwal et al. 2015
1889:2022NatCo..13.2719K
1665:2020Natur.583..711V
1468:Nature Neuroscience
965:10.7554/eLife.71892
934:Darnell et al. 2011
835:Sanford et al. 2009
505:2003Sci...302.1212U
499:(5648): 1212–1215.
161:immunoprecipitation
91:RNA-binding protein
39:immunoprecipitation
3508:10.1038/nmeth.3810
3463:10.1093/nar/gkx152
3098:10.1093/nar/gkr330
1422:10.1038/nmeth.3453
1279:Burger et al. 2013
412:interactions from
305:
260:Other CLIP methods
216:
186:
148:
64:
3457:(10): 6074–6086.
3451:Nucleic Acids Res
3420:10.1038/nsmb.1745
3330:10.1038/nsmb.1545
3275:Nucleic Acids Res
3092:(16): 6845–6853.
2875:(14): 1626–1642.
2824:(7221): 464–469.
2682:10.1038/nsmb.1838
2213:(7254): 479–486.
2132:10.4161/rna.26214
2126:(10): 1623–1630.
1737:(19): 2037–2053.
1659:(7818): 711–719.
1252:König et al. 2012
1149:Leung et al. 2011
895:10.1038/nsmb.2230
865:Moore et al. 2014
757:10.1038/nsmb.1838
361:starBase database
100:and NOVA2 in the
45:binding sites of
28:molecular biology
24:CLIP, or CLIP-seq
3556:
3529:
3519:
3484:
3474:
3441:
3431:
3394:
3384:
3373:10.1038/nbt.1873
3351:
3341:
3308:
3298:
3265:
3255:
3222:
3193:
3148:
3119:
3109:
3076:
3066:
3056:
3031:
3021:
2988:
2978:
2955:Nature Protocols
2945:
2935:
2902:
2892:
2859:
2849:
2808:
2806:
2773:
2763:
2730:
2703:
2693:
2660:
2650:
2633:(10): 990–1006.
2617:
2607:
2574:
2564:
2547:(6): 1067–1080.
2531:
2521:
2488:
2478:
2449:
2439:
2406:
2381:(6): 1394–1404.
2369:
2359:
2334:
2324:
2291:
2281:
2248:
2238:
2197:
2187:
2153:
2143:
2110:
2100:
2067:
2057:
2047:
2013:
2007:
1998:
1997:
1987:
1969:
1945:
1939:
1933:
1927:
1926:
1916:
1868:
1862:
1856:
1850:
1849:
1831:
1791:
1785:
1779:
1773:
1772:
1762:
1721:
1715:
1709:
1703:
1702:
1692:
1643:
1637:
1631:
1625:
1624:
1614:
1573:
1567:
1566:
1556:
1516:
1510:
1509:
1499:
1458:
1452:
1451:
1441:
1401:
1392:
1391:
1381:
1340:
1331:
1330:
1320:
1309:10.1038/nbt.1873
1288:
1282:
1276:
1270:
1264:
1255:
1249:
1234:
1228:
1215:
1214:
1204:
1163:
1152:
1146:
1140:
1134:
1128:
1127:
1117:
1077:
1071:
1068:Yang et al. 2011
1065:
1056:
1050:
1044:
1038:
1032:
1026:
1020:
1014:
1008:
1002:
996:
995:
985:
967:
943:
937:
931:
925:
924:
914:
874:
868:
862:
853:
847:
838:
832:
826:
820:
814:
808:
802:
796:
787:
786:
776:
736:
725:
719:
710:
709:
691:
667:
658:
657:
647:
607:
601:
600:
582:
558:
541:
540:
484:
95:splicing factors
3564:
3563:
3559:
3558:
3557:
3555:
3554:
3553:
3534:
3533:
3532:
3489:
3444:
3397:
3354:
3311:
3268:
3238:(6): 996–1006.
3225:
3196:
3151:
3122:
3079:
3034:
2998:Genome Research
2991:
2948:
2905:
2862:
2811:
2776:
2752:10.1038/nrg3141
2733:
2727:
2711:. p. 153.
2706:
2663:
2620:
2590:(19): 2037–53.
2577:
2534:
2504:(9): 991–1011.
2491:
2452:
2409:
2372:
2350:(11): 1146–60.
2337:
2313:10.1002/wrna.31
2294:
2251:
2200:
2156:
2113:
2070:
2025:
2021:
2016:
2008:
2001:
1947:
1946:
1942:
1934:
1930:
1870:
1869:
1865:
1859:Ule et al. 2005
1857:
1853:
1793:
1792:
1788:
1780:
1776:
1723:
1722:
1718:
1710:
1706:
1645:
1644:
1640:
1632:
1628:
1575:
1574:
1570:
1518:
1517:
1513:
1480:10.1038/nn.2778
1460:
1459:
1455:
1403:
1402:
1395:
1342:
1341:
1334:
1290:
1289:
1285:
1277:
1273:
1265:
1258:
1250:
1237:
1229:
1218:
1165:
1164:
1155:
1147:
1143:
1135:
1131:
1079:
1078:
1074:
1066:
1059:
1051:
1047:
1039:
1035:
1027:
1023:
1015:
1011:
1003:
999:
945:
944:
940:
932:
928:
876:
875:
871:
863:
856:
850:Chi et al. 2009
848:
841:
833:
829:
823:Yeo et al. 2009
821:
817:
811:Xue et al. 2009
809:
805:
797:
790:
738:
737:
728:
720:
713:
669:
668:
661:
609:
608:
604:
560:
559:
544:
486:
485:
478:
474:
469:
410:microRNA-target
356:
354:Further reading
327:
325:Similar methods
297:
279:
274:
262:
255:
252:
248:
208:
178:
140:
135:
87:
56:
17:
12:
11:
5:
3562:
3560:
3552:
3551:
3546:
3536:
3535:
3531:
3530:
3502:(6): 508–514.
3496:Nature Methods
3486:
3485:
3442:
3414:(2): 173–179.
3395:
3367:(7): 607–614.
3352:
3324:(2): 130–137.
3309:
3266:
3232:Molecular Cell
3223:
3205:(4): 376–386.
3194:
3149:
3120:
3077:
3041:Genome Biology
3032:
3004:(3): 381–394.
2989:
2961:(2): 263–293.
2946:
2918:(11): 1692–4.
2903:
2860:
2809:
2774:
2731:
2726:978-3527644582
2725:
2704:
2676:(7): 909–915.
2661:
2618:
2575:
2532:
2489:
2450:
2422:(1): 129–141.
2407:
2370:
2335:
2307:(2): 266–286.
2292:
2264:(2): 247–261.
2249:
2198:
2170:(3): 437–450.
2154:
2111:
2083:(4): 583–592.
2077:Molecular Cell
2068:
2022:
2020:
2017:
2015:
2014:
1999:
1954:Genome Biology
1940:
1928:
1863:
1851:
1806:(1): 235–261.
1786:
1774:
1716:
1704:
1638:
1626:
1568:
1531:(3): 453–466.
1511:
1474:(4): 452–458.
1453:
1416:(8): 767–772.
1410:Nature Methods
1393:
1356:(2): 255–261.
1332:
1303:(7): 607–614.
1283:
1271:
1256:
1235:
1216:
1179:(1): 129–141.
1153:
1141:
1129:
1092:(2): 513–522.
1072:
1057:
1045:
1033:
1021:
1009:
997:
938:
926:
889:(3): 321–327.
869:
854:
839:
827:
815:
803:
788:
751:(7): 909–915.
726:
711:
682:(3): 354–369.
676:Molecular Cell
659:
622:(8): a032243.
602:
542:
475:
473:
470:
468:
467:
461:
455:
435:
399:
367:networks from
357:
355:
352:
351:
350:
340:
334:
326:
323:
296:
293:
278:
275:
273:
270:
261:
258:
247:
244:
207:
204:
177:
174:
139:
136:
134:
131:
86:
83:
55:
52:
30:that combines
15:
13:
10:
9:
6:
4:
3:
2:
3561:
3550:
3547:
3545:
3542:
3541:
3539:
3527:
3523:
3518:
3513:
3509:
3505:
3501:
3497:
3493:
3488:
3487:
3482:
3478:
3473:
3468:
3464:
3460:
3456:
3452:
3448:
3443:
3439:
3435:
3430:
3425:
3421:
3417:
3413:
3409:
3405:
3403:
3396:
3392:
3388:
3383:
3378:
3374:
3370:
3366:
3362:
3358:
3353:
3349:
3345:
3340:
3335:
3331:
3327:
3323:
3319:
3315:
3310:
3306:
3302:
3297:
3292:
3288:
3284:
3280:
3276:
3272:
3267:
3263:
3259:
3254:
3249:
3245:
3241:
3237:
3233:
3229:
3224:
3220:
3216:
3212:
3208:
3204:
3200:
3195:
3191:
3187:
3183:
3179:
3175:
3171:
3167:
3163:
3159:
3155:
3150:
3146:
3142:
3138:
3134:
3130:
3126:
3121:
3117:
3113:
3108:
3103:
3099:
3095:
3091:
3087:
3083:
3078:
3074:
3070:
3065:
3060:
3055:
3050:
3046:
3042:
3038:
3033:
3029:
3025:
3020:
3015:
3011:
3007:
3003:
2999:
2995:
2990:
2986:
2982:
2977:
2972:
2968:
2964:
2960:
2956:
2952:
2947:
2943:
2939:
2934:
2929:
2925:
2921:
2917:
2913:
2909:
2904:
2900:
2896:
2891:
2886:
2882:
2878:
2874:
2870:
2866:
2861:
2857:
2853:
2848:
2843:
2839:
2835:
2831:
2827:
2823:
2819:
2815:
2810:
2805:
2800:
2796:
2792:
2788:
2784:
2780:
2775:
2771:
2767:
2762:
2757:
2753:
2749:
2745:
2741:
2737:
2732:
2728:
2722:
2718:
2714:
2710:
2705:
2701:
2697:
2692:
2687:
2683:
2679:
2675:
2671:
2667:
2662:
2658:
2654:
2649:
2644:
2640:
2636:
2632:
2628:
2624:
2619:
2615:
2611:
2606:
2601:
2597:
2593:
2589:
2585:
2581:
2576:
2572:
2568:
2563:
2558:
2554:
2550:
2546:
2542:
2538:
2533:
2529:
2525:
2520:
2515:
2511:
2507:
2503:
2499:
2495:
2490:
2486:
2482:
2477:
2472:
2468:
2464:
2461:(41): e2034.
2460:
2456:
2451:
2447:
2443:
2438:
2433:
2429:
2425:
2421:
2417:
2413:
2408:
2404:
2400:
2396:
2392:
2388:
2384:
2380:
2376:
2371:
2367:
2363:
2358:
2353:
2349:
2345:
2341:
2336:
2332:
2328:
2323:
2318:
2314:
2310:
2306:
2302:
2298:
2293:
2289:
2285:
2280:
2275:
2271:
2267:
2263:
2259:
2255:
2250:
2246:
2242:
2237:
2232:
2228:
2224:
2220:
2216:
2212:
2208:
2204:
2199:
2195:
2191:
2186:
2181:
2177:
2173:
2169:
2165:
2161:
2155:
2151:
2147:
2142:
2137:
2133:
2129:
2125:
2121:
2117:
2112:
2108:
2104:
2099:
2094:
2090:
2086:
2082:
2078:
2074:
2069:
2065:
2061:
2056:
2051:
2046:
2041:
2037:
2033:
2029:
2024:
2023:
2018:
2011:
2006:
2004:
2000:
1995:
1991:
1986:
1981:
1977:
1973:
1968:
1963:
1959:
1955:
1951:
1944:
1941:
1937:
1932:
1929:
1924:
1920:
1915:
1910:
1906:
1902:
1898:
1894:
1890:
1886:
1882:
1878:
1874:
1867:
1864:
1860:
1855:
1852:
1847:
1843:
1839:
1835:
1830:
1825:
1821:
1817:
1813:
1809:
1805:
1801:
1797:
1790:
1787:
1783:
1778:
1775:
1770:
1766:
1761:
1756:
1752:
1748:
1744:
1740:
1736:
1732:
1728:
1720:
1717:
1713:
1708:
1705:
1700:
1696:
1691:
1686:
1682:
1678:
1674:
1670:
1666:
1662:
1658:
1654:
1650:
1642:
1639:
1635:
1630:
1627:
1622:
1618:
1613:
1608:
1604:
1600:
1596:
1592:
1588:
1584:
1580:
1572:
1569:
1564:
1560:
1555:
1550:
1546:
1542:
1538:
1534:
1530:
1526:
1522:
1515:
1512:
1507:
1503:
1498:
1493:
1489:
1485:
1481:
1477:
1473:
1469:
1465:
1457:
1454:
1449:
1445:
1440:
1435:
1431:
1427:
1423:
1419:
1415:
1411:
1407:
1400:
1398:
1394:
1389:
1385:
1380:
1375:
1371:
1367:
1363:
1359:
1355:
1351:
1347:
1339:
1337:
1333:
1328:
1324:
1319:
1314:
1310:
1306:
1302:
1298:
1294:
1287:
1284:
1280:
1275:
1272:
1268:
1263:
1261:
1257:
1253:
1248:
1246:
1244:
1242:
1240:
1236:
1232:
1227:
1225:
1223:
1221:
1217:
1212:
1208:
1203:
1198:
1194:
1190:
1186:
1182:
1178:
1174:
1170:
1162:
1160:
1158:
1154:
1150:
1145:
1142:
1138:
1133:
1130:
1125:
1121:
1116:
1111:
1107:
1103:
1099:
1095:
1091:
1087:
1083:
1076:
1073:
1069:
1064:
1062:
1058:
1054:
1049:
1046:
1042:
1037:
1034:
1030:
1025:
1022:
1018:
1013:
1010:
1006:
1001:
998:
993:
989:
984:
979:
975:
971:
966:
961:
957:
953:
949:
942:
939:
935:
930:
927:
922:
918:
913:
908:
904:
900:
896:
892:
888:
884:
880:
873:
870:
866:
861:
859:
855:
851:
846:
844:
840:
836:
831:
828:
824:
819:
816:
812:
807:
804:
800:
795:
793:
789:
784:
780:
775:
770:
766:
762:
758:
754:
750:
746:
742:
735:
733:
731:
727:
723:
718:
716:
712:
707:
703:
699:
695:
690:
685:
681:
677:
673:
666:
664:
660:
655:
651:
646:
641:
637:
633:
629:
625:
621:
617:
613:
606:
603:
598:
594:
590:
586:
581:
576:
572:
568:
564:
557:
555:
553:
551:
549:
547:
543:
538:
534:
530:
526:
522:
518:
514:
510:
506:
502:
498:
494:
490:
483:
481:
477:
471:
465:
462:
459:
456:
453:
452:
447:
443:
439:
436:
433:
432:
427:
426:
421:
420:
415:
411:
407:
403:
400:
397:
395:
390:
386:
385:
380:
379:
374:
370:
366:
362:
359:
358:
353:
348:
344:
341:
338:
335:
332:
329:
328:
324:
322:
319:
315:
311:
301:
294:
292:
288:
285:
276:
271:
269:
266:
259:
257:
245:
243:
241:
237:
232:
230:
226:
222:
221:
212:
205:
203:
199:
196:
192:
191:
182:
175:
173:
171:
166:
162:
158:
157:cross-linking
154:
153:
144:
137:
132:
130:
128:
124:
123:
119:
115:
111:
107:
103:
99:
96:
92:
84:
82:
79:
74:
69:
60:
53:
51:
48:
44:
40:
36:
33:
29:
25:
21:
3499:
3495:
3454:
3450:
3411:
3407:
3401:
3364:
3360:
3321:
3317:
3278:
3274:
3235:
3231:
3202:
3198:
3157:
3153:
3128:
3124:
3089:
3085:
3044:
3040:
3001:
2997:
2958:
2954:
2915:
2911:
2872:
2868:
2821:
2817:
2786:
2782:
2746:(2): 77–83.
2743:
2739:
2708:
2673:
2669:
2630:
2626:
2587:
2583:
2544:
2540:
2501:
2497:
2467:10.3791/2034
2458:
2419:
2415:
2378:
2374:
2347:
2343:
2304:
2300:
2261:
2257:
2210:
2206:
2167:
2163:
2123:
2119:
2080:
2076:
2035:
2031:
1957:
1953:
1943:
1931:
1880:
1876:
1866:
1854:
1803:
1799:
1789:
1777:
1734:
1730:
1719:
1707:
1656:
1652:
1641:
1629:
1586:
1582:
1571:
1528:
1524:
1514:
1471:
1467:
1456:
1413:
1409:
1353:
1350:Cell Reports
1349:
1300:
1296:
1286:
1274:
1176:
1172:
1144:
1132:
1089:
1085:
1075:
1048:
1036:
1024:
1012:
1000:
955:
951:
941:
929:
886:
882:
872:
830:
818:
806:
748:
744:
722:Darnell 2010
679:
675:
619:
615:
605:
570:
566:
496:
492:
449:
445:
441:
429:
423:
417:
413:
409:
405:
392:
391:) data, and
388:
382:
376:
372:
368:
346:
313:
309:
306:
303:CLIP Summary
289:
280:
267:
263:
249:
233:
218:
217:
200:
188:
187:
155:combines UV
150:
149:
120:
117:
88:
73:Proteinase K
67:
65:
35:crosslinking
23:
19:
18:
2789:(9): 1084.
1883:(1): 2719.
573:(1): 1–23.
454:sequencing.
406:protein-RNA
295:Limitations
102:mouse brain
3538:Categories
3047:(8): R67.
2038:: e05005.
1960:(1): 191.
958:: e71892.
472:References
438:miRTarCLIP
394:TargetScan
318:TargetScan
277:Advantages
3131:: 60–72.
2869:Genes Dev
1976:1474-760X
1905:2041-1723
1820:2574-3414
1751:1549-5477
1681:1476-4687
1603:1097-4172
1545:1097-4172
1488:1546-1726
1430:1548-7105
1370:2211-1247
1193:1097-4172
1106:1943-2631
974:2050-084X
903:1545-9985
765:1545-9985
698:1097-4164
636:1943-0264
597:233834798
589:2662-8449
521:1095-9203
419:HITS-CLIP
378:HITS-CLIP
229:HITS-CLIP
152:HITS-CLIP
146:HITS-CLIP
138:HITS-CLIP
106:Argonaute
3526:27018577
3481:28334977
3438:20062054
3391:21633356
3348:19136955
3305:21037263
3262:20064465
3219:16314267
3190:23420615
3182:14615540
3145:28254606
3116:21652644
3073:22863408
3028:19116412
2985:24407355
2942:15388877
2899:22802532
2856:18978773
2770:22251872
2700:20601959
2657:28637692
2614:26404942
2571:22998874
2528:27044921
2485:20644507
2446:20371350
2403:23801945
2395:18028214
2366:23118367
2331:21935890
2288:21784246
2245:19536157
2194:22884328
2158:(2012).
2150:24025460
2120:RNA Biol
2107:19941819
2064:26267216
1994:36085079
1923:35581222
1846:90760475
1838:37123514
1769:26404942
1699:32728246
1621:34380047
1563:23374342
1506:21358640
1448:26121403
1388:23871666
1327:21633356
1211:20371350
1124:20382833
1086:Genetics
992:34939924
921:22343717
783:20601959
706:29395060
654:30068528
537:23420615
529:14615540
451:PAR-CLIP
425:PAR-CLIP
414:CLIP-Seq
373:CLIP-Seq
369:PAR-CLIP
347:in vitro
337:ChIP-Seq
331:RIP-Chip
314:in vitro
225:PAR-CLIP
190:PAR-CLIP
184:PAR-CLIP
176:PAR-CLIP
110:microRNA
54:Workflow
47:proteins
3517:4887338
3472:5449641
3429:2834287
3382:3400429
3339:2735254
3296:3013664
3253:2807993
3199:Methods
3162:Bibcode
3154:Science
3125:Methods
3107:3167600
3064:4053741
3019:2661799
2976:4156013
2933:1370654
2890:3404389
2847:2597294
2826:Bibcode
2804:3078052
2761:4962561
2691:3000544
2648:5495127
2605:4604345
2562:3517991
2519:5207318
2476:3156069
2437:2861495
2322:3222227
2279:3232425
2236:2733940
2215:Bibcode
2185:3418517
2141:3866244
2098:2806949
2055:4532895
2019:Sources
1985:9461102
1914:9114321
1885:Bibcode
1829:7614488
1760:4604345
1690:7410833
1661:Bibcode
1612:8445024
1554:3629564
1497:3108889
1439:4487409
1379:3730056
1318:3400429
1202:2861495
1115:2881133
983:8820740
912:3541676
774:3000544
645:6071486
501:Bibcode
493:Science
310:in vivo
133:Methods
68:in vivo
3524:
3514:
3479:
3469:
3436:
3426:
3389:
3379:
3346:
3336:
3303:
3293:
3260:
3250:
3217:
3188:
3180:
3143:
3114:
3104:
3071:
3061:
3026:
3016:
2983:
2973:
2940:
2930:
2897:
2887:
2854:
2844:
2818:Nature
2801:
2768:
2758:
2723:
2698:
2688:
2655:
2645:
2612:
2602:
2569:
2559:
2541:Neuron
2526:
2516:
2498:EMBO J
2483:
2473:
2444:
2434:
2401:
2393:
2364:
2329:
2319:
2286:
2276:
2243:
2233:
2207:Nature
2192:
2182:
2164:Neuron
2148:
2138:
2105:
2095:
2062:
2052:
1992:
1982:
1974:
1921:
1911:
1903:
1844:
1836:
1826:
1818:
1767:
1757:
1749:
1697:
1687:
1679:
1653:Nature
1619:
1609:
1601:
1561:
1551:
1543:
1504:
1494:
1486:
1446:
1436:
1428:
1386:
1376:
1368:
1325:
1315:
1209:
1199:
1191:
1122:
1112:
1104:
990:
980:
972:
919:
909:
901:
781:
771:
763:
704:
696:
652:
642:
634:
595:
587:
535:
527:
519:
240:iCount
3186:S2CID
2399:S2CID
2032:eLife
1842:S2CID
952:eLife
593:S2CID
533:S2CID
464:dCLIP
458:clipz
431:iCLIP
389:CLASH
384:iCLIP
365:ceRNA
345:, an
343:SELEX
246:eCLIP
220:iCLIP
214:iCLIP
206:iCLIP
163:with
98:NOVA1
78:iCLIP
37:with
3522:PMID
3477:PMID
3434:PMID
3387:PMID
3344:PMID
3301:PMID
3258:PMID
3215:PMID
3178:PMID
3141:PMID
3112:PMID
3069:PMID
3024:PMID
2981:PMID
2938:PMID
2895:PMID
2852:PMID
2766:PMID
2721:ISBN
2696:PMID
2653:PMID
2610:PMID
2567:PMID
2524:PMID
2481:PMID
2442:PMID
2416:Cell
2391:PMID
2362:PMID
2348:2012
2327:PMID
2284:PMID
2258:Cell
2241:PMID
2190:PMID
2146:PMID
2103:PMID
2060:PMID
1990:PMID
1972:ISSN
1919:PMID
1901:ISSN
1834:PMID
1816:ISSN
1765:PMID
1747:ISSN
1695:PMID
1677:ISSN
1617:PMID
1599:ISSN
1583:Cell
1559:PMID
1541:ISSN
1525:Cell
1502:PMID
1484:ISSN
1444:PMID
1426:ISSN
1384:PMID
1366:ISSN
1323:PMID
1207:PMID
1189:ISSN
1173:Cell
1120:PMID
1102:ISSN
988:PMID
970:ISSN
917:PMID
899:ISSN
779:PMID
761:ISSN
702:PMID
694:ISSN
650:PMID
632:ISSN
585:ISSN
525:PMID
517:ISSN
448:and
446:CLIP
408:and
159:and
93:and
3549:RNA
3512:PMC
3504:doi
3467:PMC
3459:doi
3424:PMC
3416:doi
3377:PMC
3369:doi
3334:PMC
3326:doi
3291:PMC
3283:doi
3248:PMC
3240:doi
3207:doi
3170:doi
3158:302
3133:doi
3129:118
3102:PMC
3094:doi
3059:PMC
3049:doi
3014:PMC
3006:doi
2971:PMC
2963:doi
2928:PMC
2920:doi
2912:RNA
2885:PMC
2877:doi
2842:PMC
2834:doi
2822:456
2799:PMC
2791:doi
2756:PMC
2748:doi
2713:doi
2686:PMC
2678:doi
2643:PMC
2635:doi
2600:PMC
2592:doi
2557:PMC
2549:doi
2514:PMC
2506:doi
2471:PMC
2463:doi
2432:PMC
2424:doi
2420:141
2383:doi
2352:doi
2317:PMC
2309:doi
2274:PMC
2266:doi
2262:146
2231:PMC
2223:doi
2211:460
2180:PMC
2172:doi
2136:PMC
2128:doi
2093:PMC
2085:doi
2050:PMC
2040:doi
1980:PMC
1962:doi
1909:PMC
1893:doi
1824:PMC
1808:doi
1755:PMC
1739:doi
1685:PMC
1669:doi
1657:583
1607:PMC
1591:doi
1587:184
1549:PMC
1533:doi
1529:152
1492:PMC
1476:doi
1434:PMC
1418:doi
1374:PMC
1358:doi
1313:PMC
1305:doi
1197:PMC
1181:doi
1177:141
1110:PMC
1094:doi
1090:185
978:PMC
960:doi
907:PMC
891:doi
769:PMC
753:doi
684:doi
640:PMC
624:doi
575:doi
509:doi
497:302
284:RIP
236:UMI
43:RNA
3540::
3520:.
3510:.
3500:13
3498:.
3494:.
3475:.
3465:.
3455:45
3453:.
3449:.
3432:.
3422:.
3412:17
3410:.
3406:.
3385:.
3375:.
3365:29
3363:.
3359:.
3342:.
3332:.
3322:16
3320:.
3316:.
3299:.
3289:.
3279:39
3277:.
3273:.
3256:.
3246:.
3236:36
3234:.
3230:.
3213:.
3203:37
3201:.
3184:.
3176:.
3168:.
3156:.
3139:.
3127:.
3110:.
3100:.
3090:39
3088:.
3084:.
3067:.
3057:.
3045:13
3043:.
3039:.
3022:.
3012:.
3002:19
3000:.
2996:.
2979:.
2969:.
2957:.
2953:.
2936:.
2926:.
2916:10
2914:.
2910:.
2893:.
2883:.
2873:26
2871:.
2867:.
2850:.
2840:.
2832:.
2820:.
2816:.
2797:.
2787:19
2785:.
2781:.
2764:.
2754:.
2744:13
2742:.
2738:.
2719:.
2694:.
2684:.
2674:17
2672:.
2668:.
2651:.
2641:.
2631:31
2629:.
2625:.
2608:.
2598:.
2588:29
2586:.
2582:.
2565:.
2555:.
2545:75
2543:.
2539:.
2522:.
2512:.
2502:35
2500:.
2496:.
2479:.
2469:.
2457:.
2440:.
2430:.
2418:.
2414:.
2397:.
2389:.
2379:83
2377:.
2360:.
2346:.
2342:.
2325:.
2315:.
2303:.
2299:.
2282:.
2272:.
2260:.
2256:.
2239:.
2229:.
2221:.
2209:.
2205:.
2188:.
2178:.
2168:75
2166:.
2162:.
2144:.
2134:.
2124:10
2122:.
2118:.
2101:.
2091:.
2081:36
2079:.
2075:.
2058:.
2048:.
2034:.
2030:.
2002:^
1988:.
1978:.
1970:.
1958:23
1956:.
1952:.
1917:.
1907:.
1899:.
1891:.
1881:13
1879:.
1875:.
1840:.
1832:.
1822:.
1814:.
1802:.
1798:.
1763:.
1753:.
1745:.
1735:29
1733:.
1729:.
1693:.
1683:.
1675:.
1667:.
1655:.
1651:.
1615:.
1605:.
1597:.
1585:.
1581:.
1557:.
1547:.
1539:.
1527:.
1523:.
1500:.
1490:.
1482:.
1472:14
1470:.
1466:.
1442:.
1432:.
1424:.
1414:12
1412:.
1408:.
1396:^
1382:.
1372:.
1364:.
1352:.
1348:.
1335:^
1321:.
1311:.
1301:29
1299:.
1295:.
1259:^
1238:^
1219:^
1205:.
1195:.
1187:.
1175:.
1171:.
1156:^
1118:.
1108:.
1100:.
1088:.
1084:.
1060:^
986:.
976:.
968:.
956:10
954:.
950:.
915:.
905:.
897:.
887:19
885:.
881:.
857:^
842:^
791:^
777:.
767:.
759:.
749:17
747:.
743:.
729:^
714:^
700:.
692:.
680:69
678:.
674:.
662:^
648:.
638:.
630:.
620:10
618:.
614:.
591:.
583:.
569:.
565:.
545:^
531:.
523:.
515:.
507:.
495:.
491:.
479:^
428:,
422:,
416:,
387:,
381:,
375:,
125:,
32:UV
3528:.
3506::
3483:.
3461::
3440:.
3418::
3404:"
3393:.
3371::
3350:.
3328::
3307:.
3285::
3264:.
3242::
3221:.
3209::
3192:.
3172::
3164::
3147:.
3135::
3118:.
3096::
3075:.
3051::
3030:.
3008::
2987:.
2965::
2959:9
2944:.
2922::
2901:.
2879::
2858:.
2836::
2828::
2807:.
2793::
2772:.
2750::
2729:.
2715::
2702:.
2680::
2659:.
2637::
2616:.
2594::
2573:.
2551::
2530:.
2508::
2487:.
2465::
2448:.
2426::
2405:.
2385::
2368:.
2354::
2333:.
2311::
2305:1
2290:.
2268::
2247:.
2225::
2217::
2196:.
2174::
2152:.
2130::
2109:.
2087::
2066:.
2042::
2036:4
2012:.
1996:.
1964::
1938:.
1925:.
1895::
1887::
1861:.
1848:.
1810::
1804:1
1784:.
1771:.
1741::
1714:.
1701:.
1671::
1663::
1636:.
1623:.
1593::
1565:.
1535::
1508:.
1478::
1450:.
1420::
1390:.
1360::
1354:4
1329:.
1307::
1281:.
1269:.
1254:.
1233:.
1213:.
1183::
1151:.
1139:.
1126:.
1096::
1070:.
1055:.
1043:.
1031:.
1019:.
1007:.
994:.
962::
936:.
923:.
893::
867:.
852:.
837:.
825:.
813:.
801:.
785:.
755::
724:.
708:.
686::
656:.
626::
599:.
577::
571:1
539:.
511::
503::
371:(
118:,
116:)
22:(
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.