1513:
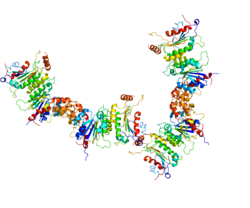
catalytic domain, occur as a heterotetramer (see Figure). These heterotetramers occur in the order: accessory protein-catalytic protein-catalytic protein-accessory protein. The particular complex shown in the Figure illustrates the heterotetramer formed by catalytic protein DNMT3A2 and accessory protein DNMT3B3. One accessory protein of the complex binds to an acidic patch on the nucleosome core (see top 3B3 in Figure). The connection of one accessory protein to the nucleosome orients the heterotetramer. The orientation places the first catalytic DNMT (closest to the accessory protein connected to the nucleosome) in an intermediate position (not close to the linker DNA). The second catalytic DNMT (lower 3A2 in Figure) is placed at the linker DNA. Methylations can take place within this linker DNA (as shown in the Figure) but not on any DNA wrapped around the nucleosome core.
1500:
340:
317:
1454:
214:
239:
591:
31:
346:
245:
1599:(AML) where they occurred in just over 25% of cases sequenced. These mutations most often occur at position R882 in the protein and this mutation may cause loss of function. DNMT3A mutations are associated with poor overall survival, suggesting that they have an important common effect on the potential of AML cells to cause lethal disease. It has also been found that
1484:, which, together, are arranged into a single globular fold. This domain can act as a reader that specifically binds to histone H3 that is unmethylated at lysine 4 (H3K4me0). The ADD domain serves as an inhibitor of the methyltransferase domain until DNMT3A binds to the unmodified lysine 4 of histone 3 (H3K4me0) for its
1553:
have reduced expression of about 1,000 genes and increased expression of about 500 genes in the hippocampus region of the brain. These changes occur 24 hours after training. At this point, there is modified expression of 9.17% of the rat hippocampal genome. Reduced expression of genes is associated
1433:
There are two main protein isoforms, DNMT3A1 and DNMT3A2 with molecular weights of about 130 kDa and 100 kDa, respectively. The DNMT3A2 protein, which lacks the N-terminal region of DNMT3A1, is encoded by a transcript initiated from a downstream promoter. These isoforms exist in different cell types.
1537:
DNMT1 is responsible for maintenance DNA methylation while DNMT3A and DNMT3B carry out both maintenance – correcting the errors of DNMT1 – and de novo DNA methylation. After DNMT1 knockout in human cancer cells, these cells were found to retain their inherited methylation pattern, which suggests
1520:
in the human genome. Many of these CpGs are located within CpG islands (regions of DNA) of relatively high density of CpG sites. Of these regions, there are 3,970 regions exclusively enriched for DNMT3A1, 3,838 regions for DNMT3A2 and 3,432 regions for DNMT3B, and there are sites that are shared
1468:
The PWWP motif is within an about 100 amino acid domain that has one area with a significant amount of basic residues (lysines and arginines), giving a positively charged surface that can bind to DNA. A separate region of the PWWP domain can bind to histone methyl-lysines through a hydrophobic
1512:
in DNA. The accompanying Figure shows a methyltransferase complex containing DNMT3A2. These enzymes, to be effective, must act in conjunction with an accessory protein (e.g. DNMT3B3, DNMT3L, or others). Two accessory proteins (which have no catalytic activity), complexed to two DNMTs with a
1538:
maintenance activity by the expressed DNMT3s. DNMT3s show equal affinity for unmethylated and hemimethylated DNA substrates while DNMT1 has a 10-40 fold preference for hemimethylated DNA. The DNMT3s can bind to both forms and hence potentially do both maintenance and de novo modifications.
1541:
De novo methylation is the main recognized activity of DNMT3A, which is essential for processes such as those mentioned in the introductory paragraphs. Genetic imprinting prevents parthenogenesis in mammals, and hence forces sexual reproduction and its multiple consequences on genetics and
1465:(see Figure of simplified domains of DNMT3A isoforms). The two accessory proteins stimulate de novo methylation by each of their interactions with the three isoforms that have a functional catalytic domain. In general, all DNMTs require accessory proteins for their biological function.
1434:
When originally established, DNMT3A2 was found to be highly expressed in testis, ovary, spleen, and thymus. It was more recently shown to be inducibly expressed in brain hippocampus and needed in the hippocampus when establishing memory. DNMT3A2 is also upregulated in the
57:
1488:
methylating activity. DNMT3A thus seems to have an inbuilt control mechanism targeting DNA for methylation only at histones that are unmethylated at histone 3 with the lysine at the 4th position from the amino end being un-methylated.
4288:
Weisenberger DJ, Velicescu M, Preciado-Lopez MA, Gonzales FA, Tsai YC, Liang G, Jones PA (September 2002). "Identification and characterization of alternatively spliced variants of DNA methyltransferase 3a in mammalian cells".
1503:
DNMTs such as DNMT3A2, with a functioning catalytic domain, require an accessory protein, such as DNMT3B3 without a functioning catalytic domain, for methylating activity in vivo, as in the heterotetrameric structure shown
3877:
Adams MD, Kerlavage AR, Fleischmann RD, Fuldner RA, Bult CJ, Lee NH, et al. (September 1995). "Initial assessment of human gene diversity and expression patterns based upon 83 million nucleotides of cDNA sequence".
4063:
Di Croce L, Raker VA, Corsaro M, Fazi F, Fanelli M, Faretta M, et al. (February 2002). "Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor".
1569:
In Dnmt3a-/- mice, many genes associated with HSC self-renewal increase in expression and some fail to be appropriately repressed during differentiation. This suggests abrogation of differentiation in
1516:
As shown by Manzo et al., there are both specific individual binding sites for the three catalytic DNMTs (3A1, 3A2 and 3B3) as well as overlapping binding sites of these enzymes. There are 28 million
1338:
DNA methylation. Such function is to be distinguished from maintenance DNA methylation which ensures the fidelity of replication of inherited epigenetic patterns. DNMT3A forms part of the family of
2822:
Gao L, Emperle M, Guo Y, Grimm SA, Ren W, Adam S, Uryu H, Zhang ZM, Chen D, Yin J, Dukatz M, Anteneh H, Jurkowska RZ, Lu J, Wang Y, Bashtrykov P, Wade PA, Wang GG, Jeltsch A, Song J (July 2020).
4451:
Yakushiji T, Uzawa K, Shibahara T, Noma H, Tanzawa H (June 2003). "Over-expression of DNA methyltransferases and CDKN2A gene methylation status in squamous cell carcinoma of the oral cavity".
4371:
Robert MF, Morin S, Beaulieu N, Gauthier F, Chute IC, Barsalou A, MacLeod AR (January 2003). "DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells".
1499:
4482:"Modification of de novo DNA methyltransferase 3a (Dnmt3a) by SUMO-1 modulates its interaction with histone deacetylases (HDACs) and its capacity to repress transcription"
3619:"Modification of de novo DNA methyltransferase 3a (Dnmt3a) by SUMO-1 modulates its interaction with histone deacetylases (HDACs) and its capacity to repress transcription"
3170:
Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, Sasaki H (June 2004). "Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting".
353:
252:
4224:
Yanagisawa Y, Ito E, Yuasa Y, Maruyama K (September 2002). "The human DNA methyltransferases DNMT3A and DNMT3B have two types of promoters with different CpG contents".
1867:"Negative regulation of DNMT3A de novo DNA methylation by frequently overexpressed UHRF family proteins as a mechanism for widespread DNA hypomethylation in cancer"
1450:
DNMT3A consists of three major protein domains: the Pro-Trp-Trp-Pro (PWWP) domain, the ATRX-DNMT3-DNMT3L (ADD) domain and the catalytic methyltransferase domain.
4117:
Hata K, Okano M, Lei H, Li E (April 2002). "Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice".
2986:
Rhee I, Jair KW, Yen RW, Lengauer C, Herman JG, Kinzler KW, et al. (April 2000). "CpG methylation is maintained in human cancer cells lacking DNMT1".
1830:
Xie S, Wang Z, Okano M, Nogami M, Li Y, He WW, et al. (August 1999). "Cloning, expression and chromosome locations of the human DNMT3 gene family".
984:
965:
175:
4148:"A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation"
2168:"A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation"
1761:
1743:
4533:
1662:
1457:
This illustrates 5 isoforms of DNMT3A showing the locations of the PWWP domain, the ADD domain and the catalytic or catalytic-like domains.
3039:"Recombinant human DNA (cytosine-5) methyltransferase. I. Expression, purification, and comparison of de novo and maintenance methylation"
2264:
Oliveira AM, Hemstedt TJ, Bading H (July 2012). "Rescue of aging-associated decline in Dnmt3a2 expression restores cognitive abilities".
339:
3417:
Shih AH, Abdel-Wahab O, Patel JP, Levine RL (September 2012). "The role of mutations in epigenetic regulators in myeloid malignancies".
1461:
The structures of DNMT3A1 and DNMT3A2 have analogies with the structure of DNMT3B1 and also with the two accessory proteins DNMT3B3 and
1508:
The three DNA methyltransferases (DNMT3A1, DNMT3A2 and DNMT3B) catalyze reactions placing a methyl group onto a cytosine, usually at a
1782:
Okano M, Xie S, Li E (July 1998). "Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases".
1521:
between the de novo DNMT proteins. In addition, whether the DNA methyltransferase (DNMT3A1, DNMT3A2 or DNMT3B) acts on an available
1248:
316:
3668:"Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin"
1612:
1255:
3936:"The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors"
2065:"The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors"
1573:(HSCs) and an increase in self-renewal cell-division instead. Indeed, it was found that differentiation was partially rescued if
2824:"Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms"
1730:
1709:
1726:
238:
213:
54:
1705:
155:
1624:
352:
251:
1370:
345:
244:
1604:
3511:"Abnormal RNA splicing and genomic instability after induction of DNMT3A mutations by CRISPR/Cas9 gene editing"
1362:
1029:
163:
4183:"Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases"
3563:"Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases"
146:, DNMT3A2, M.HsaIIIA, TBRS, DNA (cytosine-5-)-methyltransferase 3 alpha, DNA methyltransferase 3 alpha, HESJAS
4022:"Functional and physical interaction between the histone methyl transferase Suv39H1 and histone deacetylases"
1566:
In mice, this gene has shown reduced expression in ageing animals causes cognitive long-term memory decline.
1596:
1570:
1396:
1010:
1453:
2656:
Zeng Y, Ren R, Kaur G, Hardikar S, Ying Z, Babcock L, Gupta E, Zhang X, Chen T, Cheng X (November 2020).
2458:"Nucleosomal DNA binding drives the recognition of H3K36-methylated nucleosomes by the PSIP1-PWWP domain"
2609:"Molecular and enzymatic profiles of mammalian DNA methyltransferases: structures and targets for drugs"
1339:
1591:
This gene is frequently mutated in cancer, being one of 127 frequently mutated genes identified in the
3979:"Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription"
3728:"Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription"
3080:"Baculovirus-mediated expression and characterization of the full-length murine DNA methyltransferase"
4331:
4073:
3679:
3373:
3179:
2995:
2835:
2775:
2718:
1611:
as compared to their isogenic wildtype counterparts. Mutations in this gene are also associated with
1392:
227:
3666:
Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, et al. (July 2003).
2884:"Isoform-specific localization of DNMT3A regulates DNA methylation fidelity at bivalent CpG islands"
2016:"Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes"
142:
2456:
van Nuland R, van Schaik FM, Simonis M, van Heesch S, Cuppen E, Boelens R, et al. (May 2013).
2112:
Del
Castillo Falconi VM, Torres-Arciga K, Matus-Ortega G, Díaz-Chávez J, Herrera LA (August 2022).
1592:
1223:
1202:
1176:
1172:
1144:
1123:
1119:
1109:
4396:
4105:
3705:
3442:
3203:
3152:
3019:
2338:
2289:
1807:
1423:
1382:
1318:
that catalyzes the transfer of methyl groups to specific CpG structures in DNA, a process called
187:
1357:
DNA methylation modifies the information passed on by the parent to the progeny, it enables key
1231:
1227:
1198:
1188:
1184:
1180:
1152:
1148:
1105:
1101:
1097:
1093:
2356:
Cannella N, Oliveira AM, Hemstedt T, Lissek T, Buechler E, Bading H, Spanagel R (August 2018).
2014:
Steensma DP, Bejar R, Jaiswal S, Lindsley RC, Sekeres MA, Hasserjian RP, Ebert BL (July 2015).
590:
4511:
4468:
4439:
4388:
4359:
4306:
4276:
4241:
4212:
4169:
4134:
4097:
4051:
4008:
3965:
3922:
3887:
3855:
3806:
3757:
3697:
3648:
3592:
3540:
3491:
3434:
3399:
3342:
3293:
3244:
3195:
3144:
3109:
3060:
3011:
2965:
2913:
2861:
2801:
2744:
2687:
2638:
2589:
2538:
2489:
2438:
2387:
2330:
2281:
2243:
2189:
2145:
2094:
2045:
1996:
1947:
1896:
1847:
1799:
1550:
1435:
135:
47:
3460:
Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, et al. (December 2010).
1967:"The genetics of myelodysplastic syndrome: from clonal haematopoiesis to secondary leukaemia"
4501:
4493:
4460:
4429:
4421:
4380:
4349:
4339:
4298:
4266:
4233:
4202:
4194:
4159:
4126:
4089:
4081:
4041:
4033:
3998:
3990:
3955:
3947:
3934:
Robertson KD, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, Gonzales FA, Jones PA (June 1999).
3912:
3845:
3837:
3796:
3788:
3747:
3739:
3687:
3638:
3630:
3582:
3574:
3530:
3522:
3481:
3473:
3426:
3389:
3381:
3332:
3324:
3283:
3275:
3234:
3187:
3136:
3099:
3091:
3050:
3003:
2955:
2947:
2903:
2895:
2851:
2843:
2791:
2783:
2734:
2726:
2677:
2669:
2628:
2620:
2579:
2569:
2528:
2520:
2479:
2469:
2428:
2418:
2377:
2369:
2320:
2273:
2233:
2225:
2179:
2135:
2125:
2084:
2076:
2063:
Robertson KD, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, Gonzales FA, Jones PA (June 1999).
2035:
2027:
1986:
1978:
1937:
1927:
1886:
1878:
1839:
1791:
1473:
432:
363:
307:
262:
183:
1546:
1400:
1374:
1319:
407:
4410:"The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase"
3826:"The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase"
3775:
Brenner C, Deplus R, Didelot C, Loriot A, Viré E, De Smet C, et al. (January 2005).
3127:
Reik W, Walter J (January 2001). "Genomic imprinting: parental influence on the genome".
2358:"Dnmt3a2 in the Nucleus Accumbens Shell Is Required for Reinstatement of Cocaine Seeking"
1583:
knockdown – Ctnb1 codes for β-catenin, which participates in self-renewal cell division.
4335:
4320:"The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a"
4077:
3683:
3377:
3183:
2999:
2839:
2779:
2722:
30:
3535:
3510:
3486:
3461:
3394:
3361:
3337:
3312:
3288:
3263:
2960:
2935:
2908:
2883:
2856:
2823:
2796:
2763:
2739:
2706:
2682:
2657:
2633:
2608:
2584:
2557:
2533:
2508:
2484:
2457:
2433:
2407:"The Roles of Human DNA Methyltransferases and Their Isoforms in Shaping the Epigenome"
2406:
2382:
2357:
2238:
2213:
2140:
2113:
2040:
2015:
1991:
1966:
1942:
1932:
1915:
1891:
1866:
1419:
1378:
4506:
4481:
4434:
4409:
4354:
4319:
4302:
4237:
4207:
4182:
4003:
3978:
3960:
3935:
3850:
3825:
3801:
3777:"Myc represses transcription through recruitment of DNA methyltransferase corepressor"
3776:
3752:
3727:
3692:
3667:
3643:
3618:
3587:
3562:
3104:
3079:
2658:"The inactive Dnmt3b3 isoform preferentially enhances Dnmt3b-mediated DNA methylation"
2089:
2064:
1843:
1545:
Research on long-term memory storage in humans indicates that memory is maintained by
899:
894:
889:
884:
879:
874:
869:
864:
859:
854:
849:
844:
839:
834:
829:
824:
819:
814:
809:
804:
799:
794:
789:
784:
779:
774:
769:
764:
759:
743:
738:
733:
728:
723:
718:
713:
708:
692:
687:
682:
677:
672:
667:
662:
657:
652:
647:
642:
637:
632:
4527:
4271:
4254:
4046:
4021:
3078:
Pradhan S, Talbot D, Sha M, Benner J, Hornstra L, Li E, et al. (November 1997).
1366:
619:
4109:
3446:
3311:
Challen GA, Sun D, Jeong M, Luo M, Jelinek J, Berg JS, et al. (December 2011).
3156:
2342:
2293:
1492:
The catalytic domain (the methyltransferase domain) is highly conserved, even among
167:
3709:
3509:
Banaszak LG, Giudice V, Zhao X, Wu Z, Gao S, Hosokawa K, et al. (March 2018).
3207:
3023:
2762:
Xu TH, Liu M, Zhou XE, Liang G, Zhao G, Xu HE, Melcher K, Jones PA (October 2020).
2373:
1608:
1579:
425:
204:
4400:
2882:
Manzo M, Wirz J, Ambrosi C, Villaseñor R, Roschitzki B, Baubec T (December 2017).
2707:"Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation"
1811:
3360:
Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. (October 2013).
3239:
3222:
2031:
191:
1766:
National Center for
Biotechnology Information, U.S. National Library of Medicine
1748:
National Center for
Biotechnology Information, U.S. National Library of Medicine
1481:
1358:
4324:
Proceedings of the
National Academy of Sciences of the United States of America
3994:
3743:
3526:
2847:
2624:
508:
4480:
Ling Y, Sankpal UT, Robertson AK, McNally JG, Karpova T, Robertson KD (2004).
3617:
Ling Y, Sankpal UT, Robertson AK, McNally JG, Karpova T, Robertson KD (2004).
2787:
2524:
1493:
1477:
1404:
324:
221:
171:
3951:
3792:
3095:
3055:
3038:
2080:
4085:
3901:"Normalization and subtraction: two approaches to facilitate gene discovery"
2936:"DNA methylation in human epigenomes depends on local topology of CpG sites"
2899:
2509:"PWWP domains and their modes of sensing DNA and histone methylated lysines"
1882:
929:
568:
446:
391:
378:
290:
277:
179:
4515:
4472:
4443:
4392:
4363:
4344:
4310:
4280:
4255:"Dnmt3a and Dnmt1 functionally cooperate during de novo methylation of DNA"
4245:
4216:
4198:
4173:
4164:
4147:
4138:
4130:
4101:
4055:
4037:
4012:
3969:
3859:
3810:
3761:
3701:
3652:
3596:
3578:
3544:
3495:
3438:
3403:
3346:
3297:
3248:
3199:
3148:
3064:
3015:
2969:
2917:
2865:
2805:
2748:
2691:
2673:
2642:
2593:
2542:
2493:
2474:
2442:
2391:
2334:
2285:
2247:
2193:
2184:
2167:
2149:
2098:
2049:
2000:
1951:
1900:
1851:
4464:
3926:
3891:
3477:
3279:
3113:
2423:
1803:
1334:
1295:
1290:
4497:
4425:
4093:
3841:
3634:
2951:
2574:
2130:
1982:
1526:
1522:
1517:
1509:
1279:
1074:
1055:
3385:
3191:
2764:"Structure of nucleosome-bound DNA methyltransferases DNMT3A and DNMT3B"
2730:
2325:
2308:
3917:
3900:
1668:
1439:
1427:
1403:
contribute to the formation of a genetically distinct subpopulation of
1041:
996:
112:
108:
104:
100:
96:
92:
88:
84:
80:
76:
3140:
3007:
1865:
Jia Y, Li P, Fang L, Zhu H, Xu L, Cheng H, et al. (2016-04-12).
1680:
1638:
1462:
1347:
1315:
1263:
951:
683:
RNA polymerase II cis-regulatory region sequence-specific DNA binding
678:
DNA-binding transcription factor activity, RNA polymerase II-specific
3430:
3362:"Mutational landscape and significance across 12 major cancer types"
3328:
2277:
2229:
4384:
4226:
Biochimica et
Biophysica Acta (BBA) - Gene Structure and Expression
3264:"Experience-dependent epigenomic reorganization in the hippocampus"
2705:
Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X (September 2007).
2607:
Xu F, Mao C, Ding Y, Rui C, Wu L, Shi A, et al. (2010-01-01).
1674:
1656:
1644:
1632:
1549:, Rats in which a new, strong long-term memory is induced due to
1343:
3313:"Dnmt3a is essential for hematopoietic stem cell differentiation"
2114:"DNA Methyltransferases: From Evolution to Clinical Applications"
914:
910:
4181:
Kim GD, Ni J, Kelesoglu N, Roberts RJ, Pradhan S (August 2002).
3561:
Kim GD, Ni J, Kelesoglu N, Roberts RJ, Pradhan S (August 2002).
1795:
1326:
159:
3977:
Fuks F, Burgers WA, Godin N, Kasai M, Kouzarides T (May 2001).
3726:
Fuks F, Burgers WA, Godin N, Kasai M, Kouzarides T (May 2001).
2558:"Structural Basis of DNMT1 and DNMT3A-Mediated DNA Methylation"
599:
3262:
Duke CG, Kennedy AJ, Gavin CF, Day JJ, Sweatt JD (July 2017).
2307:
Oliveira AM, Hemstedt TJ, Freitag HE, Bading H (August 2016).
1650:
3037:
Pradhan S, Bacolla A, Wells RD, Roberts RJ (November 1999).
1542:
phylogenesis. DNMT3A is essential for genetic imprinting.
4253:
Fatemi M, Hermann A, Gowher H, Jeltsch A (October 2002).
3223:"Covalent modification of DNA regulates memory formation"
795:
negative regulation of transcription by RNA polymerase II
4020:
Vaute O, Nicolas E, Vandel L, Trouche D (January 2002).
2934:
Lövkvist C, Dodd IB, Sneppen K, Haerter JO (June 2016).
1422:
found on chromosome 2p23 in humans. There exists a 98%
415:
1430:
homologues. DNMT3A is widely expressed among mammals.
580:
4408:
Fuks F, Hurd PJ, Deplus R, Kouzarides T (May 2003).
3824:
Fuks F, Hurd PJ, Deplus R, Kouzarides T (May 2003).
1595:
project DNMT3A mutations were most commonly seen in
2405:Gujar H, Weisenberger DJ, Liang G (February 2019).
1216:
1171:
1137:
1092:
765:
regulation of gene expression by genetic imprinting
3899:Bonaldo MF, Lennon G, Soares MB (September 1996).
2507:Rona GB, Eleutherio EC, Pinheiro AS (March 2016).
2309:"Dnmt3a2: a hub for enhancing cognitive functions"
1722:
1720:
1718:
1701:
1699:
1697:
835:DNA methylation-dependent heterochromatin assembly
820:negative regulation of gene expression, epigenetic
2981:
2979:
1965:Sperling AS, Gibson CJ, Ebert BL (January 2017).
362:
261:
4318:Chedin F, Lieber MR, Hsieh CL (December 2002).
3721:
3719:
3612:
3610:
3608:
3606:
3556:
3554:
2259:
2257:
2161:
2159:
16:Protein-coding gene in the species Homo sapiens
1727:GRCm38: Ensembl release 89: ENSMUSG00000020661
1361:modifications essential for processes such as
865:DNA methylation involved in embryo development
1395:, a common aging-related phenomenon in which
800:DNA methylation involved in gamete generation
8:
4146:Chen T, Ueda Y, Xie S, Li E (October 2002).
3462:"DNMT3A mutations in acute myeloid leukemia"
2207:
2205:
2203:
2166:Chen T, Ueda Y, Xie S, Li E (October 2002).
1777:
1775:
1469:pocket that includes the PWWP motif itself.
1342:enzymes, which consists of the protagonists
673:DNA (cytosine-5-)-methyltransferase activity
2929:
2927:
2118:International Journal of Molecular Sciences
1914:Jan M, Ebert BL, Jaiswal S (January 2017).
1706:GRCh38: Ensembl release 89: ENSG00000119772
1472:The ADD domain of DNMT3A is composed of an
1391:is the gene most commonly found mutated in
1825:
1823:
1821:
1322:. The enzyme is encoded in humans by the
1168:
1089:
925:
615:
489:Skeletal muscle tissue of rectus abdominis
403:
302:
199:
65:
4505:
4433:
4353:
4343:
4270:
4206:
4163:
4045:
4002:
3959:
3916:
3849:
3800:
3751:
3691:
3642:
3586:
3534:
3485:
3393:
3336:
3287:
3238:
3103:
3054:
2959:
2907:
2855:
2795:
2738:
2681:
2632:
2583:
2573:
2532:
2483:
2473:
2432:
2422:
2381:
2324:
2237:
2183:
2139:
2129:
2088:
2039:
1990:
1941:
1931:
1890:
2877:
2875:
2212:Yang L, Rau R, Goodell MA (March 2015).
1607:, in that they have much more erroneous
1498:
1452:
855:cellular response to amino acid stimulus
469:Skeletal muscle tissue of biceps brachii
2214:"DNMT3A in haematological malignancies"
1693:
2817:
2815:
2556:Ren W, Gao L, Song J (December 2018).
20:
3515:Blood Cells, Molecules & Diseases
1525:depends on the sequence flanking the
1308:DNA (cytosine-5)-methyltransferase 3A
367:
328:
323:
266:
225:
220:
7:
4152:The Journal of Biological Chemistry
3466:The New England Journal of Medicine
3221:Miller CA, Sweatt JD (March 2007).
3043:The Journal of Biological Chemistry
2172:The Journal of Biological Chemistry
1418:is a 130 kDa protein encoded by 23
1933:10.1053/j.seminhematol.2016.10.002
1213:
1134:
1065:
1046:
1020:
1001:
975:
956:
688:transcription corepressor activity
585:
503:
441:
420:
14:
4453:International Journal of Oncology
780:positive regulation of cell death
4272:10.1046/j.1432-1033.2002.03198.x
4259:European Journal of Biochemistry
1474:N-terminal GATA-like zinc finger
589:
351:
344:
338:
315:
250:
243:
237:
212:
29:
1577:HSCs experienced an additional
1397:hematopoietic stem cells (HSCs)
1332:This enzyme is responsible for
2374:10.1523/JNEUROSCI.0600-18.2018
760:response to ionizing radiation
739:chromosome, centromeric region
648:DNA-methyltransferase activity
600:More reference expression data
569:More reference expression data
1:
4303:10.1016/S0378-1119(02)00976-9
4238:10.1016/S0167-4781(02)00482-7
3693:10.1016/s0960-9822(03)00432-9
1844:10.1016/S0378-1119(99)00252-8
860:regulation of gene expression
336:
235:
3240:10.1016/j.neuron.2007.02.022
2032:10.1182/blood-2015-03-631747
1613:Tatton-Brown–Rahman syndrome
1603:-mutated cell lines exhibit
1551:contextual fear conditioning
885:hepatocyte apoptotic process
880:cellular response to hypoxia
785:cellular response to ethanol
693:transcription factor binding
4534:Genes on human chromosome 2
2613:Current Medicinal Chemistry
2462:Epigenetics & Chromatin
2362:The Journal of Neuroscience
1558:methylations of the genes.
895:DNA methylation on cytosine
875:response to toxic substance
825:response to nutrient levels
770:C-5 methylation of cytosine
4550:
3527:10.1016/j.bcmd.2017.12.002
2848:10.1038/s41467-020-17109-4
2625:10.2174/092986710793205372
1615:, an overgrowth disorder.
1371:transcriptional regulation
633:methyltransferase activity
2788:10.1038/s41586-020-2747-1
2525:10.1007/s12551-015-0190-6
1762:"Mouse PubMed Reference:"
1744:"Human PubMed Reference:"
1623:DNMT3A has been shown to
1605:transcriptome instability
1294:
1289:
1285:
1278:
1262:
1243:
1220:
1195:
1164:
1141:
1116:
1085:
1072:
1068:
1053:
1049:
1040:
1027:
1023:
1008:
1004:
995:
982:
978:
963:
959:
950:
935:
928:
924:
908:
668:identical protein binding
618:
614:
597:
588:
579:
566:
547:medullary collecting duct
527:lamina propria of urethra
515:
506:
453:
444:
414:
406:
402:
385:
372:
335:
314:
305:
301:
284:
271:
234:
211:
202:
198:
153:
150:
140:
133:
128:
73:
68:
51:
46:
41:
37:
28:
23:
3995:10.1093/emboj/20.10.2536
3793:10.1038/sj.emboj.7600509
3744:10.1093/emboj/20.10.2536
3129:Nature Reviews. Genetics
3056:10.1074/jbc.274.46.33002
1571:hematopoietic stem cells
1363:cellular differentiation
555:Rostral migratory stream
4086:10.1126/science.1065173
2900:10.15252/embj.201797038
1883:10.1038/celldisc.2016.7
1597:acute myeloid leukaemia
1529:within the linker DNA.
1436:nucleus accumbens shell
1249:Chr 2: 25.23 – 25.34 Mb
551:muscle layer of urethra
4486:Nucleic Acids Research
4414:Nucleic Acids Research
4345:10.1073/pnas.262443999
4165:10.1074/jbc.M205312200
4131:10.1242/dev.129.8.1983
4026:Nucleic Acids Research
3952:10.1093/nar/27.11.2291
3940:Nucleic Acids Research
3830:Nucleic Acids Research
3623:Nucleic Acids Research
3419:Nature Reviews. Cancer
3096:10.1093/nar/25.22.4666
3084:Nucleic Acids Research
2674:10.1101/gad.341925.120
2475:10.1186/1756-8935-6-12
2218:Nature Reviews. Cancer
2185:10.1074/jbc.M205312200
2081:10.1093/nar/27.11.2291
2069:Nucleic Acids Research
1971:Nature Reviews. Cancer
1920:Seminars in Hematology
1916:"Clonal hematopoiesis"
1505:
1458:
1385:and genome stability.
1256:Chr 12: 3.86 – 3.96 Mb
900:chromatin organization
845:neuron differentiation
4465:10.3892/ijo.22.6.1201
3886:(6547 Suppl): 3–174.
3478:10.1056/NEJMoa1005143
3280:10.1101/lm.045112.117
2424:10.3390/genes10020172
1502:
1456:
1399:or other early blood
1367:embryonic development
1340:DNA methyltransferase
805:response to vitamin A
775:response to estradiol
539:epithelium of urethra
330:Chromosome 12 (mouse)
4199:10.1093/emboj/cdf401
4038:10.1093/nar/30.2.475
3579:10.1093/emboj/cdf401
2668:(21–22): 1546–1558.
2575:10.3390/genes9120620
2313:Molecular Psychiatry
2131:10.3390/ijms23168994
1983:10.1038/nrc.2016.112
1393:clonal hematopoiesis
830:response to lead ion
638:transferase activity
228:Chromosome 2 (human)
69:List of PDB id codes
42:Available structures
4336:2002PNAS...9916916C
4330:(26): 16916–16921.
4158:(41): 38746–38754.
4078:2002Sci...295.1079D
4072:(5557): 1079–1082.
3684:2003CBio...13.1192L
3386:10.1038/nature12634
3378:2013Natur.502..333K
3192:10.1038/nature02633
3184:2004Natur.429..900K
3049:(46): 33002–33010.
3000:2000Natur.404.1003R
2994:(6781): 1003–1007.
2840:2020NatCo..11.3355G
2780:2020Natur.586..151X
2731:10.1038/nature06146
2723:2007Natur.449..248J
2513:Biophysical Reviews
2326:10.1038/mp.2015.175
2266:Nature Neuroscience
2178:(41): 38746–38754.
1593:Cancer Genome Atlas
890:response to cocaine
870:response to ethanol
461:ganglionic eminence
4498:10.1093/nar/gkh195
4426:10.1093/nar/gkg332
3918:10.1101/gr.6.9.791
3842:10.1093/nar/gkg332
3635:10.1093/nar/gkh195
2952:10.1093/nar/gkw124
1587:Clinical relevance
1506:
1459:
1426:between human and
1030:ENSMUSG00000020661
850:mitotic cell cycle
753:Biological process
702:Cellular component
626:Molecular function
192:DNMT3A - orthologs
4265:(20): 4981–4984.
4193:(15): 4183–4195.
3989:(10): 2536–2544.
3946:(11): 2291–2298.
3738:(10): 2536–2544.
3678:(14): 1192–1200.
3573:(15): 4183–4195.
3472:(25): 2424–2433.
3372:(7471): 333–339.
3178:(6994): 900–903.
3090:(22): 4666–4673.
2940:Nucleic Acids Res
2894:(23): 3421–3434.
2774:(7827): 151–155.
2619:(33): 4052–4071.
2368:(34): 7516–7528.
2075:(11): 2291–2298.
1480:and a C-terminal
1446:Protein structure
1305:
1304:
1301:
1300:
1274:
1273:
1239:
1238:
1210:
1209:
1160:
1159:
1131:
1130:
1081:
1080:
1062:
1061:
1036:
1035:
1017:
1016:
991:
990:
972:
971:
920:
919:
658:metal ion binding
653:chromatin binding
610:
609:
606:
605:
575:
574:
562:
561:
500:
499:
398:
397:
297:
296:
124:
123:
120:
119:
52:Ortholog search:
4541:
4519:
4509:
4476:
4459:(6): 1201–1207.
4447:
4437:
4420:(9): 2305–2312.
4404:
4367:
4357:
4347:
4314:
4284:
4274:
4249:
4220:
4210:
4187:The EMBO Journal
4177:
4167:
4142:
4125:(8): 1983–1993.
4113:
4059:
4049:
4016:
4006:
3983:The EMBO Journal
3973:
3963:
3930:
3920:
3895:
3864:
3863:
3853:
3836:(9): 2305–2312.
3821:
3815:
3814:
3804:
3781:The EMBO Journal
3772:
3766:
3765:
3755:
3732:The EMBO Journal
3723:
3714:
3713:
3695:
3663:
3657:
3656:
3646:
3614:
3601:
3600:
3590:
3567:The EMBO Journal
3558:
3549:
3548:
3538:
3506:
3500:
3499:
3489:
3457:
3451:
3450:
3414:
3408:
3407:
3397:
3357:
3351:
3350:
3340:
3308:
3302:
3301:
3291:
3259:
3253:
3252:
3242:
3218:
3212:
3211:
3167:
3161:
3160:
3141:10.1038/35047554
3124:
3118:
3117:
3107:
3075:
3069:
3068:
3058:
3034:
3028:
3027:
3008:10.1038/35010000
2983:
2974:
2973:
2963:
2931:
2922:
2921:
2911:
2879:
2870:
2869:
2859:
2819:
2810:
2809:
2799:
2759:
2753:
2752:
2742:
2717:(7159): 248–51.
2702:
2696:
2695:
2685:
2653:
2647:
2646:
2636:
2604:
2598:
2597:
2587:
2577:
2553:
2547:
2546:
2536:
2504:
2498:
2497:
2487:
2477:
2453:
2447:
2446:
2436:
2426:
2402:
2396:
2395:
2385:
2353:
2347:
2346:
2328:
2319:(8): 1130–1136.
2304:
2298:
2297:
2272:(8): 1111–1113.
2261:
2252:
2251:
2241:
2209:
2198:
2197:
2187:
2163:
2154:
2153:
2143:
2133:
2109:
2103:
2102:
2092:
2060:
2054:
2053:
2043:
2011:
2005:
2004:
1994:
1962:
1956:
1955:
1945:
1935:
1911:
1905:
1904:
1894:
1862:
1856:
1855:
1827:
1816:
1815:
1779:
1770:
1769:
1758:
1752:
1751:
1740:
1734:
1724:
1713:
1703:
1401:cell progenitors
1287:
1286:
1258:
1251:
1234:
1214:
1205:
1169:
1165:RefSeq (protein)
1155:
1135:
1126:
1090:
1066:
1047:
1021:
1002:
976:
957:
926:
616:
602:
593:
586:
571:
535:genital tubercle
531:ejaculatory duct
511:
509:Top expressed in
504:
465:ventricular zone
449:
447:Top expressed in
442:
421:
404:
394:
381:
370:
355:
348:
342:
331:
319:
303:
293:
280:
269:
254:
247:
241:
230:
216:
200:
194:
145:
138:
115:
66:
60:
39:
38:
33:
21:
4549:
4548:
4544:
4543:
4542:
4540:
4539:
4538:
4524:
4523:
4522:
4479:
4450:
4407:
4373:Nature Genetics
4370:
4317:
4287:
4252:
4223:
4180:
4145:
4116:
4062:
4019:
3976:
3933:
3905:Genome Research
3898:
3876:
3872:
3870:Further reading
3867:
3823:
3822:
3818:
3774:
3773:
3769:
3725:
3724:
3717:
3672:Current Biology
3665:
3664:
3660:
3616:
3615:
3604:
3560:
3559:
3552:
3508:
3507:
3503:
3459:
3458:
3454:
3431:10.1038/nrc3343
3416:
3415:
3411:
3359:
3358:
3354:
3329:10.1038/ng.1009
3317:Nature Genetics
3310:
3309:
3305:
3261:
3260:
3256:
3220:
3219:
3215:
3169:
3168:
3164:
3126:
3125:
3121:
3077:
3076:
3072:
3036:
3035:
3031:
2985:
2984:
2977:
2946:(11): 5123–32.
2933:
2932:
2925:
2881:
2880:
2873:
2821:
2820:
2813:
2761:
2760:
2756:
2704:
2703:
2699:
2655:
2654:
2650:
2606:
2605:
2601:
2555:
2554:
2550:
2506:
2505:
2501:
2455:
2454:
2450:
2404:
2403:
2399:
2355:
2354:
2350:
2306:
2305:
2301:
2278:10.1038/nn.3151
2263:
2262:
2255:
2230:10.1038/nrc3895
2211:
2210:
2201:
2165:
2164:
2157:
2111:
2110:
2106:
2062:
2061:
2057:
2013:
2012:
2008:
1964:
1963:
1959:
1913:
1912:
1908:
1864:
1863:
1859:
1829:
1828:
1819:
1784:Nature Genetics
1781:
1780:
1773:
1760:
1759:
1755:
1742:
1741:
1737:
1725:
1716:
1704:
1695:
1691:
1686:
1621:
1589:
1564:
1547:DNA methylation
1535:
1448:
1438:in response to
1413:
1375:heterochromatin
1320:DNA methylation
1296:View/Edit Mouse
1291:View/Edit Human
1254:
1247:
1244:Location (UCSC)
1230:
1226:
1222:
1201:
1197:
1191:
1187:
1183:
1179:
1175:
1151:
1147:
1143:
1122:
1118:
1112:
1108:
1104:
1100:
1096:
1011:ENSG00000119772
904:
840:spermatogenesis
810:DNA methylation
748:
729:heterochromatin
697:
663:protein binding
598:
567:
558:
553:
549:
545:
541:
537:
533:
529:
525:
521:
507:
496:
491:
487:
485:muscle of thigh
483:
479:
475:
471:
467:
463:
459:
445:
389:
376:
369:12|12 A1.1
368:
358:
357:
356:
349:
329:
306:Gene location (
288:
275:
267:
257:
256:
255:
248:
226:
203:Gene location (
154:
141:
134:
75:
53:
17:
12:
11:
5:
4547:
4545:
4537:
4536:
4526:
4525:
4521:
4520:
4492:(2): 598–610.
4477:
4448:
4405:
4385:10.1038/ng1068
4368:
4315:
4285:
4250:
4232:(3): 457–465.
4221:
4178:
4143:
4114:
4060:
4032:(2): 475–481.
4017:
3974:
3931:
3911:(9): 791–806.
3896:
3873:
3871:
3868:
3866:
3865:
3816:
3787:(2): 336–346.
3767:
3715:
3658:
3629:(2): 598–610.
3602:
3550:
3501:
3452:
3425:(9): 599–612.
3409:
3352:
3303:
3274:(7): 278–288.
3254:
3233:(6): 857–869.
3213:
3162:
3119:
3070:
3029:
2975:
2923:
2871:
2811:
2754:
2697:
2648:
2599:
2548:
2499:
2448:
2397:
2348:
2299:
2253:
2224:(3): 152–165.
2199:
2155:
2104:
2055:
2006:
1957:
1906:
1871:Cell Discovery
1857:
1817:
1790:(3): 219–220.
1771:
1753:
1735:
1714:
1692:
1690:
1687:
1685:
1684:
1678:
1672:
1666:
1660:
1654:
1648:
1642:
1636:
1629:
1620:
1617:
1588:
1585:
1563:
1562:Animal studies
1560:
1534:
1531:
1447:
1444:
1412:
1409:
1379:X-inactivation
1303:
1302:
1299:
1298:
1293:
1283:
1282:
1276:
1275:
1272:
1271:
1269:
1267:
1260:
1259:
1252:
1245:
1241:
1240:
1237:
1236:
1218:
1217:
1211:
1208:
1207:
1193:
1192:
1166:
1162:
1161:
1158:
1157:
1139:
1138:
1132:
1129:
1128:
1114:
1113:
1087:
1083:
1082:
1079:
1078:
1070:
1069:
1063:
1060:
1059:
1051:
1050:
1044:
1038:
1037:
1034:
1033:
1025:
1024:
1018:
1015:
1014:
1006:
1005:
999:
993:
992:
989:
988:
980:
979:
973:
970:
969:
961:
960:
954:
948:
947:
942:
937:
933:
932:
922:
921:
918:
917:
906:
905:
903:
902:
897:
892:
887:
882:
877:
872:
867:
862:
857:
852:
847:
842:
837:
832:
827:
822:
817:
812:
807:
802:
797:
792:
787:
782:
777:
772:
767:
762:
756:
754:
750:
749:
747:
746:
741:
736:
731:
726:
721:
719:nuclear matrix
716:
711:
705:
703:
699:
698:
696:
695:
690:
685:
680:
675:
670:
665:
660:
655:
650:
645:
640:
635:
629:
627:
623:
622:
612:
611:
608:
607:
604:
603:
595:
594:
583:
577:
576:
573:
572:
564:
563:
560:
559:
557:
556:
552:
548:
544:
543:female urethra
540:
536:
532:
528:
524:
520:
519:tail of embryo
516:
513:
512:
501:
498:
497:
495:
494:
490:
486:
482:
478:
474:
470:
466:
462:
458:
454:
451:
450:
438:
437:
429:
418:
412:
411:
408:RNA expression
400:
399:
396:
395:
387:
383:
382:
374:
371:
366:
360:
359:
350:
343:
337:
333:
332:
327:
321:
320:
312:
311:
299:
298:
295:
294:
286:
282:
281:
273:
270:
265:
259:
258:
249:
242:
236:
232:
231:
224:
218:
217:
209:
208:
196:
195:
152:
148:
147:
139:
131:
130:
126:
125:
122:
121:
118:
117:
71:
70:
62:
61:
50:
44:
43:
35:
34:
26:
25:
15:
13:
10:
9:
6:
4:
3:
2:
4546:
4535:
4532:
4531:
4529:
4517:
4513:
4508:
4503:
4499:
4495:
4491:
4487:
4483:
4478:
4474:
4470:
4466:
4462:
4458:
4454:
4449:
4445:
4441:
4436:
4431:
4427:
4423:
4419:
4415:
4411:
4406:
4402:
4398:
4394:
4390:
4386:
4382:
4378:
4374:
4369:
4365:
4361:
4356:
4351:
4346:
4341:
4337:
4333:
4329:
4325:
4321:
4316:
4312:
4308:
4304:
4300:
4296:
4292:
4286:
4282:
4278:
4273:
4268:
4264:
4260:
4256:
4251:
4247:
4243:
4239:
4235:
4231:
4227:
4222:
4218:
4214:
4209:
4204:
4200:
4196:
4192:
4188:
4184:
4179:
4175:
4171:
4166:
4161:
4157:
4153:
4149:
4144:
4140:
4136:
4132:
4128:
4124:
4120:
4115:
4111:
4107:
4103:
4099:
4095:
4094:11576/2506625
4091:
4087:
4083:
4079:
4075:
4071:
4067:
4061:
4057:
4053:
4048:
4043:
4039:
4035:
4031:
4027:
4023:
4018:
4014:
4010:
4005:
4000:
3996:
3992:
3988:
3984:
3980:
3975:
3971:
3967:
3962:
3957:
3953:
3949:
3945:
3941:
3937:
3932:
3928:
3924:
3919:
3914:
3910:
3906:
3902:
3897:
3893:
3889:
3885:
3881:
3875:
3874:
3869:
3861:
3857:
3852:
3847:
3843:
3839:
3835:
3831:
3827:
3820:
3817:
3812:
3808:
3803:
3798:
3794:
3790:
3786:
3782:
3778:
3771:
3768:
3763:
3759:
3754:
3749:
3745:
3741:
3737:
3733:
3729:
3722:
3720:
3716:
3711:
3707:
3703:
3699:
3694:
3689:
3685:
3681:
3677:
3673:
3669:
3662:
3659:
3654:
3650:
3645:
3640:
3636:
3632:
3628:
3624:
3620:
3613:
3611:
3609:
3607:
3603:
3598:
3594:
3589:
3584:
3580:
3576:
3572:
3568:
3564:
3557:
3555:
3551:
3546:
3542:
3537:
3532:
3528:
3524:
3520:
3516:
3512:
3505:
3502:
3497:
3493:
3488:
3483:
3479:
3475:
3471:
3467:
3463:
3456:
3453:
3448:
3444:
3440:
3436:
3432:
3428:
3424:
3420:
3413:
3410:
3405:
3401:
3396:
3391:
3387:
3383:
3379:
3375:
3371:
3367:
3363:
3356:
3353:
3348:
3344:
3339:
3334:
3330:
3326:
3322:
3318:
3314:
3307:
3304:
3299:
3295:
3290:
3285:
3281:
3277:
3273:
3269:
3265:
3258:
3255:
3250:
3246:
3241:
3236:
3232:
3228:
3224:
3217:
3214:
3209:
3205:
3201:
3197:
3193:
3189:
3185:
3181:
3177:
3173:
3166:
3163:
3158:
3154:
3150:
3146:
3142:
3138:
3134:
3130:
3123:
3120:
3115:
3111:
3106:
3101:
3097:
3093:
3089:
3085:
3081:
3074:
3071:
3066:
3062:
3057:
3052:
3048:
3044:
3040:
3033:
3030:
3025:
3021:
3017:
3013:
3009:
3005:
3001:
2997:
2993:
2989:
2982:
2980:
2976:
2971:
2967:
2962:
2957:
2953:
2949:
2945:
2941:
2937:
2930:
2928:
2924:
2919:
2915:
2910:
2905:
2901:
2897:
2893:
2889:
2885:
2878:
2876:
2872:
2867:
2863:
2858:
2853:
2849:
2845:
2841:
2837:
2833:
2829:
2825:
2818:
2816:
2812:
2807:
2803:
2798:
2793:
2789:
2785:
2781:
2777:
2773:
2769:
2765:
2758:
2755:
2750:
2746:
2741:
2736:
2732:
2728:
2724:
2720:
2716:
2712:
2708:
2701:
2698:
2693:
2689:
2684:
2679:
2675:
2671:
2667:
2663:
2659:
2652:
2649:
2644:
2640:
2635:
2630:
2626:
2622:
2618:
2614:
2610:
2603:
2600:
2595:
2591:
2586:
2581:
2576:
2571:
2567:
2563:
2559:
2552:
2549:
2544:
2540:
2535:
2530:
2526:
2522:
2518:
2514:
2510:
2503:
2500:
2495:
2491:
2486:
2481:
2476:
2471:
2467:
2463:
2459:
2452:
2449:
2444:
2440:
2435:
2430:
2425:
2420:
2416:
2412:
2408:
2401:
2398:
2393:
2389:
2384:
2379:
2375:
2371:
2367:
2363:
2359:
2352:
2349:
2344:
2340:
2336:
2332:
2327:
2322:
2318:
2314:
2310:
2303:
2300:
2295:
2291:
2287:
2283:
2279:
2275:
2271:
2267:
2260:
2258:
2254:
2249:
2245:
2240:
2235:
2231:
2227:
2223:
2219:
2215:
2208:
2206:
2204:
2200:
2195:
2191:
2186:
2181:
2177:
2173:
2169:
2162:
2160:
2156:
2151:
2147:
2142:
2137:
2132:
2127:
2123:
2119:
2115:
2108:
2105:
2100:
2096:
2091:
2086:
2082:
2078:
2074:
2070:
2066:
2059:
2056:
2051:
2047:
2042:
2037:
2033:
2029:
2025:
2021:
2017:
2010:
2007:
2002:
1998:
1993:
1988:
1984:
1980:
1976:
1972:
1968:
1961:
1958:
1953:
1949:
1944:
1939:
1934:
1929:
1925:
1921:
1917:
1910:
1907:
1902:
1898:
1893:
1888:
1884:
1880:
1876:
1872:
1868:
1861:
1858:
1853:
1849:
1845:
1841:
1837:
1833:
1826:
1824:
1822:
1818:
1813:
1809:
1805:
1801:
1797:
1793:
1789:
1785:
1778:
1776:
1772:
1767:
1763:
1757:
1754:
1749:
1745:
1739:
1736:
1732:
1728:
1723:
1721:
1719:
1715:
1711:
1707:
1702:
1700:
1698:
1694:
1688:
1682:
1679:
1676:
1673:
1670:
1667:
1664:
1661:
1658:
1655:
1652:
1649:
1646:
1643:
1640:
1637:
1634:
1631:
1630:
1628:
1626:
1618:
1616:
1614:
1610:
1606:
1602:
1598:
1594:
1586:
1584:
1582:
1581:
1576:
1572:
1567:
1561:
1559:
1557:
1552:
1548:
1543:
1539:
1532:
1530:
1528:
1524:
1519:
1514:
1511:
1501:
1497:
1495:
1490:
1487:
1483:
1479:
1475:
1470:
1466:
1464:
1455:
1451:
1445:
1443:
1441:
1437:
1431:
1429:
1425:
1421:
1417:
1410:
1408:
1406:
1402:
1398:
1394:
1390:
1386:
1384:
1380:
1376:
1372:
1368:
1364:
1360:
1356:
1351:
1349:
1346:, DNMT3A and
1345:
1341:
1337:
1336:
1330:
1328:
1325:
1321:
1317:
1313:
1309:
1297:
1292:
1288:
1284:
1281:
1277:
1270:
1268:
1265:
1261:
1257:
1253:
1250:
1246:
1242:
1235:
1233:
1229:
1225:
1219:
1215:
1212:
1206:
1204:
1200:
1194:
1190:
1186:
1182:
1178:
1174:
1170:
1167:
1163:
1156:
1154:
1150:
1146:
1140:
1136:
1133:
1127:
1125:
1121:
1115:
1111:
1107:
1103:
1099:
1095:
1091:
1088:
1086:RefSeq (mRNA)
1084:
1077:
1076:
1071:
1067:
1064:
1058:
1057:
1052:
1048:
1045:
1043:
1039:
1032:
1031:
1026:
1022:
1019:
1013:
1012:
1007:
1003:
1000:
998:
994:
987:
986:
981:
977:
974:
968:
967:
962:
958:
955:
953:
949:
946:
943:
941:
938:
934:
931:
927:
923:
916:
912:
907:
901:
898:
896:
893:
891:
888:
886:
883:
881:
878:
876:
873:
871:
868:
866:
863:
861:
858:
856:
853:
851:
848:
846:
843:
841:
838:
836:
833:
831:
828:
826:
823:
821:
818:
816:
813:
811:
808:
806:
803:
801:
798:
796:
793:
791:
788:
786:
783:
781:
778:
776:
773:
771:
768:
766:
763:
761:
758:
757:
755:
752:
751:
745:
742:
740:
737:
735:
732:
730:
727:
725:
722:
720:
717:
715:
712:
710:
707:
706:
704:
701:
700:
694:
691:
689:
686:
684:
681:
679:
676:
674:
671:
669:
666:
664:
661:
659:
656:
654:
651:
649:
646:
644:
641:
639:
636:
634:
631:
630:
628:
625:
624:
621:
620:Gene ontology
617:
613:
601:
596:
592:
587:
584:
582:
578:
570:
565:
554:
550:
546:
542:
538:
534:
530:
526:
522:
518:
517:
514:
510:
505:
502:
492:
488:
484:
480:
476:
472:
468:
464:
460:
456:
455:
452:
448:
443:
440:
439:
436:
434:
430:
428:
427:
423:
422:
419:
417:
413:
409:
405:
401:
393:
388:
384:
380:
375:
365:
361:
354:
347:
341:
334:
326:
322:
318:
313:
309:
304:
300:
292:
287:
283:
279:
274:
264:
260:
253:
246:
240:
233:
229:
223:
219:
215:
210:
206:
201:
197:
193:
189:
185:
181:
177:
173:
169:
165:
161:
157:
149:
144:
137:
132:
127:
116:
114:
110:
106:
102:
98:
94:
90:
86:
82:
78:
72:
67:
64:
63:
59:
56:
49:
45:
40:
36:
32:
27:
22:
19:
4489:
4485:
4456:
4452:
4417:
4413:
4379:(1): 61–65.
4376:
4372:
4327:
4323:
4297:(1): 91–99.
4294:
4290:
4262:
4258:
4229:
4225:
4190:
4186:
4155:
4151:
4122:
4118:
4069:
4065:
4029:
4025:
3986:
3982:
3943:
3939:
3908:
3904:
3883:
3879:
3833:
3829:
3819:
3784:
3780:
3770:
3735:
3731:
3675:
3671:
3661:
3626:
3622:
3570:
3566:
3518:
3514:
3504:
3469:
3465:
3455:
3422:
3418:
3412:
3369:
3365:
3355:
3323:(1): 23–31.
3320:
3316:
3306:
3271:
3267:
3257:
3230:
3226:
3216:
3175:
3171:
3165:
3135:(1): 21–32.
3132:
3128:
3122:
3087:
3083:
3073:
3046:
3042:
3032:
2991:
2987:
2943:
2939:
2891:
2887:
2831:
2827:
2771:
2767:
2757:
2714:
2710:
2700:
2665:
2661:
2651:
2616:
2612:
2602:
2565:
2561:
2551:
2519:(1): 63–74.
2516:
2512:
2502:
2465:
2461:
2451:
2414:
2410:
2400:
2365:
2361:
2351:
2316:
2312:
2302:
2269:
2265:
2221:
2217:
2175:
2171:
2124:(16): 8994.
2121:
2117:
2107:
2072:
2068:
2058:
2023:
2019:
2009:
1974:
1970:
1960:
1926:(1): 43–50.
1923:
1919:
1909:
1874:
1870:
1860:
1838:(1): 87–95.
1835:
1831:
1787:
1783:
1765:
1756:
1747:
1738:
1622:
1619:Interactions
1609:RNA splicing
1600:
1590:
1578:
1574:
1568:
1565:
1555:
1544:
1540:
1536:
1515:
1507:
1491:
1485:
1471:
1467:
1460:
1449:
1432:
1415:
1414:
1388:
1387:
1354:
1352:
1333:
1331:
1323:
1311:
1307:
1306:
1224:NP_001258682
1221:
1203:NP_001362748
1196:
1177:NP_001307822
1173:NP_001307821
1145:NM_001271753
1142:
1124:NM_001375819
1120:NM_001320893
1117:
1110:NM_001320892
1073:
1054:
1028:
1009:
983:
964:
944:
939:
790:human ageing
523:male urethra
431:
424:
151:External IDs
74:
18:
4119:Development
2834:(1): 3355.
2568:(12): 620.
2026:(1): 9–16.
1977:(1): 5–19.
1796:10.1038/890
1494:prokaryotes
1482:alpha helix
1405:blood cells
1377:formation,
815:methylation
724:nucleoplasm
714:euchromatin
643:DNA binding
481:right ovary
457:sural nerve
289:25,342,590
276:25,227,855
129:Identifiers
2828:Nat Commun
2417:(2): 172.
1733:, May 2017
1712:, May 2017
1689:References
1478:PHD finger
1383:imprinting
1359:epigenetic
493:left ovary
435:(ortholog)
390:3,964,443
377:3,856,007
172:HomoloGene
3521:: 10–22.
3268:Learn Mem
2662:Genes Dev
2468:(1): 12.
1877:: 16007.
1575:Dnmt3a-/-
1518:CpG sites
1232:NP_714965
1228:NP_031898
1199:NP_783329
1189:NP_783328
1185:NP_715640
1181:NP_072046
1153:NM_153743
1149:NM_007872
1106:NM_175630
1102:NM_175629
1098:NM_153759
1094:NM_022552
930:Orthologs
709:cytoplasm
180:GeneCards
4528:Category
4516:14752048
4473:12738984
4444:12711675
4393:12496760
4364:12481029
4311:12406579
4281:12383256
4246:12359337
4217:12145218
4174:12138111
4139:11934864
4110:29532358
4102:11834837
4056:11788710
4013:11350943
3970:10325416
3860:12711675
3811:15616584
3762:11350943
3702:12867029
3653:14752048
3597:12145218
3545:29324392
3496:21067377
3447:20214444
3439:22898539
3404:24132290
3347:22138693
3298:28620075
3249:17359920
3200:15215868
3157:12050251
3149:11253064
3065:10551868
3016:10801130
2970:26932361
2918:29074627
2866:32620778
2806:32968275
2749:17713477
2692:33004415
2643:20939822
2594:30544982
2543:28510146
2494:23656834
2443:30813436
2392:30030395
2343:25308306
2335:26598069
2294:10590208
2286:22751036
2248:25693834
2194:12138111
2150:36012258
2099:10325416
2050:25931582
2001:27834397
1952:28088988
1901:27462454
1852:10433969
1729:–
1708:–
1625:interact
1533:Function
1527:CpG site
1523:CpG site
1510:CpG site
1424:homology
1314:) is an
1280:Wikidata
909:Sources:
477:placenta
4332:Bibcode
4074:Bibcode
4066:Science
3927:8889548
3892:7566098
3710:2320997
3680:Bibcode
3536:6728079
3487:3201818
3395:3927368
3374:Bibcode
3338:3637952
3289:5473107
3208:4344982
3180:Bibcode
3114:9358180
3024:4425037
2996:Bibcode
2961:4914085
2909:5709737
2857:7335073
2836:Bibcode
2797:7540737
2776:Bibcode
2740:2712830
2719:Bibcode
2683:7608744
2634:3003592
2585:6316889
2534:5425739
2485:3663649
2434:6409524
2383:6596133
2239:5814392
2141:9409253
2041:4624443
1992:5470392
1943:8045769
1892:4849474
1804:9662389
1731:Ensembl
1710:Ensembl
1669:SUV39H1
1556:de novo
1486:de novo
1440:cocaine
1355:de novo
1335:de novo
1042:UniProt
997:Ensembl
936:Species
915:QuickGO
744:nucleus
734:XY body
410:pattern
168:1261827
136:Aliases
4514:
4507:373322
4504:
4471:
4442:
4435:154218
4432:
4401:561490
4399:
4391:
4362:
4355:139244
4352:
4309:
4279:
4244:
4215:
4208:126147
4205:
4172:
4137:
4108:
4100:
4054:
4044:
4011:
4004:125250
4001:
3968:
3961:148793
3958:
3925:
3890:
3880:Nature
3858:
3851:154218
3848:
3809:
3802:545804
3799:
3760:
3753:125250
3750:
3708:
3700:
3651:
3644:373322
3641:
3595:
3588:126147
3585:
3543:
3533:
3494:
3484:
3445:
3437:
3402:
3392:
3366:Nature
3345:
3335:
3296:
3286:
3247:
3227:Neuron
3206:
3198:
3172:Nature
3155:
3147:
3112:
3105:147102
3102:
3063:
3022:
3014:
2988:Nature
2968:
2958:
2916:
2906:
2888:EMBO J
2864:
2854:
2804:
2794:
2768:Nature
2747:
2737:
2711:Nature
2690:
2680:
2641:
2631:
2592:
2582:
2541:
2531:
2492:
2482:
2441:
2431:
2390:
2380:
2341:
2333:
2292:
2284:
2246:
2236:
2192:
2148:
2138:
2097:
2090:148793
2087:
2048:
2038:
1999:
1989:
1950:
1940:
1899:
1889:
1850:
1812:256263
1810:
1802:
1681:ZNF238
1639:DNMT3B
1627:with:
1601:DNMT3A
1463:DNMT3L
1428:murine
1416:DNMT3A
1389:DNMT3a
1353:While
1348:DNMT3B
1324:DNMT3A
1316:enzyme
1312:DNMT3A
1266:search
1264:PubMed
1075:O88508
1056:Q9Y6K1
952:Entrez
581:BioGPS
473:oocyte
268:2p23.3
184:DNMT3A
160:602769
143:DNMT3A
24:DNMT3A
4397:S2CID
4106:S2CID
4047:99834
3706:S2CID
3443:S2CID
3204:S2CID
3153:S2CID
3020:S2CID
2562:Genes
2411:Genes
2339:S2CID
2290:S2CID
2020:Blood
1808:S2CID
1675:UBE2I
1663:PIAS2
1657:PIAS1
1645:HDAC1
1633:DNMT1
1580:Ctnb1
1554:with
1504:here.
1420:exons
1344:DNMT1
985:13435
945:Mouse
940:Human
911:Amigo
433:Mouse
426:Human
373:Start
308:Mouse
272:Start
205:Human
4512:PMID
4469:PMID
4440:PMID
4389:PMID
4360:PMID
4307:PMID
4291:Gene
4277:PMID
4242:PMID
4230:1577
4213:PMID
4170:PMID
4135:PMID
4098:PMID
4052:PMID
4009:PMID
3966:PMID
3923:PMID
3888:PMID
3856:PMID
3807:PMID
3758:PMID
3698:PMID
3649:PMID
3593:PMID
3541:PMID
3492:PMID
3435:PMID
3400:PMID
3343:PMID
3294:PMID
3245:PMID
3196:PMID
3145:PMID
3110:PMID
3061:PMID
3012:PMID
2966:PMID
2914:PMID
2862:PMID
2802:PMID
2745:PMID
2688:PMID
2639:PMID
2590:PMID
2539:PMID
2490:PMID
2439:PMID
2388:PMID
2331:PMID
2282:PMID
2244:PMID
2190:PMID
2146:PMID
2095:PMID
2046:PMID
1997:PMID
1948:PMID
1897:PMID
1848:PMID
1832:Gene
1800:PMID
1476:, a
1411:Gene
1365:and
1327:gene
966:1788
416:Bgee
364:Band
325:Chr.
263:Band
222:Chr.
176:7294
156:OMIM
113:2QRV
109:3A1A
105:3SVM
101:4QBQ
97:3LLR
93:3A1B
89:4QBS
85:4U7P
81:4QBR
77:4U7T
58:RCSB
55:PDBe
4502:PMC
4494:doi
4461:doi
4430:PMC
4422:doi
4381:doi
4350:PMC
4340:doi
4299:doi
4295:298
4267:doi
4263:269
4234:doi
4203:PMC
4195:doi
4160:doi
4156:277
4127:doi
4123:129
4090:hdl
4082:doi
4070:295
4042:PMC
4034:doi
3999:PMC
3991:doi
3956:PMC
3948:doi
3913:doi
3884:377
3846:PMC
3838:doi
3797:PMC
3789:doi
3748:PMC
3740:doi
3688:doi
3639:PMC
3631:doi
3583:PMC
3575:doi
3531:PMC
3523:doi
3482:PMC
3474:doi
3470:363
3427:doi
3390:PMC
3382:doi
3370:502
3333:PMC
3325:doi
3284:PMC
3276:doi
3235:doi
3188:doi
3176:429
3137:doi
3100:PMC
3092:doi
3051:doi
3047:274
3004:doi
2992:404
2956:PMC
2948:doi
2904:PMC
2896:doi
2852:PMC
2844:doi
2792:PMC
2784:doi
2772:586
2735:PMC
2727:doi
2715:449
2678:PMC
2670:doi
2629:PMC
2621:doi
2580:PMC
2570:doi
2529:PMC
2521:doi
2480:PMC
2470:doi
2429:PMC
2419:doi
2378:PMC
2370:doi
2321:doi
2274:doi
2234:PMC
2226:doi
2180:doi
2176:277
2136:PMC
2126:doi
2085:PMC
2077:doi
2036:PMC
2028:doi
2024:126
1987:PMC
1979:doi
1938:PMC
1928:doi
1887:PMC
1879:doi
1840:doi
1836:236
1792:doi
1677:and
1651:Myc
386:End
285:End
188:OMA
164:MGI
48:PDB
4530::
4510:.
4500:.
4490:32
4488:.
4484:.
4467:.
4457:22
4455:.
4438:.
4428:.
4418:31
4416:.
4412:.
4395:.
4387:.
4377:33
4375:.
4358:.
4348:.
4338:.
4328:99
4326:.
4322:.
4305:.
4293:.
4275:.
4261:.
4257:.
4240:.
4228:.
4211:.
4201:.
4191:21
4189:.
4185:.
4168:.
4154:.
4150:.
4133:.
4121:.
4104:.
4096:.
4088:.
4080:.
4068:.
4050:.
4040:.
4030:30
4028:.
4024:.
4007:.
3997:.
3987:20
3985:.
3981:.
3964:.
3954:.
3944:27
3942:.
3938:.
3921:.
3907:.
3903:.
3882:.
3854:.
3844:.
3834:31
3832:.
3828:.
3805:.
3795:.
3785:24
3783:.
3779:.
3756:.
3746:.
3736:20
3734:.
3730:.
3718:^
3704:.
3696:.
3686:.
3676:13
3674:.
3670:.
3647:.
3637:.
3627:32
3625:.
3621:.
3605:^
3591:.
3581:.
3571:21
3569:.
3565:.
3553:^
3539:.
3529:.
3519:69
3517:.
3513:.
3490:.
3480:.
3468:.
3464:.
3441:.
3433:.
3423:12
3421:.
3398:.
3388:.
3380:.
3368:.
3364:.
3341:.
3331:.
3321:44
3319:.
3315:.
3292:.
3282:.
3272:24
3270:.
3266:.
3243:.
3231:53
3229:.
3225:.
3202:.
3194:.
3186:.
3174:.
3151:.
3143:.
3131:.
3108:.
3098:.
3088:25
3086:.
3082:.
3059:.
3045:.
3041:.
3018:.
3010:.
3002:.
2990:.
2978:^
2964:.
2954:.
2944:44
2942:.
2938:.
2926:^
2912:.
2902:.
2892:36
2890:.
2886:.
2874:^
2860:.
2850:.
2842:.
2832:11
2830:.
2826:.
2814:^
2800:.
2790:.
2782:.
2770:.
2766:.
2743:.
2733:.
2725:.
2713:.
2709:.
2686:.
2676:.
2666:34
2664:.
2660:.
2637:.
2627:.
2617:17
2615:.
2611:.
2588:.
2578:.
2564:.
2560:.
2537:.
2527:.
2515:.
2511:.
2488:.
2478:.
2464:.
2460:.
2437:.
2427:.
2415:10
2413:.
2409:.
2386:.
2376:.
2366:38
2364:.
2360:.
2337:.
2329:.
2317:21
2315:.
2311:.
2288:.
2280:.
2270:15
2268:.
2256:^
2242:.
2232:.
2222:15
2220:.
2216:.
2202:^
2188:.
2174:.
2170:.
2158:^
2144:.
2134:.
2122:23
2120:.
2116:.
2093:.
2083:.
2073:27
2071:.
2067:.
2044:.
2034:.
2022:.
2018:.
1995:.
1985:.
1975:17
1973:.
1969:.
1946:.
1936:.
1924:54
1922:.
1918:.
1895:.
1885:.
1873:.
1869:.
1846:.
1834:.
1820:^
1806:.
1798:.
1788:19
1786:.
1774:^
1764:.
1746:.
1717:^
1696:^
1496:.
1442:.
1407:.
1381:,
1373:,
1369:,
1350:.
1329:.
913:/
392:bp
379:bp
291:bp
278:bp
186:;
182::
178:;
174::
170:;
166::
162:;
158::
111:,
107:,
103:,
99:,
95:,
91:,
87:,
83:,
79:,
4518:.
4496::
4475:.
4463::
4446:.
4424::
4403:.
4383::
4366:.
4342::
4334::
4313:.
4301::
4283:.
4269::
4248:.
4236::
4219:.
4197::
4176:.
4162::
4141:.
4129::
4112:.
4092::
4084::
4076::
4058:.
4036::
4015:.
3993::
3972:.
3950::
3929:.
3915::
3909:6
3894:.
3862:.
3840::
3813:.
3791::
3764:.
3742::
3712:.
3690::
3682::
3655:.
3633::
3599:.
3577::
3547:.
3525::
3498:.
3476::
3449:.
3429::
3406:.
3384::
3376::
3349:.
3327::
3300:.
3278::
3251:.
3237::
3210:.
3190::
3182::
3159:.
3139::
3133:2
3116:.
3094::
3067:.
3053::
3026:.
3006::
2998::
2972:.
2950::
2920:.
2898::
2868:.
2846::
2838::
2808:.
2786::
2778::
2751:.
2729::
2721::
2694:.
2672::
2645:.
2623::
2596:.
2572::
2566:9
2545:.
2523::
2517:8
2496:.
2472::
2466:6
2445:.
2421::
2394:.
2372::
2345:.
2323::
2296:.
2276::
2250:.
2228::
2196:.
2182::
2152:.
2128::
2101:.
2079::
2052:.
2030::
2003:.
1981::
1954:.
1930::
1903:.
1881::
1875:2
1854:.
1842::
1814:.
1794::
1768:.
1750:.
1683:.
1671:,
1665:,
1659:,
1653:,
1647:,
1641:,
1635:,
1310:(
310:)
207:)
190::
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.