1286:, can be assessed by the length of time that a labelled dsDNA probe with a short homologous sequence remains bound to a DNA containing a short region of homology to it. A study of this type has shown that a mismatch in any of the three positions at the end of a stretch of homology will not increase the length of time that the probe remains bound, and in Rad51 or RecA constructs an internal mismatch will cause a similar reduction in binding time. All of the enzymes are capable of "stepping over" a mismatch and continuing to bind the probe more firmly if a longer region of homology exists. However, with Dmc1 a triplet with a single internal (but not terminal) mismatch will contribute to the stability of probe binding to a similar extent as one without a mismatch. In this way, Dmc1 is specially suited to its role as a meiosis-specific recombinase, as this activity permits it more effectively to catalyze recombination between sequences that are not perfectly matched.
328:
305:
202:
227:
586:
579:
1221:. DMC1 and RAD51 share over 50% amino acid similarity. In budding yeast, Rad51 serves as a strand exchange protein in mitosis where it is critical for the repair of DNA breaks. Rad51 is converted to an accessory factor for Dmc1 during meiosis by inhibition of its strand exchange activity. Homologs of DMC1 are well conserved and have been identified in many organisms including divergent fungi, plants and mammals including humans.
334:
233:
3047:
1246:
center. hDMC1 can conduct D-loop formation in supercoiled DNA. DMC1 has weak ATPase activity and is able to promote heteroduplex formation in the presence of a non-hydrolysable analog of ATP, indicating a requirement for ATP binding over ATP hydrolysis. DMC1 also shows enhanced binding to nucleosomes with histone tails removed, indicating that chromosome architecture may play a role in DMC1 binding, but not Rad51.
47:
1334:. Both Dmc1 and Rad51 have an intrinsic ability to self-aggregate. The presence of Rad51 filaments stabilizes adjacent Dmc1 filaments and conversely Dmc1 stabilizes adjacent Rad51 filaments. A model was proposed in which Dmc1 and Rad51 form separate filaments on the same single stranded DNA and cross-talk between the two recombinases affects their biochemical properties.
1266:(SSBs) to protect the ssDNA and prevent the formation of secondary structures. DMC1 is loaded onto the 3' ssDNA to form a right-handed helical nucleoprotein filament. Subsequently, this nucleoprotein filament conducts a homology search in a homologous DNA region. Single-strand invasion in a complementary region in the homologous chromosome by the 3'-ended DNA strand forms a
1254:
The protein encoded by this gene is essential for meiotic homologous recombination. Genetic recombination in meiosis plays an important role in generating diversity of genetic information and facilitates the reductional segregation of chromosomes that must occur for formation of gametes during sexual
1302:
and the
Structural Maintenance of Chromosome 5/6 (SMC5/6) complex. The protein has also been shown to bind Tid1(Rdh54), Mei5/Sae3, and Hop2/Mnd1. All of these interacting proteins act to enhance Dmc1's activity in purified systems and are also implicated as being required for Dmc1 function in cells.
1281:
Like other members of the Rad51/RecA family, Dmc1 stabilizes strand exchange intermediates (Rad1/RecA-stretched DNA, or RS-DNA) in stretched triplets similar to B form DNA. Each molecule of the protein binds a triplet of nucleotides, and the strength of that binding, as assessed by the change in
1245:
Human DMC1 is a homomultimer in the form of an octameric ring with a diameter of 140 Å and a hole in the middle of 45 Å. DMC1 binds preferentially to ssDNA over dsDNA. Unlike RecA and Rad51, DMC1 does not appear to form a helical filament on DNA, instead forming rings with DNA passing through the
2701:
Moens PB, Kolas NK, Tarsounas M, Marcon E, Cohen PE, Spyropoulos B (April 2002). "The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA-DNA interactions without reciprocal recombination".
1361:
DMC1 interacts with the
Structural Maintenance of Chromosomes 5/6 (SMC5/6) complex. SCM5/6 complex inhibits the formation of DNA intermediates and is involved in their resolution. There is evidence that SCM5/6 interacts with and inhibit meiotic localization of DMC1. Rad51 can inhibit this
73:
1214:. The name "Dmc" stands for "disrupted meiotic cDNA" and refers to the method used for its discovery which involved using clones from a meiosis-specific cDNA library to direct knock-out mutations of abundantly expressed meiotic genes
1753:
Bishop DK, Park D, Xu L, Kleckner N (May 1992). "DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression".
1262:. DSBs are lengthened through the actions of exonucleases to trim the 5' ends and form long 3' single-stranded DNA (ssDNA) overhangs. These 3' overhangs are stabilized by the effects of
1374:, where the testes produce little to no sperm. In mice, a single amino acid change can prevent crossing over and lead to meiotic arrest through an autosomal dominant mechanism.
341:
240:
1278:. Resolution of this Holiday junction results in crossover or non-crossover product. Crossover products are generated to a lesser extent than non-crossover products.
1349:
Hop2 and Mnd1 associate into a heterodimer which itself has affinity for dsDNA, and to a lesser extent, ssDNA. Hop2/Mnd1 stimulates strand-exchange activity of DMC1
1337:
During meiosis, even in the absence of Rad51 strand exchange activity, Dmc1 appears to be able to repair all meiotic DNA breaks, and this absence does not affect
1474:"The mouse and human homologs of DMC1, the yeast meiosis-specific homologous recombination gene, have a common unique form of exon-skipped transcript in meiosis"
2801:
Sehorn MG, Sigurdsson S, Bussen W, Unger VM, Sung P (May 2004). "Human meiotic recombinase Dmc1 promotes ATP-dependent homologous DNA strand exchange".
894:
875:
163:
3075:
3026:
1451:
1433:
1353:
The interaction of Hop2/Mnd1 and DMC1 are thought to promote preferential binding of DMC1 to ssDNA and bring homologs into close proximity.
327:
1118:
1125:
304:
3046:
1620:
1420:
1399:
2434:"Heterodimeric complexes of Hop2 and Mnd1 function with Dmc1 to promote meiotic homolog juxtaposition and strand assimilation"
1306:
DMC1 has also been shown to interact with FIGNL1. FIGNL1 is believed to promote the disassembly of DMC1 during male meiosis.
226:
201:
1416:
70:
1395:
143:
1295:
340:
239:
1263:
31:
3019:
333:
232:
2768:"Structural basis for octameric ring formation and DNA interaction of the human homologous-pairing protein Dmc1"
2207:"FIGNL1 AAA+ ATPase remodels RAD51 and DMC1 filaments in pre-meiotic DNA replication and meiotic recombination"
1331:
1199:
939:
151:
1526:"Expression profiles of a human gene identified as a structural homologue of meiosis-specific recA-like genes"
2516:
Bannister LA, Pezza RJ, Donaldson JR, de Rooij DG, Schimenti KJ, Camerini-Otero RD, Schimenti JC (May 2007).
1621:"Entrez Gene: DMC1 DMC1 dosage suppressor of mck1 homolog, meiosis-specific homologous recombination (yeast)"
920:
1949:"Chromatin architecture may dictate the target site for DMC1, but not for RAD51, during homologous pairing"
2733:"p53 Protein interacts specifically with the meiosis-specific mammalian RecA-like protein DMC1 in meiosis"
1801:
Masson JY, West SC (February 2001). "The Rad51 and Dmc1 recombinases: a non-identical twin relationship".
1338:
215:
1258:
During meiosis, programmed DNA double strand breaks (DSBs) are introduced by topoisomerase-like enzyme
1362:
interaction, and this may be its role as an accessory factor during meiotic homologous recombination.
3012:
2810:
2671:
2615:
Masson JY, Davies AA, Hajibagheri N, Van Dyck E, Benson FE, Stasiak AZ, et al. (November 1999).
2445:
2388:
2329:
2218:
1960:
1898:
Masson JY, Davies AA, Hajibagheri N, Van Dyck E, Benson FE, Stasiak AZ, et al. (November 1999).
1849:
1709:
1650:
2267:"Spontaneous self-segregation of Rad51 and Dmc1 DNA recombinases within mixed recombinase filaments"
1230:
130:
1207:
1097:
1076:
1068:
1038:
1017:
1009:
2834:
1778:
175:
2574:"Interaction of human rad51 recombination protein with single-stranded DNA binding protein, RPA"
2375:
Bugreev DV, Huang F, Mazina OM, Pezza RJ, Voloshin ON, Camerini-Otero RD, Mazin AV (June 2014).
2318:"Analysis of the impact of the absence of RAD51 strand exchange activity in Arabidopsis meiosis"
1101:
1072:
1042:
1013:
1947:
Kobayashi W, Takaku M, Machida S, Tachiwana H, Maehara K, Ohkawa Y, Kurumizaka H (April 2016).
2992:
2949:
2914:
2893:"Role of the N-terminal domain of the human DMC1 protein in octamer formation and DNA binding"
2879:
2826:
2789:
2754:
2719:
2689:
2646:
2603:
2549:
2473:
2414:
2357:
2298:
2244:
2187:
2135:
2086:
2037:
1986:
1929:
1877:
1818:
1770:
1735:
1678:
1599:
1547:
1503:
1283:
1275:
123:
63:
2982:
2974:
2939:
2904:
2869:
2859:
2818:
2779:
2744:
2711:
2679:
2636:
2628:
2593:
2585:
2539:
2529:
2463:
2453:
2404:
2396:
2347:
2337:
2288:
2278:
2234:
2226:
2177:
2169:
2125:
2117:
2076:
2068:
2027:
2017:
1976:
1968:
1919:
1911:
1867:
1857:
1810:
1762:
1725:
1717:
1668:
1658:
1589:
1581:
1537:
1493:
1485:
585:
578:
420:
351:
295:
250:
2766:
Kinebuchi T, Kagawa W, Enomoto R, Tanaka K, Miyagawa K, Shibata T, et al. (May 2004).
1217:
The Dmc1 protein is one of two homologs of RecA found in eukaryotic cells, the other being
2658:
Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, et al. (December 1999).
395:
171:
1194:
Meiotic recombination protein Dmc1 is a homolog of the bacterial strand exchange protein
2814:
2675:
2617:"The meiosis-specific recombinase hDmc1 forms ring structures and interacts with hRad51"
2449:
2392:
2333:
2239:
2222:
2206:
1964:
1900:"The meiosis-specific recombinase hDmc1 forms ring structures and interacts with hRad51"
1853:
1713:
1698:"Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis"
1654:
2987:
2962:
2641:
2616:
2544:
2517:
2409:
2376:
2352:
2317:
2293:
2266:
2182:
2157:
2130:
2105:
2081:
2056:
2032:
2005:
1981:
1948:
1924:
1899:
1838:"Recombination activities of HsDmc1 protein, the meiotic human homolog of RecA protein"
1730:
1697:
1594:
1569:
1370:
Mutations in the DMC1 gene are associated with male infertility, due to nonobstructive
1234:
2874:
2847:
2846:
Collins JE, Wright CL, Edwards CA, Davis MP, Grinham JA, Cole CG, et al. (2004).
2784:
2767:
2598:
2573:
2468:
2433:
2104:
Lee JY, Terakawa T, Qi Z, Steinfeld JB, Redding S, Kwon Y, et al. (August 2015).
1814:
1498:
1473:
809:
804:
799:
794:
789:
784:
779:
774:
769:
764:
759:
754:
749:
744:
739:
734:
729:
724:
708:
703:
698:
693:
688:
672:
667:
662:
657:
652:
647:
642:
637:
632:
627:
46:
3069:
1872:
1837:
1766:
1673:
1638:
614:
2518:"A dominant, recombination-defective allele of Dmc1 causing male-specific sterility"
1782:
1570:"Interactions between human BRCA2 protein and the meiosis-specific recombinase DMC1"
2838:
2106:"DNA RECOMBINATION. Base triplet stepping by the Rad51/RecA family of recombinases"
1637:
Passy SI, Yu X, Li Z, Radding CM, Masson JY, West SC, Egelman EH (September 1999).
1319:
1267:
413:
192:
155:
2432:
Chen YK, Leng CH, Olivares H, Lee MH, Chang YC, Kung WM, et al. (July 2004).
2731:
Habu T, Wakabayashi N, Yoshida K, Yomogida K, Nishimune Y, Morita T (June 2004).
2534:
2342:
2205:
Ito M, Furukohri A, Matsuzaki K, Fujita Y, Toyoda A, Shinohara A (October 2023).
2022:
179:
2072:
1456:
National Center for
Biotechnology Information, U.S. National Library of Medicine
1438:
National Center for
Biotechnology Information, U.S. National Library of Medicine
1371:
1229:
The DMC1 gene and protein were discovered in the budding yeast S. cerevisiae by
2632:
2438:
Proceedings of the
National Academy of Sciences of the United States of America
2230:
1915:
1842:
Proceedings of the
National Academy of Sciences of the United States of America
1643:
Proceedings of the
National Academy of Sciences of the United States of America
496:
312:
209:
159:
2864:
2589:
2283:
2173:
1862:
1663:
1585:
1542:
1525:
2749:
2732:
2458:
2121:
1721:
1274:(D-loop). This D-Loop is extended as DNA repair synthesis occurs, forming a
1211:
839:
556:
434:
379:
366:
278:
265:
167:
2996:
2953:
2944:
2927:
2918:
2909:
2892:
2883:
2830:
2793:
2758:
2723:
2715:
2693:
2650:
2553:
2477:
2418:
2361:
2302:
2248:
2191:
2139:
2090:
2041:
1990:
1933:
1822:
1739:
1682:
1603:
1489:
2963:"Hop2/Mnd1 acts on two critical steps in Dmc1-promoted homologous pairing"
2607:
2491:
1881:
1774:
1551:
1507:
1210:
and carrying out a search for allelic DNA sequences located on homologous
1165:
1160:
1149:
984:
965:
2978:
2822:
2156:
Chen H, He C, Wang C, Wang X, Ruan F, Yan J, et al. (August 2021).
2400:
1315:
1203:
1181:
951:
906:
2492:"DMC1 DNA meiotic recombinase 1 [Homo sapiens (human)] - Gene"
1972:
100:
96:
92:
2158:"RAD51 supports DMC1 by inhibiting the SMC5/6 complex during meiosis"
1271:
1133:
861:
17:
2926:
Bugreev DV, Golub EI, Stasiak AZ, Stasiak A, Mazin AV (July 2005).
2684:
2659:
2961:
Pezza RJ, Voloshin ON, Vanevski F, Camerini-Otero RD (July 2007).
2848:"A genome annotation-driven approach to cloning the human ORFeome"
1323:
1299:
1259:
1218:
2572:
Golub EI, Gupta RC, Haaf T, Wold MS, Radding CM (December 1998).
2265:
Crickard JB, Kaniecki K, Kwon Y, Sung P, Greene EC (March 2018).
824:
820:
1696:
Cloud V, Chan YL, Grubb J, Budke B, Bishop DK (September 2012).
1330:
to form specialized filaments that are adapted for facilitating
1195:
1188:
594:
147:
3008:
2928:"Activation of human meiosis-specific recombinase Dmc1 by Ca2+"
2891:
Kinebuchi T, Kagawa W, Kurumizaka H, Yokoyama S (August 2005).
1472:
Habu T, Taki T, West A, Nishimune Y, Morita T (February 1996).
1233:
in 1992 when he was a postdoctoral fellow in the laboratory of
3004:
1327:
2006:"DMC1 attenuates RAD51-mediated recombination in Arabidopsis"
2377:"HOP2-MND1 modulates RAD51 binding to nucleotides and DNA"
2004:
Da Ines O, Bazile J, Gallego ME, White CI (August 2022).
403:
790:
chromosome organization involved in meiotic cell cycle
568:
134:, DMC1H, LIM15, dJ199H16.1, DNA meiotic recombinase 1
2496:
1836:
Li Z, Golub EI, Gupta R, Radding CM (October 1997).
1639:"Human Dmc1 protein binds DNA as an octameric ring"
1090:
1061:
1031:
1002:
1412:
1410:
1408:
1391:
1389:
1387:
2316:Singh G, Da Ines O, Gallego ME, White CI (2017).
1524:Sato S, Seki N, Hotta Y, Tabata S (August 1995).
350:
249:
2260:
2258:
2057:"Meiotic Recombination: The Essence of Heredity"
1563:
1561:
1178:Meiotic recombination protein DMC1/LIM15 homolog
27:Protein-coding gene in the species Homo sapiens
1893:
1891:
1796:
1794:
1792:
1417:GRCm38: Ensembl release 89: ENSMUSG00000022429
3055:: Crystal structure of the human Dmc1 protein
3020:
1206:by assembling at the sites of programmed DNA
8:
2151:
2149:
1632:
1630:
1568:Thorslund T, Esashi F, West SC (June 2007).
1332:recombination between homologous chromosomes
1519:
1517:
1467:
1465:
1396:GRCh38: Ensembl release 89: ENSG00000100206
3027:
3013:
3005:
2061:Cold Spring Harbor Perspectives in Biology
835:
610:
391:
290:
187:
81:
2986:
2943:
2908:
2873:
2863:
2783:
2748:
2683:
2660:"The DNA sequence of human chromosome 22"
2640:
2597:
2543:
2533:
2467:
2457:
2408:
2351:
2341:
2292:
2282:
2238:
2181:
2129:
2080:
2031:
2021:
1980:
1923:
1871:
1861:
1729:
1672:
1662:
1593:
1541:
1497:
1615:
1613:
1326:and Dmc1, interact with single-stranded
760:homologous chromosome pairing at meiosis
3042:
1383:
36:
638:ATP-dependent activity, acting on DNA
355:
316:
311:
254:
213:
208:
7:
2932:The Journal of Biological Chemistry
2897:The Journal of Biological Chemistry
2498:. U.S. National Library of Medicine
2271:The Journal of Biological Chemistry
1087:
1058:
1028:
999:
975:
956:
930:
911:
885:
866:
573:
491:
429:
408:
25:
1198:. Dmc1 plays the central role in
1184:that in humans is encoded by the
3045:
805:reciprocal meiotic recombination
584:
577:
339:
332:
326:
303:
238:
231:
225:
200:
45:
1803:Trends in Biochemical Sciences
1294:DMC1 (gene) has been shown to
1264:single strand binding proteins
730:response to ionizing radiation
668:endodeoxyribonuclease activity
595:More reference expression data
557:More reference expression data
1:
2785:10.1016/S1097-2765(04)00218-7
1815:10.1016/S0968-0004(00)01742-4
658:four-way junction DNA binding
324:
223:
3076:Genes on human chromosome 22
2535:10.1371/journal.pbio.0050105
2343:10.1371/journal.pone.0183006
2023:10.1371/journal.pgen.1010322
1767:10.1016/0092-8674(92)90446-j
780:ovarian follicle development
709:condensed nuclear chromosome
643:DNA strand exchange activity
2073:10.1101/cshperspect.a016618
663:double-stranded DNA binding
648:single-stranded DNA binding
3092:
2231:10.1038/s41467-023-42576-w
32:Devil May Cry (video game)
29:
3040:
2055:Hunter N (October 2015).
1452:"Mouse PubMed Reference:"
1434:"Human PubMed Reference:"
1164:
1159:
1155:
1148:
1132:
1113:
1094:
1065:
1054:
1035:
1006:
995:
982:
978:
963:
959:
950:
937:
933:
918:
914:
905:
892:
888:
873:
869:
860:
845:
838:
834:
818:
613:
609:
592:
576:
567:
554:
503:
494:
441:
432:
402:
394:
390:
373:
360:
323:
302:
293:
289:
272:
259:
222:
199:
190:
186:
141:
138:
128:
121:
116:
89:
84:
67:
62:
57:
53:
44:
39:
2865:10.1186/gb-2004-5-10-r84
2633:10.1093/emboj/18.22.6552
2284:10.1074/jbc.RA117.001143
1916:10.1093/emboj/18.22.6552
1863:10.1073/pnas.94.21.11221
1664:10.1073/pnas.96.19.10684
1586:10.1038/sj.emboj.7601739
1200:homologous recombination
1126:Chr 15: 79.45 – 79.49 Mb
1119:Chr 22: 38.52 – 38.57 Mb
740:female gamete generation
735:DNA recombinase assembly
30:For the video game, see
2967:Genes & Development
2704:Journal of Cell Science
2459:10.1073/pnas.0404195101
2122:10.1126/science.aab2666
1722:10.1126/science.1219379
1237:at Harvard University.
2945:10.1074/jbc.M502248200
2910:10.1074/jbc.M503372200
2716:10.1242/jcs.115.8.1611
2590:10.1093/nar/26.23.5388
2578:Nucleic Acids Research
2174:10.1093/plcell/koab136
1543:10.1093/dnares/2.4.183
1478:Nucleic Acids Research
357:15 E1|15 37.79 cM
2750:10.1093/carcin/bgh099
2381:Nature Communications
2211:Nature Communications
1366:Clinical significance
1339:meiotic crossing over
795:mitotic recombination
775:spermatid development
750:DNA metabolic process
318:Chromosome 15 (mouse)
216:Chromosome 22 (human)
1490:10.1093/nar/24.3.470
1208:double strand breaks
85:List of PDB id codes
58:Available structures
2979:10.1101/gad.1562907
2938:(29): 26886–26895.
2903:(31): 28382–28387.
2823:10.1038/nature02563
2815:2004Natur.429..433S
2710:(Pt 8): 1611–1622.
2676:1999Natur.402..489D
2450:2004PNAS..10110572C
2444:(29): 10572–10577.
2393:2014NatCo...5.4198B
2334:2017PLoSO..1283006S
2223:2023NatCo..14.6857I
1965:2016NatSR...624228K
1854:1997PNAS...9411221L
1848:(21): 11221–11226.
1714:2012Sci...337.1222C
1708:(6099): 1222–1225.
1655:1999PNAS...9610684P
1649:(19): 10684–10688.
511:seminiferous tubule
473:ganglionic eminence
2401:10.1038/ncomms5198
1953:Scientific Reports
940:ENSMUSG00000022429
718:Biological process
682:Cellular component
633:nucleotide binding
621:Molecular function
445:buccal mucosa cell
3063:
3062:
2973:(14): 1758–1766.
2809:(6990): 433–437.
2670:(6761): 489–495.
2627:(22): 6552–6560.
2584:(23): 5388–5393.
2277:(11): 4191–4200.
2116:(6251): 977–981.
1973:10.1038/srep24228
1910:(22): 6552–6560.
1580:(12): 2915–2922.
1284:Gibbs free energy
1276:Holliday junction
1272:displacement loop
1270:in the form of a
1175:
1174:
1171:
1170:
1144:
1143:
1109:
1108:
1084:
1083:
1050:
1049:
1025:
1024:
991:
990:
972:
971:
946:
945:
927:
926:
901:
900:
882:
881:
830:
829:
765:oocyte maturation
755:gamete generation
605:
604:
601:
600:
563:
562:
550:
549:
488:
487:
386:
385:
285:
284:
112:
111:
108:
107:
68:Ortholog search:
16:(Redirected from
3083:
3049:
3029:
3022:
3015:
3006:
3000:
2990:
2957:
2947:
2922:
2912:
2887:
2877:
2867:
2842:
2797:
2787:
2762:
2752:
2727:
2697:
2687:
2654:
2644:
2621:The EMBO Journal
2611:
2601:
2558:
2557:
2547:
2537:
2513:
2507:
2506:
2504:
2503:
2488:
2482:
2481:
2471:
2461:
2429:
2423:
2422:
2412:
2372:
2366:
2365:
2355:
2345:
2313:
2307:
2306:
2296:
2286:
2262:
2253:
2252:
2242:
2202:
2196:
2195:
2185:
2168:(8): 2869–2882.
2153:
2144:
2143:
2133:
2101:
2095:
2094:
2084:
2052:
2046:
2045:
2035:
2025:
2001:
1995:
1994:
1984:
1944:
1938:
1937:
1927:
1904:The EMBO Journal
1895:
1886:
1885:
1875:
1865:
1833:
1827:
1826:
1798:
1787:
1786:
1750:
1744:
1743:
1733:
1693:
1687:
1686:
1676:
1666:
1634:
1625:
1624:
1617:
1608:
1607:
1597:
1574:The EMBO Journal
1565:
1556:
1555:
1545:
1521:
1512:
1511:
1501:
1469:
1460:
1459:
1448:
1442:
1441:
1430:
1424:
1414:
1403:
1393:
1157:
1156:
1128:
1121:
1104:
1088:
1079:
1059:
1055:RefSeq (protein)
1045:
1029:
1020:
1000:
976:
957:
931:
912:
886:
867:
836:
611:
597:
588:
581:
574:
559:
527:secondary oocyte
499:
497:Top expressed in
492:
461:ventricular zone
437:
435:Top expressed in
430:
409:
392:
382:
369:
358:
343:
336:
330:
319:
307:
291:
281:
268:
257:
242:
235:
229:
218:
204:
188:
182:
180:DMC1 - orthologs
133:
126:
103:
82:
76:
55:
54:
49:
37:
21:
3091:
3090:
3086:
3085:
3084:
3082:
3081:
3080:
3066:
3065:
3064:
3059:
3056:
3050:
3036:
3033:
3003:
2960:
2925:
2890:
2845:
2800:
2765:
2730:
2700:
2657:
2614:
2571:
2567:
2565:Further reading
2562:
2561:
2515:
2514:
2510:
2501:
2499:
2490:
2489:
2485:
2431:
2430:
2426:
2374:
2373:
2369:
2328:(8): e0183006.
2315:
2314:
2310:
2264:
2263:
2256:
2204:
2203:
2199:
2155:
2154:
2147:
2103:
2102:
2098:
2067:(12): a016618.
2054:
2053:
2049:
2016:(8): e1010322.
2003:
2002:
1998:
1946:
1945:
1941:
1897:
1896:
1889:
1835:
1834:
1830:
1800:
1799:
1790:
1752:
1751:
1747:
1695:
1694:
1690:
1636:
1635:
1628:
1619:
1618:
1611:
1567:
1566:
1559:
1523:
1522:
1515:
1471:
1470:
1463:
1450:
1449:
1445:
1432:
1431:
1427:
1415:
1406:
1394:
1385:
1380:
1368:
1359:
1347:
1312:
1292:
1252:
1243:
1227:
1166:View/Edit Mouse
1161:View/Edit Human
1124:
1117:
1114:Location (UCSC)
1100:
1096:
1075:
1071:
1067:
1041:
1037:
1016:
1012:
1008:
921:ENSG00000100206
814:
770:spermatogenesis
725:strand invasion
713:
677:
653:protein binding
593:
583:
582:
555:
546:
541:
537:
533:
529:
525:
521:
517:
513:
509:
495:
484:
479:
477:muscle of thigh
475:
471:
469:Achilles tendon
467:
463:
459:
455:
451:
447:
433:
377:
364:
356:
346:
345:
344:
337:
317:
294:Gene location (
276:
263:
255:
245:
244:
243:
236:
214:
191:Gene location (
142:
129:
122:
91:
69:
35:
28:
23:
22:
15:
12:
11:
5:
3089:
3087:
3079:
3078:
3068:
3067:
3061:
3060:
3058:
3057:
3051:
3044:
3041:
3038:
3037:
3034:
3032:
3031:
3024:
3017:
3009:
3002:
3001:
2958:
2923:
2888:
2852:Genome Biology
2843:
2798:
2778:(3): 363–374.
2772:Molecular Cell
2763:
2743:(6): 889–893.
2737:Carcinogenesis
2728:
2698:
2685:10.1038/990031
2655:
2612:
2568:
2566:
2563:
2560:
2559:
2508:
2483:
2424:
2367:
2308:
2254:
2197:
2162:The Plant Cell
2145:
2096:
2047:
1996:
1939:
1887:
1828:
1809:(2): 131–136.
1788:
1761:(3): 439–456.
1745:
1688:
1626:
1609:
1557:
1536:(4): 183–186.
1513:
1484:(3): 470–477.
1461:
1443:
1425:
1404:
1382:
1381:
1379:
1376:
1367:
1364:
1358:
1355:
1346:
1343:
1311:
1308:
1291:
1288:
1255:reproduction.
1251:
1248:
1242:
1239:
1235:Nancy Kleckner
1231:Douglas Bishop
1226:
1223:
1173:
1172:
1169:
1168:
1163:
1153:
1152:
1146:
1145:
1142:
1141:
1139:
1137:
1130:
1129:
1122:
1115:
1111:
1110:
1107:
1106:
1092:
1091:
1085:
1082:
1081:
1063:
1062:
1056:
1052:
1051:
1048:
1047:
1033:
1032:
1026:
1023:
1022:
1004:
1003:
997:
993:
992:
989:
988:
980:
979:
973:
970:
969:
961:
960:
954:
948:
947:
944:
943:
935:
934:
928:
925:
924:
916:
915:
909:
903:
902:
899:
898:
890:
889:
883:
880:
879:
871:
870:
864:
858:
857:
852:
847:
843:
842:
832:
831:
828:
827:
816:
815:
813:
812:
807:
802:
797:
792:
787:
782:
777:
772:
767:
762:
757:
752:
747:
745:male meiosis I
742:
737:
732:
727:
721:
719:
715:
714:
712:
711:
706:
701:
696:
691:
685:
683:
679:
678:
676:
675:
670:
665:
660:
655:
650:
645:
640:
635:
630:
624:
622:
618:
617:
607:
606:
603:
602:
599:
598:
590:
589:
571:
565:
564:
561:
560:
552:
551:
548:
547:
545:
544:
540:
539:primary oocyte
536:
532:
528:
524:
520:
516:
512:
508:
504:
501:
500:
489:
486:
485:
483:
482:
478:
474:
470:
466:
462:
458:
454:
450:
446:
442:
439:
438:
426:
425:
417:
406:
400:
399:
396:RNA expression
388:
387:
384:
383:
375:
371:
370:
362:
359:
354:
348:
347:
338:
331:
325:
321:
320:
315:
309:
308:
300:
299:
287:
286:
283:
282:
274:
270:
269:
261:
258:
253:
247:
246:
237:
230:
224:
220:
219:
212:
206:
205:
197:
196:
184:
183:
140:
136:
135:
127:
119:
118:
114:
113:
110:
109:
106:
105:
87:
86:
78:
77:
66:
60:
59:
51:
50:
42:
41:
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
3088:
3077:
3074:
3073:
3071:
3054:
3048:
3043:
3039:
3030:
3025:
3023:
3018:
3016:
3011:
3010:
3007:
2998:
2994:
2989:
2984:
2980:
2976:
2972:
2968:
2964:
2959:
2955:
2951:
2946:
2941:
2937:
2933:
2929:
2924:
2920:
2916:
2911:
2906:
2902:
2898:
2894:
2889:
2885:
2881:
2876:
2871:
2866:
2861:
2857:
2853:
2849:
2844:
2840:
2836:
2832:
2828:
2824:
2820:
2816:
2812:
2808:
2804:
2799:
2795:
2791:
2786:
2781:
2777:
2773:
2769:
2764:
2760:
2756:
2751:
2746:
2742:
2738:
2734:
2729:
2725:
2721:
2717:
2713:
2709:
2705:
2699:
2695:
2691:
2686:
2681:
2677:
2673:
2669:
2665:
2661:
2656:
2652:
2648:
2643:
2638:
2634:
2630:
2626:
2622:
2618:
2613:
2609:
2605:
2600:
2595:
2591:
2587:
2583:
2579:
2575:
2570:
2569:
2564:
2555:
2551:
2546:
2541:
2536:
2531:
2527:
2523:
2519:
2512:
2509:
2497:
2493:
2487:
2484:
2479:
2475:
2470:
2465:
2460:
2455:
2451:
2447:
2443:
2439:
2435:
2428:
2425:
2420:
2416:
2411:
2406:
2402:
2398:
2394:
2390:
2386:
2382:
2378:
2371:
2368:
2363:
2359:
2354:
2349:
2344:
2339:
2335:
2331:
2327:
2323:
2319:
2312:
2309:
2304:
2300:
2295:
2290:
2285:
2280:
2276:
2272:
2268:
2261:
2259:
2255:
2250:
2246:
2241:
2236:
2232:
2228:
2224:
2220:
2216:
2212:
2208:
2201:
2198:
2193:
2189:
2184:
2179:
2175:
2171:
2167:
2163:
2159:
2152:
2150:
2146:
2141:
2137:
2132:
2127:
2123:
2119:
2115:
2111:
2107:
2100:
2097:
2092:
2088:
2083:
2078:
2074:
2070:
2066:
2062:
2058:
2051:
2048:
2043:
2039:
2034:
2029:
2024:
2019:
2015:
2011:
2010:PLOS Genetics
2007:
2000:
1997:
1992:
1988:
1983:
1978:
1974:
1970:
1966:
1962:
1958:
1954:
1950:
1943:
1940:
1935:
1931:
1926:
1921:
1917:
1913:
1909:
1905:
1901:
1894:
1892:
1888:
1883:
1879:
1874:
1869:
1864:
1859:
1855:
1851:
1847:
1843:
1839:
1832:
1829:
1824:
1820:
1816:
1812:
1808:
1804:
1797:
1795:
1793:
1789:
1784:
1780:
1776:
1772:
1768:
1764:
1760:
1756:
1749:
1746:
1741:
1737:
1732:
1727:
1723:
1719:
1715:
1711:
1707:
1703:
1699:
1692:
1689:
1684:
1680:
1675:
1670:
1665:
1660:
1656:
1652:
1648:
1644:
1640:
1633:
1631:
1627:
1622:
1616:
1614:
1610:
1605:
1601:
1596:
1591:
1587:
1583:
1579:
1575:
1571:
1564:
1562:
1558:
1553:
1549:
1544:
1539:
1535:
1531:
1527:
1520:
1518:
1514:
1509:
1505:
1500:
1495:
1491:
1487:
1483:
1479:
1475:
1468:
1466:
1462:
1457:
1453:
1447:
1444:
1439:
1435:
1429:
1426:
1422:
1418:
1413:
1411:
1409:
1405:
1401:
1397:
1392:
1390:
1388:
1384:
1377:
1375:
1373:
1365:
1363:
1356:
1354:
1352:
1344:
1342:
1340:
1335:
1333:
1329:
1325:
1321:
1317:
1309:
1307:
1304:
1301:
1297:
1289:
1287:
1285:
1279:
1277:
1273:
1269:
1265:
1261:
1256:
1249:
1247:
1240:
1238:
1236:
1232:
1224:
1222:
1220:
1215:
1213:
1209:
1205:
1201:
1197:
1192:
1190:
1187:
1183:
1179:
1167:
1162:
1158:
1154:
1151:
1147:
1140:
1138:
1135:
1131:
1127:
1123:
1120:
1116:
1112:
1105:
1103:
1099:
1093:
1089:
1086:
1080:
1078:
1074:
1070:
1064:
1060:
1057:
1053:
1046:
1044:
1040:
1034:
1030:
1027:
1021:
1019:
1015:
1011:
1005:
1001:
998:
996:RefSeq (mRNA)
994:
987:
986:
981:
977:
974:
968:
967:
962:
958:
955:
953:
949:
942:
941:
936:
932:
929:
923:
922:
917:
913:
910:
908:
904:
897:
896:
891:
887:
884:
878:
877:
872:
868:
865:
863:
859:
856:
853:
851:
848:
844:
841:
837:
833:
826:
822:
817:
811:
808:
806:
803:
801:
798:
796:
793:
791:
788:
786:
783:
781:
778:
776:
773:
771:
768:
766:
763:
761:
758:
756:
753:
751:
748:
746:
743:
741:
738:
736:
733:
731:
728:
726:
723:
722:
720:
717:
716:
710:
707:
705:
702:
700:
697:
695:
692:
690:
687:
686:
684:
681:
680:
674:
671:
669:
666:
664:
661:
659:
656:
654:
651:
649:
646:
644:
641:
639:
636:
634:
631:
629:
626:
625:
623:
620:
619:
616:
615:Gene ontology
612:
608:
596:
591:
587:
580:
575:
572:
570:
566:
558:
553:
542:
538:
534:
530:
526:
522:
518:
514:
510:
506:
505:
502:
498:
493:
490:
480:
476:
472:
468:
464:
460:
456:
452:
448:
444:
443:
440:
436:
431:
428:
427:
424:
422:
418:
416:
415:
411:
410:
407:
405:
401:
397:
393:
389:
381:
376:
372:
368:
363:
353:
349:
342:
335:
329:
322:
314:
310:
306:
301:
297:
292:
288:
280:
275:
271:
267:
262:
252:
248:
241:
234:
228:
221:
217:
211:
207:
203:
198:
194:
189:
185:
181:
177:
173:
169:
165:
161:
157:
153:
149:
145:
137:
132:
125:
120:
115:
104:
102:
98:
94:
88:
83:
80:
79:
75:
72:
65:
61:
56:
52:
48:
43:
38:
33:
19:
3052:
2970:
2966:
2935:
2931:
2900:
2896:
2855:
2851:
2806:
2802:
2775:
2771:
2740:
2736:
2707:
2703:
2667:
2663:
2624:
2620:
2581:
2577:
2525:
2522:PLOS Biology
2521:
2511:
2500:. Retrieved
2495:
2486:
2441:
2437:
2427:
2384:
2380:
2370:
2325:
2321:
2311:
2274:
2270:
2214:
2210:
2200:
2165:
2161:
2113:
2109:
2099:
2064:
2060:
2050:
2013:
2009:
1999:
1956:
1952:
1942:
1907:
1903:
1845:
1841:
1831:
1806:
1802:
1758:
1754:
1748:
1705:
1701:
1691:
1646:
1642:
1577:
1573:
1533:
1530:DNA Research
1529:
1481:
1477:
1455:
1446:
1437:
1428:
1369:
1360:
1350:
1348:
1336:
1320:recombinases
1313:
1305:
1293:
1290:Interactions
1280:
1268:heteroduplex
1257:
1253:
1244:
1228:
1216:
1193:
1185:
1177:
1176:
1098:NP_001265155
1095:
1077:NP_001349946
1069:NP_001265137
1066:
1039:NM_001278226
1036:
1018:NM_001363017
1010:NM_001278208
1007:
983:
964:
938:
919:
893:
874:
854:
849:
453:right testis
419:
412:
139:External IDs
90:
3035:PDB gallery
2858:(10): R84.
2528:(5): e105.
2387:(1): 4198.
2217:(1): 6857.
1372:azoospermia
694:nucleoplasm
673:ATP binding
628:DNA binding
535:granulocyte
457:left testis
378:79,489,310
365:79,445,698
277:38,570,204
264:38,518,948
117:Identifiers
2502:2023-12-05
1423:, May 2017
1402:, May 2017
1378:References
1318:, the two
1212:chromatids
800:DNA repair
785:cell cycle
689:chromosome
515:blastocyst
423:(ortholog)
160:HomoloGene
1959:: 24228.
1351:in vitro.
1345:Hop2/Mnd1
1241:Structure
1225:Discovery
1102:NP_034189
1073:NP_008999
1043:NM_010059
1014:NM_007068
840:Orthologs
519:spermatid
168:GeneCards
3070:Category
2997:17639081
2954:15917244
2919:15917243
2884:15461802
2831:15164066
2794:15125839
2759:14764457
2724:11950880
2694:10591208
2651:10562567
2554:17425408
2478:15249670
2419:24943459
2362:28797117
2322:PLOS ONE
2303:29382724
2249:37891173
2240:10611733
2192:34009315
2140:26315438
2091:26511629
2042:36007010
1991:27052786
1934:10562567
1823:11166572
1783:45890186
1740:22955832
1683:10485886
1604:17541404
1419:–
1398:–
1296:interact
1250:Function
1150:Wikidata
819:Sources:
699:telomere
543:epiblast
465:testicle
2988:1920170
2839:4316803
2811:Bibcode
2672:Bibcode
2642:1171718
2608:9826763
2545:1847842
2446:Bibcode
2410:4279451
2389:Bibcode
2353:5552350
2330:Bibcode
2294:5858004
2219:Bibcode
2183:8408460
2131:4580133
2110:Science
2082:4665078
2033:9451096
1982:4823753
1961:Bibcode
1925:1171718
1882:9326590
1850:Bibcode
1775:1581960
1731:4056682
1710:Bibcode
1702:Science
1651:Bibcode
1595:1894777
1552:8590282
1508:8602360
1421:Ensembl
1400:Ensembl
1341:rates.
1316:meiosis
1314:During
1204:meiosis
1182:protein
952:UniProt
907:Ensembl
846:Species
825:QuickGO
810:meiosis
704:nucleus
398:pattern
256:22q13.1
124:Aliases
2995:
2985:
2952:
2917:
2882:
2875:545604
2872:
2837:
2829:
2803:Nature
2792:
2757:
2722:
2692:
2664:Nature
2649:
2639:
2606:
2599:148005
2596:
2552:
2542:
2476:
2469:490024
2466:
2417:
2407:
2360:
2350:
2301:
2291:
2247:
2237:
2190:
2180:
2138:
2128:
2089:
2079:
2040:
2030:
1989:
1979:
1932:
1922:
1880:
1870:
1821:
1781:
1773:
1738:
1728:
1681:
1671:
1602:
1592:
1550:
1506:
1499:145652
1496:
1357:SCM5/6
1136:search
1134:PubMed
985:Q61880
966:Q14565
862:Entrez
569:BioGPS
531:embryo
523:embryo
507:morula
481:tonsil
156:105393
148:602721
2835:S2CID
1873:23422
1779:S2CID
1674:17943
1324:Rad51
1310:Rad51
1300:RAD51
1298:with
1260:Spo11
1219:Rad51
1180:is a
895:13404
876:11144
855:Mouse
850:Human
821:Amigo
449:gonad
421:Mouse
414:Human
361:Start
296:Mouse
260:Start
193:Human
3053:1v5w
2993:PMID
2950:PMID
2915:PMID
2880:PMID
2827:PMID
2790:PMID
2755:PMID
2720:PMID
2690:PMID
2647:PMID
2604:PMID
2550:PMID
2474:PMID
2415:PMID
2358:PMID
2299:PMID
2245:PMID
2188:PMID
2136:PMID
2087:PMID
2038:PMID
1987:PMID
1930:PMID
1878:PMID
1819:PMID
1771:PMID
1755:Cell
1736:PMID
1679:PMID
1600:PMID
1548:PMID
1504:PMID
1196:RecA
1189:gene
1186:DMC1
404:Bgee
352:Band
313:Chr.
251:Band
210:Chr.
172:DMC1
164:5135
144:OMIM
131:DMC1
101:4HYY
97:2ZJB
93:1V5W
74:RCSB
71:PDBe
40:DMC1
18:Dmc1
2983:PMC
2975:doi
2940:doi
2936:280
2905:doi
2901:280
2870:PMC
2860:doi
2819:doi
2807:429
2780:doi
2745:doi
2712:doi
2708:115
2680:doi
2668:402
2637:PMC
2629:doi
2594:PMC
2586:doi
2540:PMC
2530:doi
2464:PMC
2454:doi
2442:101
2405:PMC
2397:doi
2348:PMC
2338:doi
2289:PMC
2279:doi
2275:293
2235:PMC
2227:doi
2178:PMC
2170:doi
2126:PMC
2118:doi
2114:349
2077:PMC
2069:doi
2028:PMC
2018:doi
1977:PMC
1969:doi
1920:PMC
1912:doi
1868:PMC
1858:doi
1811:doi
1763:doi
1726:PMC
1718:doi
1706:337
1669:PMC
1659:doi
1590:PMC
1582:doi
1538:doi
1494:PMC
1486:doi
1328:DNA
1202:in
374:End
273:End
176:OMA
152:MGI
64:PDB
3072::
2991:.
2981:.
2971:21
2969:.
2965:.
2948:.
2934:.
2930:.
2913:.
2899:.
2895:.
2878:.
2868:.
2854:.
2850:.
2833:.
2825:.
2817:.
2805:.
2788:.
2776:14
2774:.
2770:.
2753:.
2741:25
2739:.
2735:.
2718:.
2706:.
2688:.
2678:.
2666:.
2662:.
2645:.
2635:.
2625:18
2623:.
2619:.
2602:.
2592:.
2582:26
2580:.
2576:.
2548:.
2538:.
2524:.
2520:.
2494:.
2472:.
2462:.
2452:.
2440:.
2436:.
2413:.
2403:.
2395:.
2383:.
2379:.
2356:.
2346:.
2336:.
2326:12
2324:.
2320:.
2297:.
2287:.
2273:.
2269:.
2257:^
2243:.
2233:.
2225:.
2215:14
2213:.
2209:.
2186:.
2176:.
2166:33
2164:.
2160:.
2148:^
2134:.
2124:.
2112:.
2108:.
2085:.
2075:.
2063:.
2059:.
2036:.
2026:.
2014:18
2012:.
2008:.
1985:.
1975:.
1967:.
1955:.
1951:.
1928:.
1918:.
1908:18
1906:.
1902:.
1890:^
1876:.
1866:.
1856:.
1846:94
1844:.
1840:.
1817:.
1807:26
1805:.
1791:^
1777:.
1769:.
1759:69
1757:.
1734:.
1724:.
1716:.
1704:.
1700:.
1677:.
1667:.
1657:.
1647:96
1645:.
1641:.
1629:^
1612:^
1598:.
1588:.
1578:26
1576:.
1572:.
1560:^
1546:.
1532:.
1528:.
1516:^
1502:.
1492:.
1482:24
1480:.
1476:.
1464:^
1454:.
1436:.
1407:^
1386:^
1322:,
1191:.
823:/
380:bp
367:bp
279:bp
266:bp
174:;
170::
166:;
162::
158:;
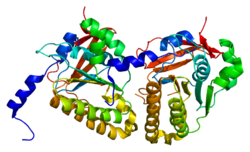
154::
150:;
146::
99:,
95:,
3028:e
3021:t
3014:v
2999:.
2977::
2956:.
2942::
2921:.
2907::
2886:.
2862::
2856:5
2841:.
2821::
2813::
2796:.
2782::
2761:.
2747::
2726:.
2714::
2696:.
2682::
2674::
2653:.
2631::
2610:.
2588::
2556:.
2532::
2526:5
2505:.
2480:.
2456::
2448::
2421:.
2399::
2391::
2385:5
2364:.
2340::
2332::
2305:.
2281::
2251:.
2229::
2221::
2194:.
2172::
2142:.
2120::
2093:.
2071::
2065:7
2044:.
2020::
1993:.
1971::
1963::
1957:6
1936:.
1914::
1884:.
1860::
1852::
1825:.
1813::
1785:.
1765::
1742:.
1720::
1712::
1685:.
1661::
1653::
1623:.
1606:.
1584::
1554:.
1540::
1534:2
1510:.
1488::
1458:.
1440:.
298:)
195:)
178::
34:.
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.