486:, dnaB is a hexameric protein of six 471-residue subunits, which form a ring-shaped structure with threefold symmetry. During DNA replication, the lagging strand of DNA binds in the central channel of dnaB, and the second DNA strand is excluded. The binding of NTPs causes a conformational change and subsequent hydrolysis allows the dnaB to translocate along the DNA, thus mechanically forcing the separation of the DNA strands.
2683:
122:
500:
550:
family (ATPases associated with diverse cellular activities). Many AAA+ ATPases, including DnaA, form oligomers and hydrolyze ATP relatively slowly. This ATP hydrolysis acts as a switch mediating interconversion of the protein between two states. In the case of DnaA, the ATP-bound form is active and the ADP-bound form is inactive.
470:
gene. DnaB is expressed as a monomer and oligomerises into hexamer through N-terminal interactions. Replicative helicases have a central ring and that feature is conserved across bacterial to eukaryotes. The energy for DnaB activity is provided by NTP hydrolysis. Mechanical energy moves the DnaB into
569:
Loading of the DnaB helicase is the key step in replication initiation. As a replicative helicase, DnaB migrates along the single-stranded DNA in the 5'→3' direction, unwinding the DNA as it travels. The DnaB helicases loaded onto the two DNA strands thus travel in opposite directions, creating two
553:
Eight DnaA protein molecules, all in the ATP-bound state, assemble to form a helical complex encompassing the R and I sites in oriC. DnaA has a higher affinity for the R sites than I sites, and binds R sites equally well in its ATP or ADP-bound form. The I sites, which bind only the ATP-bound DnaA,
565:
The DnaC protein, another AAA+ ATPase, then loads the DnaB protein onto the separated DNA strands in the denatured region. A hexamer of DnaC, each subunit bound to ATP, forms a tight complex with the hexameric, ring-shaped DnaB helicase. This DnaC-DnaB interaction opens the DnaB ring, the process
549:
At least 10 different enzymes or proteins participate in the initiation phase of replication. They open the DNA helix at the origin and establish a prepriming complex for subsequent reactions. The crucial component in the initiation process is the DnaA protein, a member of the AAA+ ATPase protein
566:
being aided by a further interaction between DnaB and DnaA. Two of the ring-shaped DnaB hexamers are loaded in the DUE, one onto each DNA strand. The ATP bound to DnaC is hydrolyzed, releasing the DnaC and leaving the DnaB bound to the DNA.
562:
in the A:T-rich 'DUE' (DNA Unwinding
Element) region. The complex formed at the replication origin also includes several DNA-binding proteins- Hu, IHF and FIS that facilitate DNA bending.
1510:
518:
359:
225:
554:
allow discrimination between the active and inactive forms of DnaA. The tight right-handed wrapping of the DNA around this complex introduces an effective positive
37:
756:
2203:
1503:
1733:
1320:
1283:
173:
1702:
1496:
1403:
2402:
1412:
978:
536:
1465:
42:
2293:
2288:
1325:
2558:
379:
245:
2310:
1135:
985:
749:
2673:
2358:
1675:
2280:
1126:
794:
769:
559:
1255:
1231:
1100:
932:
879:
861:
773:
570:
potential replication forks. All other proteins at the replication fork are linked directly or indirectly to DnaB.
460:
2543:
2659:
2646:
2633:
2620:
2607:
2594:
2581:
2339:
1880:
1778:
1603:
1599:
1576:
1549:
1536:
1523:
1309:
315:
2553:
452:
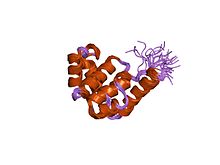
The N-terminal has a multi-helical structure that forms an orthogonal bundle. The C-terminal domain contains an
2703:
2507:
2450:
1894:
1589:
1554:
1527:
1470:
742:
700:
66:
367:
233:
2455:
2264:
2227:
1566:
872:
803:
1994:
1634:
1369:
1300:
453:
2476:
2395:
2124:
2032:
1295:
1064:
734:
2548:
363:
229:
1342:
1315:
1266:
415:
186:
320:
2512:
2329:
1072:
2708:
2445:
2344:
1910:
1488:
2713:
1584:
1278:
1095:
666:
613:
354:
220:
2491:
2486:
2460:
2388:
1974:
1920:
1902:
1768:
1758:
1712:
1235:
1084:
1079:
971:
656:
648:
603:
399:
127:
1787:
346:
212:
2538:
2522:
2435:
2321:
2210:
2024:
1724:
1544:
1448:
1240:
1114:
832:
782:
765:
696:
403:
90:
71:
2687:
2576:
2517:
1644:
1088:
1068:
661:
632:
430:
407:
608:
591:
2697:
2481:
2440:
2188:
1798:
1290:
1217:
555:
423:
178:
283:
154:
2430:
2173:
2146:
2075:
2068:
2051:
2046:
2020:
1429:
342:
208:
296:
730:
716:
308:
166:
2654:
2589:
2425:
2269:
1374:
100:
2682:
652:
2254:
1943:
1460:
1434:
1222:
1207:
427:
49:
2628:
2602:
1960:
1519:
1398:
1212:
1202:
670:
617:
592:"Crystal structure of the N-terminal domain of the DnaB hexameric helicase"
182:
121:
1475:
1456:
1028:
1019:
817:
726:
712:
395:
303:
161:
54:
2365:
2334:
2112:
2107:
1866:
1856:
1831:
1826:
1561:
1039:
1014:
1009:
841:
419:
2641:
2411:
2242:
1884:
1861:
1851:
1821:
1816:
1811:
1806:
1763:
1753:
1748:
1743:
1738:
1707:
1695:
1690:
1685:
1680:
1668:
1663:
1658:
1607:
434:
374:
240:
456:-binding site and is therefore probably the site of ATP hydrolysis.
2615:
2298:
2237:
2220:
2215:
2063:
2041:
2007:
2002:
1987:
1965:
1953:
1948:
1938:
1933:
1928:
1653:
1629:
1389:
1384:
1379:
1362:
1357:
1352:
1347:
1335:
1330:
1048:
1043:
1000:
995:
990:
2303:
2259:
2247:
2232:
2198:
2193:
2178:
2161:
2156:
2151:
2139:
2134:
2129:
2119:
2095:
2090:
2085:
2080:
1982:
1271:
1195:
1190:
1185:
1180:
1175:
1170:
1165:
1160:
1155:
1150:
1145:
1140:
1032:
962:
957:
952:
947:
942:
937:
925:
918:
909:
904:
899:
894:
889:
884:
846:
822:
810:
722:
708:
446:
442:
438:
336:
290:
278:
202:
149:
2384:
1492:
738:
2353:
2349:
493:
411:
445:, a negative regulator. After DnaC dissociates, DnaB binds
2380:
631:
Bocanegra R, Ismael Plaza GA, Pulido CR, Ibarra B (2021).
459:
In eukaryotes, helicase function is provided by the MCM (
514:
471:
the replication fork, physically splitting it in half.
721:
This article incorporates text from the public domain
707:
This article incorporates text from the public domain
2671:
414:
and denatures the duplex is unknown, a change in the
406:. Although the mechanism by which DnaB both couples
2567:
2531:
2500:
2469:
2418:
2319:
2278:
2018:
1892:
1879:
1844:
1796:
1777:
1723:
1643:
1622:
1615:
1598:
1575:
1535:
1447:
1421:
1251:
1122:
1113:
1057:
857:
790:
781:
558:. The associated strain in the nearby DNA leads to
509:
may be too technical for most readers to understand
373:
353:
335:
330:
314:
302:
289:
277:
269:
264:
259:
239:
219:
201:
196:
172:
160:
148:
140:
135:
114:
96:
86:
81:
65:
60:
48:
36:
28:
23:
18:
641:Computational and Structural Biotechnology Journal
585:
583:
2396:
1504:
750:
8:
433:has been observed and may occur during the
2403:
2389:
2381:
1889:
1619:
1612:
1511:
1497:
1489:
1119:
787:
757:
743:
735:
633:"DNA replication machinery: Insights from
590:Fass D, Bogden CE, Berger JM (June 1999).
327:
193:
126:nmr structure of the n-terminal domain of
120:
78:
699:at the U.S. National Library of Medicine
660:
607:
537:Learn how and when to remove this message
521:, without removing the technical details.
466:The DnaB helicase is the product of the
2678:
579:
490:Mechanism of initiation of replication
256:
111:
15:
683:Lehninger, Principles of Biochemistry
519:make it understandable to non-experts
7:
437:cycle. Initially when DnaB binds to
260:DnaB-like helicase C terminal domain
115:DnaB-like helicase N terminal domain
14:
1413:Control of chromosome duplication
979:Autonomously replicating sequence
2681:
498:
1:
1136:DNA polymerase III holoenzyme
986:Single-strand binding protein
609:10.1016/s0969-2126(99)80090-2
331:Available protein structures:
197:Available protein structures:
2281:Protein-synthesizing GTPase
637:single-molecule approaches"
2730:
1232:Prokaryotic DNA polymerase
933:Minichromosome maintenance
880:Origin recognition complex
720:
706:
653:10.1016/j.csbj.2021.04.013
461:Minichromosome maintenance
2559:Michaelis–Menten kinetics
2340:Guanylate-binding protein
1524:acid anhydride hydrolases
1310:Eukaryotic DNA polymerase
326:
192:
119:
77:
2451:Diffusion-limited enzyme
1895:Heterotrimeric G protein
1590:Phosphoadenylylsulfatase
701:Medical Subject Headings
441:, it is associated with
19:Replicative DNA helicase
1567:Thiamine-triphosphatase
873:Pre-replication complex
804:Pre-replication complex
410:to translocation along
2544:Eadie–Hofstee diagram
2477:Allosteric regulation
2322:Polymerization motors
2033:Rho family of GTPases
1296:Replication protein A
1065:Origin of replication
2554:Lineweaver–Burk plot
1267:Replication factor C
416:quaternary structure
2330:dynamin superfamily
2513:Enzyme superfamily
2446:Enzyme promiscuity
2669:
2668:
2378:
2377:
2374:
2373:
1875:
1874:
1840:
1839:
1585:Adenylylsulfatase
1486:
1485:
1443:
1442:
1279:Flap endonuclease
1109:
1108:
1096:Okazaki fragments
547:
546:
539:
389:
388:
385:
384:
380:structure summary
255:
254:
251:
250:
246:structure summary
110:
109:
106:
105:
2721:
2686:
2685:
2677:
2549:Hanes–Woolf plot
2492:Enzyme activator
2487:Enzyme inhibitor
2461:Enzyme catalysis
2405:
2398:
2391:
2382:
1890:
1620:
1613:
1513:
1506:
1499:
1490:
1236:DNA polymerase I
1120:
1080:Replication fork
972:Licensing factor
788:
759:
752:
745:
736:
684:
681:
675:
674:
664:
628:
622:
621:
611:
587:
542:
535:
531:
528:
522:
502:
501:
494:
400:replication fork
398:which opens the
394:is an enzyme in
328:
257:
194:
124:
112:
79:
16:
2729:
2728:
2724:
2723:
2722:
2720:
2719:
2718:
2704:Protein domains
2694:
2693:
2692:
2680:
2672:
2670:
2665:
2577:Oxidoreductases
2563:
2539:Enzyme kinetics
2527:
2523:List of enzymes
2496:
2465:
2436:Catalytic triad
2414:
2409:
2379:
2370:
2315:
2274:
2025:Ras superfamily
2014:
1998:
1978:
1924:
1914:
1906:
1871:
1836:
1792:
1773:
1719:
1676:Plasma membrane
1639:
1594:
1571:
1545:Pyrophosphatase
1531:
1517:
1487:
1482:
1439:
1417:
1257:
1253:
1247:
1241:Klenow fragment
1124:
1105:
1089:leading strands
1053:
863:
859:
853:
792:
777:
766:DNA replication
763:
733:
719:
693:
688:
687:
682:
678:
630:
629:
625:
589:
588:
581:
576:
543:
532:
526:
523:
515:help improve it
512:
503:
499:
492:
480:
404:DNA replication
131:
12:
11:
5:
2727:
2725:
2717:
2716:
2711:
2706:
2696:
2695:
2691:
2690:
2667:
2666:
2664:
2663:
2650:
2637:
2624:
2611:
2598:
2585:
2571:
2569:
2565:
2564:
2562:
2561:
2556:
2551:
2546:
2541:
2535:
2533:
2529:
2528:
2526:
2525:
2520:
2515:
2510:
2504:
2502:
2501:Classification
2498:
2497:
2495:
2494:
2489:
2484:
2479:
2473:
2471:
2467:
2466:
2464:
2463:
2458:
2453:
2448:
2443:
2438:
2433:
2428:
2422:
2420:
2416:
2415:
2410:
2408:
2407:
2400:
2393:
2385:
2376:
2375:
2372:
2371:
2369:
2368:
2363:
2362:
2361:
2356:
2347:
2342:
2337:
2326:
2324:
2317:
2316:
2314:
2313:
2308:
2307:
2306:
2301:
2296:
2285:
2283:
2276:
2275:
2273:
2272:
2267:
2262:
2257:
2252:
2251:
2250:
2245:
2240:
2235:
2225:
2224:
2223:
2218:
2208:
2207:
2206:
2201:
2196:
2184:
2183:
2182:
2181:
2176:
2166:
2165:
2164:
2159:
2154:
2144:
2143:
2142:
2137:
2132:
2122:
2117:
2116:
2115:
2110:
2100:
2099:
2098:
2093:
2088:
2083:
2073:
2072:
2071:
2066:
2056:
2055:
2054:
2049:
2044:
2029:
2027:
2016:
2015:
2013:
2012:
2011:
2010:
2005:
1996:
1992:
1991:
1990:
1985:
1976:
1972:
1971:
1970:
1969:
1968:
1958:
1957:
1956:
1951:
1941:
1936:
1931:
1922:
1918:
1917:
1916:
1912:
1904:
1899:
1897:
1887:
1877:
1876:
1873:
1872:
1870:
1869:
1864:
1859:
1854:
1848:
1846:
1842:
1841:
1838:
1837:
1835:
1834:
1829:
1824:
1819:
1814:
1809:
1803:
1801:
1794:
1793:
1791:
1790:
1784:
1782:
1775:
1774:
1772:
1771:
1766:
1761:
1756:
1751:
1746:
1741:
1736:
1730:
1728:
1721:
1720:
1718:
1717:
1716:
1715:
1710:
1700:
1699:
1698:
1693:
1688:
1683:
1673:
1672:
1671:
1666:
1661:
1650:
1648:
1641:
1640:
1638:
1637:
1632:
1626:
1624:
1623:Cu++ (3.6.3.4)
1617:
1610:
1596:
1595:
1593:
1592:
1587:
1581:
1579:
1573:
1572:
1570:
1569:
1564:
1559:
1558:
1557:
1552:
1541:
1539:
1533:
1532:
1518:
1516:
1515:
1508:
1501:
1493:
1484:
1483:
1481:
1480:
1479:
1478:
1473:
1468:
1453:
1451:
1445:
1444:
1441:
1440:
1438:
1437:
1432:
1425:
1423:
1419:
1418:
1416:
1415:
1409:
1408:
1407:
1406:
1395:
1394:
1393:
1392:
1387:
1382:
1377:
1367:
1366:
1365:
1360:
1355:
1350:
1340:
1339:
1338:
1333:
1328:
1323:
1313:
1306:
1305:
1304:
1303:
1293:
1288:
1287:
1286:
1276:
1275:
1274:
1263:
1261:
1249:
1248:
1246:
1245:
1244:
1243:
1228:
1227:
1226:
1225:
1215:
1210:
1205:
1200:
1199:
1198:
1193:
1188:
1183:
1178:
1173:
1168:
1163:
1158:
1153:
1148:
1143:
1132:
1130:
1117:
1111:
1110:
1107:
1106:
1104:
1103:
1098:
1093:
1092:
1091:
1076:
1075:
1061:
1059:
1055:
1054:
1052:
1051:
1046:
1036:
1035:
1025:
1024:
1023:
1022:
1017:
1006:
1005:
1004:
1003:
998:
993:
982:
981:
975:
974:
968:
967:
966:
965:
960:
955:
950:
945:
940:
929:
928:
922:
921:
915:
914:
913:
912:
907:
902:
897:
892:
887:
876:
875:
869:
867:
862:preparation in
855:
854:
852:
851:
850:
849:
838:
837:
836:
835:
830:
825:
814:
813:
807:
806:
800:
798:
785:
779:
778:
764:
762:
761:
754:
747:
739:
705:
704:
697:DnaB+Helicases
692:
691:External links
689:
686:
685:
676:
623:
578:
577:
575:
572:
545:
544:
506:
504:
497:
491:
488:
479:
473:
408:ATP hydrolysis
387:
386:
383:
382:
377:
371:
370:
357:
351:
350:
340:
333:
332:
324:
323:
318:
312:
311:
306:
300:
299:
294:
287:
286:
281:
275:
274:
271:
267:
266:
262:
261:
253:
252:
249:
248:
243:
237:
236:
223:
217:
216:
206:
199:
198:
190:
189:
176:
170:
169:
164:
158:
157:
152:
146:
145:
142:
138:
137:
133:
132:
125:
117:
116:
108:
107:
104:
103:
98:
94:
93:
88:
84:
83:
75:
74:
69:
63:
62:
58:
57:
52:
46:
45:
40:
34:
33:
30:
26:
25:
21:
20:
13:
10:
9:
6:
4:
3:
2:
2726:
2715:
2712:
2710:
2707:
2705:
2702:
2701:
2699:
2689:
2684:
2679:
2675:
2661:
2657:
2656:
2651:
2648:
2644:
2643:
2638:
2635:
2631:
2630:
2625:
2622:
2618:
2617:
2612:
2609:
2605:
2604:
2599:
2596:
2592:
2591:
2586:
2583:
2579:
2578:
2573:
2572:
2570:
2566:
2560:
2557:
2555:
2552:
2550:
2547:
2545:
2542:
2540:
2537:
2536:
2534:
2530:
2524:
2521:
2519:
2518:Enzyme family
2516:
2514:
2511:
2509:
2506:
2505:
2503:
2499:
2493:
2490:
2488:
2485:
2483:
2482:Cooperativity
2480:
2478:
2475:
2474:
2472:
2468:
2462:
2459:
2457:
2454:
2452:
2449:
2447:
2444:
2442:
2441:Oxyanion hole
2439:
2437:
2434:
2432:
2429:
2427:
2424:
2423:
2421:
2417:
2413:
2406:
2401:
2399:
2394:
2392:
2387:
2386:
2383:
2367:
2364:
2360:
2357:
2355:
2351:
2348:
2346:
2343:
2341:
2338:
2336:
2333:
2332:
2331:
2328:
2327:
2325:
2323:
2318:
2312:
2309:
2305:
2302:
2300:
2297:
2295:
2292:
2291:
2290:
2287:
2286:
2284:
2282:
2277:
2271:
2268:
2266:
2263:
2261:
2258:
2256:
2253:
2249:
2246:
2244:
2241:
2239:
2236:
2234:
2231:
2230:
2229:
2226:
2222:
2219:
2217:
2214:
2213:
2212:
2209:
2205:
2202:
2200:
2197:
2195:
2192:
2191:
2190:
2186:
2185:
2180:
2177:
2175:
2172:
2171:
2170:
2167:
2163:
2160:
2158:
2155:
2153:
2150:
2149:
2148:
2145:
2141:
2138:
2136:
2133:
2131:
2128:
2127:
2126:
2123:
2121:
2118:
2114:
2111:
2109:
2106:
2105:
2104:
2101:
2097:
2094:
2092:
2089:
2087:
2084:
2082:
2079:
2078:
2077:
2074:
2070:
2067:
2065:
2062:
2061:
2060:
2057:
2053:
2050:
2048:
2045:
2043:
2040:
2039:
2038:
2034:
2031:
2030:
2028:
2026:
2022:
2017:
2009:
2006:
2004:
2001:
2000:
1999:
1993:
1989:
1986:
1984:
1981:
1980:
1979:
1973:
1967:
1964:
1963:
1962:
1959:
1955:
1952:
1950:
1947:
1946:
1945:
1942:
1940:
1937:
1935:
1932:
1930:
1927:
1926:
1925:
1919:
1915:
1909:
1908:
1907:
1901:
1900:
1898:
1896:
1891:
1888:
1886:
1882:
1878:
1868:
1865:
1863:
1860:
1858:
1855:
1853:
1850:
1849:
1847:
1843:
1833:
1830:
1828:
1825:
1823:
1820:
1818:
1815:
1813:
1810:
1808:
1805:
1804:
1802:
1800:
1799:P-type ATPase
1795:
1789:
1786:
1785:
1783:
1780:
1776:
1770:
1767:
1765:
1762:
1760:
1757:
1755:
1752:
1750:
1747:
1745:
1742:
1740:
1737:
1735:
1732:
1731:
1729:
1726:
1722:
1714:
1711:
1709:
1706:
1705:
1704:
1701:
1697:
1694:
1692:
1689:
1687:
1684:
1682:
1679:
1678:
1677:
1674:
1670:
1667:
1665:
1662:
1660:
1657:
1656:
1655:
1652:
1651:
1649:
1646:
1642:
1636:
1633:
1631:
1628:
1627:
1625:
1621:
1618:
1614:
1611:
1609:
1605:
1601:
1597:
1591:
1588:
1586:
1583:
1582:
1580:
1578:
1574:
1568:
1565:
1563:
1560:
1556:
1553:
1551:
1548:
1547:
1546:
1543:
1542:
1540:
1538:
1534:
1529:
1525:
1521:
1514:
1509:
1507:
1502:
1500:
1495:
1494:
1491:
1477:
1474:
1472:
1469:
1467:
1464:
1463:
1462:
1458:
1455:
1454:
1452:
1450:
1446:
1436:
1433:
1431:
1427:
1426:
1424:
1420:
1414:
1411:
1410:
1405:
1402:
1401:
1400:
1397:
1396:
1391:
1388:
1386:
1383:
1381:
1378:
1376:
1373:
1372:
1371:
1368:
1364:
1361:
1359:
1356:
1354:
1351:
1349:
1346:
1345:
1344:
1341:
1337:
1334:
1332:
1329:
1327:
1324:
1322:
1319:
1318:
1317:
1314:
1311:
1308:
1307:
1302:
1299:
1298:
1297:
1294:
1292:
1291:Topoisomerase
1289:
1285:
1282:
1281:
1280:
1277:
1273:
1270:
1269:
1268:
1265:
1264:
1262:
1259:
1250:
1242:
1239:
1238:
1237:
1233:
1230:
1229:
1224:
1221:
1220:
1219:
1218:Topoisomerase
1216:
1214:
1211:
1209:
1206:
1204:
1201:
1197:
1194:
1192:
1189:
1187:
1184:
1182:
1179:
1177:
1174:
1172:
1169:
1167:
1164:
1162:
1159:
1157:
1154:
1152:
1149:
1147:
1144:
1142:
1139:
1138:
1137:
1134:
1133:
1131:
1128:
1121:
1118:
1116:
1112:
1102:
1099:
1097:
1094:
1090:
1086:
1083:
1082:
1081:
1078:
1077:
1074:
1070:
1066:
1063:
1062:
1060:
1056:
1050:
1047:
1045:
1041:
1038:
1037:
1034:
1030:
1027:
1026:
1021:
1018:
1016:
1013:
1012:
1011:
1008:
1007:
1002:
999:
997:
994:
992:
989:
988:
987:
984:
983:
980:
977:
976:
973:
970:
969:
964:
961:
959:
956:
954:
951:
949:
946:
944:
941:
939:
936:
935:
934:
931:
930:
927:
924:
923:
920:
917:
916:
911:
908:
906:
903:
901:
898:
896:
893:
891:
888:
886:
883:
882:
881:
878:
877:
874:
871:
870:
868:
865:
856:
848:
845:
844:
843:
840:
839:
834:
831:
829:
826:
824:
821:
820:
819:
816:
815:
812:
809:
808:
805:
802:
801:
799:
796:
789:
786:
784:
780:
775:
771:
767:
760:
755:
753:
748:
746:
741:
740:
737:
732:
728:
724:
718:
714:
710:
702:
698:
695:
694:
690:
680:
677:
672:
668:
663:
658:
654:
650:
647:: 2057–2069.
646:
642:
638:
636:
627:
624:
619:
615:
610:
605:
601:
597:
593:
586:
584:
580:
573:
571:
567:
563:
561:
557:
551:
541:
538:
530:
520:
516:
510:
507:This article
505:
496:
495:
489:
487:
485:
477:
474:
472:
469:
464:
462:
457:
455:
450:
448:
444:
440:
436:
432:
429:
425:
421:
417:
413:
409:
405:
401:
397:
393:
392:DnaB helicase
381:
378:
376:
372:
369:
365:
361:
358:
356:
352:
348:
344:
341:
338:
334:
329:
325:
322:
319:
317:
313:
310:
307:
305:
301:
298:
295:
292:
288:
285:
282:
280:
276:
272:
268:
263:
258:
247:
244:
242:
238:
235:
231:
227:
224:
222:
218:
214:
210:
207:
204:
200:
195:
191:
188:
184:
180:
177:
175:
171:
168:
165:
163:
159:
156:
153:
151:
147:
143:
139:
134:
130:Dnab helicase
129:
123:
118:
113:
102:
99:
95:
92:
89:
85:
80:
76:
73:
70:
68:
64:
59:
56:
53:
51:
47:
44:
41:
39:
35:
31:
27:
22:
17:
2655:Translocases
2652:
2639:
2626:
2613:
2600:
2590:Transferases
2587:
2574:
2431:Binding site
2168:
2102:
2058:
2036:
2021:Small GTPase
1635:Wilson/ATP7B
1630:Menkes/ATP7A
1430:Processivity
1256:synthesis in
827:
679:
644:
640:
634:
626:
602:(6): 691–8.
599:
595:
568:
564:
560:denaturation
552:
548:
533:
527:January 2018
524:
508:
483:
481:
475:
467:
465:
458:
451:
424:dimerisation
391:
390:
2426:Active site
2345:Mitofusin-1
2320:3.6.5.5-6:
2289:Prokaryotic
1449:Termination
1123:Prokaryotic
1115:Replication
791:Prokaryotic
770:prokaryotic
768:(comparing
463:) complex.
265:Identifiers
136:Identifiers
91:Swiss-model
24:Identifiers
2698:Categories
2629:Isomerases
2603:Hydrolases
2470:Regulation
2311:Eukaryotic
1944:Transducin
1781:(3.6.3.10)
1520:Hydrolases
1461:Telomerase
1435:DNA ligase
1428:Movement:
1252:Eukaryotic
1223:DNA gyrase
1208:DNA ligase
1127:elongation
858:Eukaryotic
795:initiation
783:Initiation
774:eukaryotic
574:References
428:N-terminal
422:involving
343:structures
209:structures
87:Structures
82:Search for
61:Other data
2709:Helicases
2508:EC number
2279:3.6.5.3:
2019:3.6.5.2:
1961:Gustducin
1893:3.6.5.1:
1727:(3.6.3.9)
1647:(3.6.3.8)
1550:Inorganic
1399:DNA clamp
1213:DNA clamp
1203:Replisome
731:IPR007694
717:IPR007693
596:Structure
556:supercoil
435:enzymatic
309:IPR007694
167:IPR007693
67:EC number
38:NCBI gene
2714:EC 3.6.1
2532:Kinetics
2456:Cofactor
2419:Activity
1555:Thiamine
1457:Telomere
1073:Replicon
1029:Helicase
1020:RNASEH2A
864:G1 phase
818:Helicase
727:InterPro
713:InterPro
671:33995902
635:in vitro
618:10404598
396:bacteria
360:RCSB PDB
304:InterPro
226:RCSB PDB
162:InterPro
101:InterPro
2688:Biology
2642:Ligases
2412:Enzymes
2366:Tubulin
2335:Dynamin
2187:other:
1867:Katanin
1857:Kinesin
1832:ATP13A3
1827:ATP13A2
1562:Apyrase
1370:epsilon
1258:S phase
1085:Lagging
1040:Primase
1015:RNASEH1
1010:RNase H
842:Primase
662:8085672
513:Please
484:E. coli
476:E. coli
426:of the
420:protein
418:of the
402:during
321:cd00984
284:PF03796
155:PF00772
128:E. coli
97:Domains
72:3.6.1.-
50:UniProt
2674:Portal
2616:Lyases
2243:ARL13B
2103:RhoBTB
1997:α12/13
1885:GTPase
1862:Myosin
1852:Dynein
1822:ATP12A
1817:ATP11B
1812:ATP10A
1807:ATP8B1
1797:Other
1769:ATP1B4
1764:ATP1B3
1759:ATP1B2
1754:ATP1B1
1749:ATP1A4
1744:ATP1A3
1739:ATP1A2
1734:ATP1A1
1725:Na+/K+
1713:ATP2C2
1708:ATP2C1
1696:ATP2B4
1691:ATP2B3
1686:ATP2B2
1681:ATP2B1
1669:ATP2A3
1664:ATP2A2
1659:ATP2A1
1608:ATPase
1101:Primer
703:(MeSH)
669:
659:
616:
431:domain
375:PDBsum
349:
339:
297:CL0023
273:DnaB_C
270:Symbol
241:PDBsum
215:
205:
187:SUPFAM
141:Symbol
55:P0ACB0
43:948555
29:Symbol
2568:Types
2299:EF-Tu
2238:SAR1B
2221:RAB27
2216:RAB23
2169:RhoDF
2059:RhoUV
2042:CDC42
2037:Cdc42
2023:>
2008:GNA13
2003:GNA12
1988:GNA11
1977:αq/11
1966:GNAT3
1954:GNAT2
1949:GNAT1
1939:GNAI3
1934:GNAI2
1929:GNAI1
1881:3.6.5
1845:3.6.4
1788:ATP4A
1779:H+/K+
1654:SERCA
1616:3.6.3
1600:3.6.3
1577:3.6.2
1537:3.6.1
1390:POLE4
1385:POLE3
1380:POLE2
1363:POLD4
1358:POLD3
1353:POLD2
1348:POLD1
1343:delta
1336:PRIM2
1331:PRIM1
1326:POLA2
1321:POLA1
1316:alpha
1049:PRIM2
1044:PRIM1
1001:SSBP4
996:SSBP3
991:SSBP2
183:SCOPe
174:SCOP2
2660:list
2653:EC7
2647:list
2640:EC6
2634:list
2627:EC5
2621:list
2614:EC4
2608:list
2601:EC3
2595:list
2588:EC2
2582:list
2575:EC1
2359:OPA1
2352:and
2304:EF-G
2294:IF-2
2260:Rheb
2248:ARL6
2233:ARF6
2204:NRAS
2199:KRAS
2194:HRAS
2179:RhoD
2174:RhoF
2120:RhoH
2096:RhoG
2081:Rac1
2069:RhoV
2064:RhoU
2047:TC10
1983:GNAQ
1703:SPCA
1530:3.6)
1476:DKC1
1471:TERC
1466:TERT
1422:Both
1404:PCNA
1375:POLE
1301:RPA1
1284:FEN1
1272:RFC1
1196:holE
1191:holD
1186:holC
1181:holB
1176:holA
1171:dnaX
1166:dnaT
1161:dnaQ
1156:dnaN
1151:dnaH
1146:dnaE
1141:dnaC
1087:and
1058:Both
1033:HFM1
963:MCM7
958:MCM6
953:MCM5
948:MCM4
943:MCM3
938:MCM2
926:Cdt1
919:Cdc6
910:ORC6
905:ORC5
900:ORC4
895:ORC3
890:ORC2
885:ORC1
847:dnaG
828:dnaB
823:dnaA
811:dnaC
725:and
723:Pfam
711:and
709:Pfam
667:PMID
614:PMID
478:dnaB
468:dnaB
447:dnaG
443:dnaC
439:dnaA
368:PDBj
364:PDBe
347:ECOD
337:Pfam
293:clan
291:Pfam
279:Pfam
234:PDBj
230:PDBe
213:ECOD
203:Pfam
179:1jwe
150:Pfam
144:DnaB
32:DnaB
2354:MX2
2350:MX1
2270:RGK
2265:Rap
2255:Ran
2228:Arf
2211:Rab
2189:Ras
2147:Rnd
2125:Rho
2076:Rac
2052:TCL
1913:olf
1645:Ca+
1069:Ori
772:to
657:PMC
649:doi
604:doi
517:to
482:In
454:ATP
412:DNA
355:PDB
316:CDD
221:PDB
2700::
2035::
1923:αi
1905:αs
1883::
1606::
1528:EC
1522::
1459::
1234::
1042::
1031::
833:T7
729::
715::
665:.
655:.
645:19
643:.
639:.
612:.
598:.
594:.
582:^
449:.
366:;
362:;
345:/
232:;
228:;
211:/
185:/
181:/
2676::
2662:)
2658:(
2649:)
2645:(
2636:)
2632:(
2623:)
2619:(
2610:)
2606:(
2597:)
2593:(
2584:)
2580:(
2404:e
2397:t
2390:v
2162:3
2157:2
2152:1
2140:C
2135:B
2130:A
2113:2
2108:1
2091:3
2086:2
1995:G
1975:G
1921:G
1911:G
1903:G
1604:4
1602:-
1526:(
1512:e
1505:t
1498:v
1312::
1260:)
1254:(
1129:)
1125:(
1071:/
1067:/
866:)
860:(
797:)
793:(
776:)
758:e
751:t
744:v
673:.
651::
620:.
606::
600:7
540:)
534:(
529:)
525:(
511:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.