207:, a middle periplasmic protein, an inner membrane protein, and a transmembrane duct. The transmembrane duct is located in the outer membrane of the cell. The duct is also bound to two other proteins: a periplasmic membrane protein and an integral membrane transporter. The periplasmic membrane protein and the inner membrane protein of the system are coupled to control the opening and closing of the duct (channel). When a toxin binds to this inner membrane protein, the inner membrane proteins gives rise to a biochemical cascade that transmits signals to the periplasmic membrane protein and outer membrane protein to open the channel and move the toxin out of the cell. This mechanism uses an energy-dependent, protein-protein interaction that is generated by the transfer of the toxin for an H+ ion by the inner membrane transporter. The fully assembled
34:
22:
414:
proteins (MDRs)- also referred as P-glycoprotein, multidrug resistance-associated proteins (MRPs), peptide transporters (PEPTs), and Na+ phosphate transporters (NPTs). These transporters are distributed along particular portions of the renal proximal tubule, intestine, liver, blood–brain barrier, and
423:
Several trials are currently being conducted to develop drugs that can be co-administered with antibiotics to act as inhibitors for the efflux-mediated extrusion of antibiotics. As yet, no efflux inhibitor has been approved for therapeutic use, but some are being used to determine the prevalence of
69:. This active efflux mechanism is responsible for various types of resistance to bacterial pathogens within bacterial species - the most concerning being antibiotic resistance because microorganisms can have adapted efflux pumps to divert toxins out of the cytoplasm and into extracellular media.
366:, thus contributing to both intrinsic (natural) and acquired resistance respectively. As an intrinsic mechanism of resistance, efflux pump genes can survive a hostile environment (for example in the presence of antibiotics) which allows for the selection of
392:
Expression of several efflux pumps in a given bacterial species may lead to a broad spectrum of resistance when considering the shared substrates of some multi-drug efflux pumps, where one efflux pump may confer resistance to a wide range of
76:) to pump out unwanted toxic substances through specific efflux pumps. Some efflux systems are drug-specific, whereas others may accommodate multiple drugs with small multidrug resistance (SMR) transporters.
61:. All microorganisms, with a few exceptions, have highly conserved DNA sequences in their genome that encode efflux pumps. Efflux pumps actively move substances out of a microorganism, in a process known as
57:
in their ability to remove antibiotics. The efflux could also be the movement of heavy metals, organic pollutants, plant-produced compounds, quorum sensing signals, bacterial metabolites and
223:
Although antibiotics are the most clinically important substrates of efflux systems, it is probable that most efflux pumps have other natural physiological functions. Examples include:
162:
298:
formation. However, the substrates for such pumps, and whether changes in their efflux activity affect biofilm formation directly or indirectly, remain to be determined.
302:
The ability of efflux systems to recognize a large number of compounds other than their natural substrates is probably because substrate recognition is based on
1370:
Molnár J, Engi H, Hohmann J, Molnár P, Deli J, Wesolowska O, Michalak K, Wang Q (2010). "Reversal of multidrug resistance by natural substances from plants".
1195:"Antibiotic inducibility of the MexXY multidrug efflux system of Pseudomonas aeruginosa: involvement of the antibiotic-inducible PA5471 gene product"
410:. Efflux pumps are one of the major causes of anticancer drug resistance in eukaryotic cells. They include monocarboxylate transporters (MCTs),
87:
of all kinds of cells. They are active transporters, meaning that they require a source of chemical energy to perform their function. Some are
945:"AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants"
614:
168:
1141:"Characterization of AcrD, a resistance-nodulation-cell division-type multidrug efflux pump from the fire blight pathogen Erwinia amylovora"
554:
Blanco P, Hernando-Amado S, Reales-Calderon JA, Corona F, Lira F, Alcalde-Rico M, Bernardini A, Sanchez MB, Martinez JL (February 2016).
1406:
1332:
Juliano RL, Ling V (November 1976). "A surface glycoprotein modulating drug permeability in
Chinese hamster ovary cell mutants".
884:
Shi X, Chen M, Yu Z, Bell JM, Wang H, Forrester I, Villarreal H, Jakana J, Du D, Luisi BF, Ludtke SJ, Wang Z (14 June 2019).
994:"Two distinct major facilitator superfamily drug efflux pumps mediate chloramphenicol resistance in Streptomyces coelicolor"
204:
146:
1243:
1457:"ZnO nanoparticles enhanced antibacterial activity of ciprofloxacin against Staphylococcus aureus and Escherichia coli"
338:
molecules - possessing both hydrophilic and hydrophobic characters - they are easily recognized by many efflux pumps.
382:
is also advantageous for the microorganisms as it allows for the easy spread of efflux genes between distant species.
233:
AcrAB efflux system, which has a physiologic role of pumping out bile acids and fatty acids to lower their toxicity.
1455:
Banoee M, Seif S, Nazari ZE, Jafari-Fesharaki P, Shahverdi HR, Moballegh A, Moghaddam KM, Shahverdi AR (May 2010).
191:, the RND family was once thought to be unique to Gram negative bacteria. They have since been found in all major
1502:
1100:"Induction of the mtrCDE-encoded efflux pump system of Neisseria gonorrhoeae requires MtrA, an AraC-like protein"
112:
411:
347:
323:
331:
265:
The MtrCDE system plays a protective role by providing resistance to faecal lipids in rectal isolates of
188:
176:
92:
481:
327:
267:
66:
1456:
735:"Distribution and physiology of ABC-type transporters contributing to multidrug resistance in bacteria"
1507:
1054:
897:
84:
833:
Wang Z, Fan G, Hryc CF, Blaza JN, Serysheva II, Schmid MF, Chiu W, Luisi BF, Du D (29 March 2017).
1407:"Alkaloids: an overview of their antibacterial, antibiotic-enhancing and antivirulence activities"
1041:
Du D, Wang Z, James NR, Voss JE, Klimont E, Ohene-Agyei T, Venter H, Chiu W, Luisi BF (May 2014).
1437:
303:
192:
1479:
1429:
1387:
1349:
1314:
1265:
1224:
1172:
1121:
1080:
1023:
974:
925:
866:
815:
764:
715:
653:
610:
587:
533:
276:
1471:
1421:
1379:
1341:
1304:
1296:
1255:
1214:
1206:
1162:
1152:
1111:
1070:
1062:
1013:
1005:
964:
956:
915:
905:
856:
846:
805:
795:
754:
746:
705:
697:
643:
632:"Bacterial multidrug efflux pumps: mechanisms, physiology and pharmacological exploitations"
577:
567:
523:
513:
229:
138:
96:
88:
73:
58:
50:
701:
441:
386:
371:
153:
402:
In eukaryotic cells, the existence of efflux pumps has been known since the discovery of
1058:
901:
1309:
1284:
1219:
1194:
1167:
1140:
1075:
1042:
1018:
993:
920:
885:
861:
834:
810:
783:
759:
734:
710:
685:
582:
555:
528:
501:
433:
403:
307:
1425:
969:
944:
258:
is suspected to have a role in the transport of the calcium-channel components in the
53:
in cells that moves out unwanted material. Efflux pumps are an important component in
1496:
1441:
1345:
1116:
1099:
556:"Bacterial Multidrug Efflux Pumps: Much More Than Antibiotic Resistance Determinants"
248:
134:
Bacterial efflux pumps are classified into five major superfamilies, based on their
1210:
681:
465:
437:
425:
244:
238:
184:
1285:"The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria"
960:
572:
407:
335:
311:
1383:
910:
648:
631:
518:
469:
449:
445:
379:
359:
315:
291:
is inducible by antibiotics that target ribosomes via the PA5471 gene product.
135:
108:
16:
Protein complexes that move compounds, generally toxic, out of bacterial cells
800:
1157:
835:"An allosteric transport mechanism for the AcrAB-TolC multidrug efflux pump"
444:
have been shown to inhibit bacterial efflux pumps including the carotenoids
429:
281:
175:
Of these, only the ABC superfamily are primary transporters, the rest being
104:
100:
1483:
1433:
1391:
1318:
1269:
1228:
1176:
1125:
1084:
1027:
929:
870:
819:
768:
719:
657:
591:
537:
978:
750:
25:
Protein TolC, the outer membrane component of a tripartite efflux pump in
1353:
1300:
1260:
1009:
461:
453:
440:, thereby facilitating fluorescent cell sorting for DNA content. Various
375:
363:
116:
54:
1475:
1066:
886:"In situ structure and assembly of the multidrug efflux pump AcrAB-TolC"
851:
502:"Efflux pump inhibitors for bacterial pathogens: From bench to bedside"
457:
295:
80:
318:
character rather than on defined chemical properties, as in classical
1464:
Journal of
Biomedical Materials Research Part B: Applied Biomaterials
549:
547:
367:
319:
215:
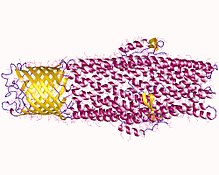
structures of AcrAB-TolC pump have been solved by cryoEM and cryoET.
180:
120:
32:
20:
287:
The MexXY component of the MexXY-OprM multidrug efflux system of
374:
these genes. Being located on transportable genetic elements as
355:
123:
33:
21:
609:. Switzerland: Springer International Publishing. p. 45.
284:, plant (host) colonization, and resistance to plant toxins.
72:
Efflux systems function via an energy-dependent mechanism (
350:
is large; this is usually attributed to the following:
141:
and the energy source used to export their substrates:
733:
Lubelski J, Konings WN, Driessen AJ (September 2007).
385:
Antibiotics can act as inducers and regulators of the
247:
pump for this organism when it turns on production of
1244:"Role of bacterial efflux pumps in biofilm formation"
992:
Vecchione JJ, Alexander B, Sello JK (November 2009).
163:
resistance-nodulation-cell division superfamily (RND)
95:
hydrolysis as a source of energy, whereas others are
294:
Efflux pumps have also been shown to play a role in
1043:"Structure of the AcrAB-TolC multidrug efflux pump"
636:
1334:Biochimica et Biophysica Acta (BBA) - Biomembranes
1098:Rouquette C, Harmon JB, Shafer WM (August 1999).
784:"Efflux pump-mediated resistance in chemotherapy"
358:elements encoding efflux pumps may be encoded on
1405:Cushnie TP, Cushnie B, Lamb AJ (November 2014).
500:Sharma A, Gupta VK, Pathania R (February 2019).
187:as a source of energy. Whereas MFS dominates in
1188:
1186:
1242:Alav I, Sutton JM, Rahman KM (February 2018).
788:Annals of Medical and Health Sciences Research
1414:International Journal of Antimicrobial Agents
8:
158:The small multidrug resistance family (SMR)
1365:
1363:
1283:Li XZ, Plésiat P, Nikaido H (April 2015).
1193:Morita Y, Sobel ML, Poole K (March 2006).
739:Microbiology and Molecular Biology Reviews
334:recognition. Because most antibiotics are
1308:
1259:
1218:
1166:
1156:
1115:
1074:
1017:
968:
943:Okusu H, Ma D, Nikaido H (January 1996).
919:
909:
860:
850:
809:
799:
758:
709:
686:"Bacterial multidrug efflux transporters"
647:
581:
571:
527:
517:
424:efflux pumps in clinical isolates and in
607:Small Multidrug Resistance Efflux Pumps
492:
472:, also inhibit bacterial efflux pumps.
1139:Pletzer D, Weingart H (January 2014).
111:) in which transport is coupled to an
1372:Current Topics in Medicinal Chemistry
1248:Journal of Antimicrobial Chemotherapy
998:Antimicrobial Agents and Chemotherapy
702:10.1146/annurev-biophys-051013-022855
630:Sun J, Deng Z, Yan A (October 2014).
203:Efflux pumps generally consist of an
169:multi antimicrobial extrusion protein
7:
782:Ughachukwu P, Unekwe P (July 2012).
675:
673:
671:
669:
667:
113:electrochemical potential difference
346:The impact of efflux mechanisms on
37:AcrB, the other component of pump,
342:Impact on antimicrobial resistance
14:
1426:10.1016/j.ijantimicag.2014.06.001
280:is important for this organism's
1117:10.1046/j.1365-2958.1999.01517.x
436:-mediated efflux of DNA-binding
432:, for example, is used to block
1211:10.1128/JB.188.5.1847-1855.2006
83:transporters localized in the
1:
1289:Clinical Microbiology Reviews
961:10.1128/jb.178.1.306-308.1996
573:10.3390/microorganisms4010014
415:other portions of the brain.
205:outer membrane efflux protein
147:major facilitator superfamily
97:secondary active transporters
1346:10.1016/0005-2736(76)90160-7
690:Annual Review of Biophysics
274:The AcrAB efflux system of
236:The MFS family Ptr pump in
89:primary active transporters
1524:
1384:10.2174/156802610792928103
911:10.1038/s41467-019-10512-6
649:10.1016/j.bbrc.2014.05.090
605:Bay DC, Turner RJ (2016).
519:10.4103/ijmr.IJMR_2079_17
254:The AcrAB–TolC system in
65:which is a vital part of
801:10.4103/2141-9248.105671
412:multiple drug resistance
348:antimicrobial resistance
1199:Journal of Bacteriology
1158:10.1186/1471-2180-14-13
949:Journal of Bacteriology
406:in 1976 by Juliano and
249:pristinamycins I and II
1104:Molecular Microbiology
189:Gram positive bacteria
177:secondary transporters
93:adenosine triphosphate
42:
30:
890:Nature Communications
751:10.1128/MMBR.00001-07
482:Antibiotic resistance
389:of some efflux pumps.
268:Neisseria gonorrhoeae
67:xenobiotic metabolism
36:
24:
1301:10.1128/CMR.00117-14
1010:10.1128/AAC.00853-09
306:properties, such as
85:cytoplasmic membrane
1476:10.1002/jbm.b.31615
1067:10.1038/nature13205
1059:2014Natur.509..512D
902:2019NatCo..10.2635S
852:10.7554/eLife.24905
460:, and the alkaloid
115:created by pumping
1261:10.1093/jac/dky042
680:Delmar JA, Su CC,
51:active transporter
43:
31:
616:978-3-319-39658-3
452:, the flavonoids
277:Erwinia amylovora
243:appears to be an
241:pristinaespiralis
79:Efflux pumps are
59:neurotransmitters
1515:
1503:Membrane biology
1488:
1487:
1461:
1452:
1446:
1445:
1411:
1402:
1396:
1395:
1367:
1358:
1357:
1329:
1323:
1322:
1312:
1280:
1274:
1273:
1263:
1254:(8): 2003–2020.
1239:
1233:
1232:
1222:
1190:
1181:
1180:
1170:
1160:
1145:BMC Microbiology
1136:
1130:
1129:
1119:
1095:
1089:
1088:
1078:
1038:
1032:
1031:
1021:
989:
983:
982:
972:
940:
934:
933:
923:
913:
881:
875:
874:
864:
854:
830:
824:
823:
813:
803:
779:
773:
772:
762:
730:
724:
723:
713:
677:
662:
661:
651:
627:
621:
620:
602:
596:
595:
585:
575:
551:
542:
541:
531:
521:
506:Indian J Med Res
497:
442:natural products
154:ABC transporters
74:active transport
39:Escherichia coli
27:Escherichia coli
1523:
1522:
1518:
1517:
1516:
1514:
1513:
1512:
1493:
1492:
1491:
1459:
1454:
1453:
1449:
1409:
1404:
1403:
1399:
1378:(17): 1757–68.
1369:
1368:
1361:
1331:
1330:
1326:
1282:
1281:
1277:
1241:
1240:
1236:
1192:
1191:
1184:
1138:
1137:
1133:
1097:
1096:
1092:
1053:(7501): 512–5.
1040:
1039:
1035:
991:
990:
986:
942:
941:
937:
883:
882:
878:
832:
831:
827:
781:
780:
776:
732:
731:
727:
679:
678:
665:
629:
628:
624:
617:
604:
603:
599:
553:
552:
545:
499:
498:
494:
490:
478:
421:
400:
393:antimicrobials.
344:
304:physicochemical
221:
201:
132:
126:into the cell.
17:
12:
11:
5:
1521:
1519:
1511:
1510:
1505:
1495:
1494:
1490:
1489:
1447:
1397:
1359:
1324:
1295:(2): 337–418.
1275:
1234:
1205:(5): 1847–55.
1182:
1131:
1090:
1033:
1004:(11): 4673–7.
984:
935:
876:
825:
774:
725:
663:
622:
615:
597:
560:Microorganisms
543:
512:(2): 129–145.
491:
489:
486:
485:
484:
477:
474:
468:, for example
434:P-glycoprotein
420:
417:
404:P-glycoprotein
399:
396:
395:
394:
390:
383:
343:
340:
308:hydrophobicity
300:
299:
292:
285:
272:
263:
252:
234:
220:
217:
200:
197:
173:
172:
171:family (MATE).
165:
159:
156:
150:
131:
128:
63:active efflux,
15:
13:
10:
9:
6:
4:
3:
2:
1520:
1509:
1506:
1504:
1501:
1500:
1498:
1485:
1481:
1477:
1473:
1470:(2): 557–61.
1469:
1465:
1458:
1451:
1448:
1443:
1439:
1435:
1431:
1427:
1423:
1420:(5): 377–86.
1419:
1415:
1408:
1401:
1398:
1393:
1389:
1385:
1381:
1377:
1373:
1366:
1364:
1360:
1355:
1351:
1347:
1343:
1340:(1): 152–62.
1339:
1335:
1328:
1325:
1320:
1316:
1311:
1306:
1302:
1298:
1294:
1290:
1286:
1279:
1276:
1271:
1267:
1262:
1257:
1253:
1249:
1245:
1238:
1235:
1230:
1226:
1221:
1216:
1212:
1208:
1204:
1200:
1196:
1189:
1187:
1183:
1178:
1174:
1169:
1164:
1159:
1154:
1150:
1146:
1142:
1135:
1132:
1127:
1123:
1118:
1113:
1109:
1105:
1101:
1094:
1091:
1086:
1082:
1077:
1072:
1068:
1064:
1060:
1056:
1052:
1048:
1044:
1037:
1034:
1029:
1025:
1020:
1015:
1011:
1007:
1003:
999:
995:
988:
985:
980:
976:
971:
966:
962:
958:
954:
950:
946:
939:
936:
931:
927:
922:
917:
912:
907:
903:
899:
895:
891:
887:
880:
877:
872:
868:
863:
858:
853:
848:
844:
840:
836:
829:
826:
821:
817:
812:
807:
802:
797:
793:
789:
785:
778:
775:
770:
766:
761:
756:
752:
748:
745:(3): 463–76.
744:
740:
736:
729:
726:
721:
717:
712:
707:
703:
699:
695:
691:
687:
683:
676:
674:
672:
670:
668:
664:
659:
655:
650:
645:
642:(2): 254–67.
641:
637:
633:
626:
623:
618:
612:
608:
601:
598:
593:
589:
584:
579:
574:
569:
565:
561:
557:
550:
548:
544:
539:
535:
530:
525:
520:
515:
511:
507:
503:
496:
493:
487:
483:
480:
479:
475:
473:
471:
467:
466:nanoparticles
463:
459:
455:
451:
447:
443:
439:
435:
431:
427:
418:
416:
413:
409:
405:
397:
391:
388:
384:
381:
377:
373:
369:
365:
361:
357:
353:
352:
351:
349:
341:
339:
337:
333:
329:
325:
321:
317:
313:
309:
305:
297:
293:
290:
289:P. aeruginosa
286:
283:
279:
278:
273:
270:
269:
264:
261:
257:
253:
250:
246:
242:
240:
235:
232:
231:
226:
225:
224:
218:
216:
214:
210:
206:
198:
196:
194:
190:
186:
182:
178:
170:
166:
164:
160:
157:
155:
151:
148:
144:
143:
142:
140:
137:
129:
127:
125:
122:
118:
114:
110:
106:
102:
98:
94:
90:
86:
82:
81:proteinaceous
77:
75:
70:
68:
64:
60:
56:
52:
48:
40:
35:
28:
23:
19:
1467:
1463:
1450:
1417:
1413:
1400:
1375:
1371:
1337:
1333:
1327:
1292:
1288:
1278:
1251:
1247:
1237:
1202:
1198:
1148:
1144:
1134:
1110:(3): 651–8.
1107:
1103:
1093:
1050:
1046:
1036:
1001:
997:
987:
955:(1): 306–8.
952:
948:
938:
893:
889:
879:
842:
838:
828:
794:(2): 191–8.
791:
787:
777:
742:
738:
728:
693:
689:
639:
635:
625:
606:
600:
563:
559:
509:
505:
495:
438:fluorophores
426:cell biology
422:
401:
345:
301:
288:
275:
266:
259:
255:
245:autoimmunity
239:Streptomyces
237:
228:
222:
212:
208:
202:
174:
133:
78:
71:
62:
46:
44:
38:
26:
18:
1508:Antibiotics
896:(1): 2635.
380:transposons
360:chromosomes
336:amphiphilic
312:aromaticity
109:antiporters
47:efflux pump
1497:Categories
696:: 93–117.
488:References
470:zinc oxide
450:capsorubin
446:capsanthin
428:research.
419:Inhibitors
398:Eukaryotic
387:expression
370:that over-
183:or sodium
179:utilizing
136:amino acid
105:symporters
101:uniporters
91:utilizing
1442:205171789
566:(1): 14.
430:Verapamil
324:substrate
316:ionizable
282:virulence
262:membrane.
199:Structure
130:Bacterial
1484:20225250
1434:25130096
1392:20645919
1319:25788514
1270:29506149
1229:16484195
1177:24443882
1126:10417654
1085:24747401
1028:19687245
930:31201302
871:28355133
820:23439914
769:17804667
720:24702006
684:(2014).
658:24878531
592:27681908
538:31219077
476:See also
464:. Some
462:lysergol
454:rotenone
376:plasmids
364:plasmids
332:receptor
219:Function
209:in vitro
193:kingdoms
185:gradient
139:sequence
117:hydrogen
55:bacteria
1310:4402952
1220:1426571
1168:3915751
1076:4361902
1055:Bibcode
1019:2772354
979:8550435
921:6570770
898:Bibcode
862:5404916
811:3573517
760:2168643
711:4769028
583:5029519
529:6563736
458:chrysin
372:express
368:mutants
362:and/or
356:genetic
296:biofilm
260:E. coli
256:E. coli
230:E. coli
213:in vivo
1482:
1440:
1432:
1390:
1354:990323
1352:
1317:
1307:
1268:
1227:
1217:
1175:
1165:
1151:: 13.
1124:
1083:
1073:
1047:Nature
1026:
1016:
977:
970:177656
967:
928:
918:
869:
859:
818:
808:
767:
757:
718:
708:
656:
613:
590:
580:
536:
526:
328:ligand
320:enzyme
181:proton
121:sodium
49:is an
1460:(PDF)
1438:S2CID
1410:(PDF)
839:eLife
682:Yu EW
149:(MFS)
107:, or
1480:PMID
1430:PMID
1388:PMID
1350:PMID
1315:PMID
1266:PMID
1225:PMID
1173:PMID
1122:PMID
1081:PMID
1024:PMID
975:PMID
926:PMID
867:PMID
816:PMID
765:PMID
716:PMID
654:PMID
611:ISBN
588:PMID
534:PMID
456:and
448:and
408:Ling
354:The
314:and
227:The
211:and
167:The
161:The
152:The
145:The
124:ions
1472:doi
1422:doi
1380:doi
1342:doi
1338:455
1305:PMC
1297:doi
1256:doi
1215:PMC
1207:doi
1203:188
1163:PMC
1153:doi
1112:doi
1071:PMC
1063:doi
1051:509
1014:PMC
1006:doi
965:PMC
957:doi
953:178
916:PMC
906:doi
857:PMC
847:doi
806:PMC
796:doi
755:PMC
747:doi
706:PMC
698:doi
644:doi
640:453
578:PMC
568:doi
524:PMC
514:doi
510:149
378:or
326:or
119:or
45:An
1499::
1478:.
1468:93
1466:.
1462:.
1436:.
1428:.
1418:44
1416:.
1412:.
1386:.
1376:10
1374:.
1362:^
1348:.
1336:.
1313:.
1303:.
1293:28
1291:.
1287:.
1264:.
1252:73
1250:.
1246:.
1223:.
1213:.
1201:.
1197:.
1185:^
1171:.
1161:.
1149:14
1147:.
1143:.
1120:.
1108:33
1106:.
1102:.
1079:.
1069:.
1061:.
1049:.
1045:.
1022:.
1012:.
1002:53
1000:.
996:.
973:.
963:.
951:.
947:.
924:.
914:.
904:.
894:10
892:.
888:.
865:.
855:.
845:.
841:.
837:.
814:.
804:.
790:.
786:.
763:.
753:.
743:71
741:.
737:.
714:.
704:.
694:43
692:.
688:.
666:^
652:.
638:.
634:.
586:.
576:.
562:.
558:.
546:^
532:.
522:.
508:.
504:.
310:,
195:.
103:,
1486:.
1474::
1444:.
1424::
1394:.
1382::
1356:.
1344::
1321:.
1299::
1272:.
1258::
1231:.
1209::
1179:.
1155::
1128:.
1114::
1087:.
1065::
1057::
1030:.
1008::
981:.
959::
932:.
908::
900::
873:.
849::
843:6
822:.
798::
792:2
771:.
749::
722:.
700::
660:.
646::
619:.
594:.
570::
564:4
540:.
516::
330:-
322:-
271:.
251:.
99:(
41:.
29:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.