369:
216:
26:
906:
Chishti AH, Kim AC, Marfatia SM, Lutchman M, Hanspal M, Jindal H, Liu SC, Low PS, Rouleau GA, Mohandas N, Chasis JA, Conboy JG, Gascard P, Takakuwa Y, Huang SC, Benz EJ, Bretscher A, Fehon RG, Gusella JF, Ramesh V, Solomon F, Marchesi VT, Tsukita S, Tsukita S, Hoover KB (August 1998). "The FERM
671:
The FERM domain has also been called the amino-terminal domain, the 30kDa domain, 4.1N30, the membrane-cytoskeletal-linking domain, the ERM-like domain, the ezrin-like domain of the band 4.1 superfamily, the
593:), but the other proteins in which the FERM domain is found do not share any region of similarity outside of this domain. ERM proteins are made of three domains, the FERM domain, a central
625:. For cytoskeleton-membrane cross-linking, the dormant molecules becomes activated and the FERM domain attaches to the membrane by binding specific membrane proteins, while the last 34
493:
327:
174:
118:
106:
429:
263:
86:
513:
347:
194:
854:
944:"Structure of the ERM protein moesin reveals the FERM domain fold masked by an extended actin binding tail domain"
610:
449:
283:
1088:
501:
335:
182:
833:
812:
644:(RhoGDI), which suggests that in addition to functioning as a cross-linker, ERM proteins may influence Rho
879:
805:
497:
331:
178:
442:
276:
99:
454:
288:
657:
653:
626:
130:
973:
673:
645:
602:
590:
30:
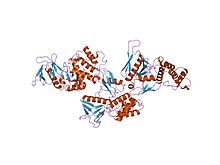
crystal structure of the ferm domain of merlin, the neurofibromatosis 2 tumor suppressor protein.
993:"Structural basis of the membrane-targeting and unmasking mechanisms of the radixin FERM domain"
633:. Aside from binding to membranes, the activated FERM domain of ERM proteins can also bind the
1022:
965:
924:
757:
732:
649:
613:. ERM proteins are regulated by an intramolecular association of the FERM and C-terminal tail
488:
322:
169:
1012:
1004:
955:
916:
774:
480:
314:
161:
907:
domain: a unique module involved in the linkage of cytoplasmic proteins to the membrane".
738:
728:
707:
630:
614:
570:
554:
558:
1017:
992:
960:
943:
920:
111:
1082:
764:
606:
434:
268:
91:
397:
373:
crystal structure of the radixin ferm domain complexed with the nep cytoplasmic tail
244:
54:
977:
742:
721:
699:
618:
574:
562:
476:
310:
157:
410:
135:
123:
67:
1074:
1060:
1046:
605:
of the FERM domain is highly conserved among ERM proteins and is responsible for
422:
256:
79:
823:
788:
753:
746:
703:
1008:
796:
768:
714:
637:
578:
816:
809:
665:
661:
1026:
969:
928:
868:
that appear to act at junctions between the membrane and the cytoskeleton.
438:
368:
272:
215:
95:
25:
1070:
1056:
1042:
841:
837:
826:
799:
792:
692:
688:
622:
566:
417:
251:
74:
991:
Hamada K, Shimizu T, Matsui T, Tsukita S, Hakoshima T (September 2000).
865:
634:
598:
550:
546:
538:
872:
829:
784:
641:
542:
508:
342:
189:
656:
modules (F1, F2, and F3) that together form a compact clover-shaped
745:
protein concentrated in regions of cell-substratum contact and, in
609:
association by direct binding to the cytoplasmic domain or tail of
861:
857:
760:
696:
594:
569:
that associate with various proteins at the interface between the
534:
530:
832:
that are non-covalently associated with the cytoplasmic tails of
1066:
1052:
1038:
848:
470:
404:
392:
304:
239:
151:
61:
49:
727:
Radixin, which is involved in the binding of the barbed end of
220:
crystal structure of the protein 4.1r membrane binding domain
942:
Pearson MA, Reczek D, Bretscher A, Karplus PA (April 2000).
731:
to the plasma membrane in the undercoat of the cell-to-cell
652:
of the FERM domain reveals that it is composed of three
589:
Ezrin, moesin, and radixin are highly related proteins (
676:
N-terminal region, and the membrane attachment domain.
1065:
This article incorporates text from the public domain
1051:
This article incorporates text from the public domain
1037:
This article incorporates text from the public domain
720:
Moesin, which is probably involved in binding major
507:
487:
469:
464:
448:
428:
416:
403:
391:
383:
378:
361:
341:
321:
303:
298:
282:
262:
250:
238:
230:
225:
208:
188:
168:
150:
145:
129:
117:
105:
85:
73:
60:
48:
40:
35:
18:
581:in the majority of proteins in which it is found.
597:domain and a C-terminal tail domain, which binds
901:
899:
897:
8:
713:Ezrin, a component of the undercoat of the
461:
367:
295:
214:
142:
24:
1016:
959:
684:FERM domain containing proteins include:
763:and which is involved in the control of
893:
358:
205:
15:
847:Non-receptor tyrosine-protein kinase
7:
577:. The FERM domain is located at the
664:-like. The C-terminal module is a
14:
875:and PTP-D1, PTP-RL10 and PTP2E.
822:Janus tyrosine kinases (JAKs),
815:involved in signalling through
871:Protein-tyrosine phosphatases
640:dissociation inhibitor of Rho
549:module involved in localising
362:FERM C-terminal PH-like domain
1:
961:10.1016/S0092-8674(00)80836-3
921:10.1016/S0968-0004(98)01237-7
855:Protein-tyrosine phosphatases
465:Available protein structures:
299:Available protein structures:
146:Available protein structures:
660:. The N-terminal module is
1105:
1064:
1050:
1036:
883:protein phosphatase ptp-1.
611:integral membrane proteins
525:In molecular biology, the
561:are found in a number of
460:
366:
294:
213:
141:
23:
1009:10.1093/emboj/19.17.4449
813:protein tyrosine kinases
749:, of cell-cell contacts.
724:to the plasma membrane.
722:cytoskeletal structures
880:Caenorhabditis elegans
806:Focal-adhesion kinases
585:Structure and function
19:FERM N-terminal domain
909:Trends Biochem. Sci
646:signalling pathways
209:FERM central domain
791:and XV, which are
691:, which links the
591:ERM protein family
545:) is a widespread
777:(or schwannomin).
733:Adherens junction
650:crystal structure
629:of the tail bind
617:that masks their
601:. The amino-acid
523:
522:
519:
518:
514:structure summary
357:
356:
353:
352:
348:structure summary
204:
203:
200:
199:
195:structure summary
1096:
1031:
1030:
1020:
988:
982:
981:
963:
939:
933:
932:
903:
840:or polypeptidic
717:plasma membrane.
462:
371:
359:
296:
218:
206:
143:
28:
16:
1104:
1103:
1099:
1098:
1097:
1095:
1094:
1093:
1089:Protein domains
1079:
1078:
1077:
1063:
1049:
1035:
1034:
1003:(17): 4449–62.
990:
989:
985:
941:
940:
936:
905:
904:
895:
890:
783:Unconventional
729:actin filaments
708:plasma membrane
682:
631:actin filaments
587:
571:plasma membrane
555:plasma membrane
374:
221:
107:OPM superfamily
31:
12:
11:
5:
1102:
1100:
1092:
1091:
1081:
1080:
1033:
1032:
983:
934:
892:
891:
889:
886:
885:
884:
876:
869:
852:
845:
820:
803:
781:
778:
772:
750:
736:
725:
718:
711:
681:
678:
668:-like domain.
586:
583:
521:
520:
517:
516:
511:
505:
504:
491:
485:
484:
474:
467:
466:
458:
457:
452:
446:
445:
432:
426:
425:
420:
414:
413:
408:
401:
400:
395:
389:
388:
385:
381:
380:
376:
375:
372:
364:
363:
355:
354:
351:
350:
345:
339:
338:
325:
319:
318:
308:
301:
300:
292:
291:
286:
280:
279:
266:
260:
259:
254:
248:
247:
242:
236:
235:
232:
228:
227:
223:
222:
219:
211:
210:
202:
201:
198:
197:
192:
186:
185:
172:
166:
165:
155:
148:
147:
139:
138:
133:
127:
126:
121:
115:
114:
109:
103:
102:
89:
83:
82:
77:
71:
70:
65:
58:
57:
52:
46:
45:
42:
38:
37:
33:
32:
29:
21:
20:
13:
10:
9:
6:
4:
3:
2:
1101:
1090:
1087:
1086:
1084:
1076:
1072:
1068:
1062:
1058:
1054:
1048:
1044:
1040:
1028:
1024:
1019:
1014:
1010:
1006:
1002:
998:
994:
987:
984:
979:
975:
971:
967:
962:
957:
954:(3): 259–70.
953:
949:
945:
938:
935:
930:
926:
922:
918:
914:
910:
902:
900:
898:
894:
887:
882:
881:
877:
874:
870:
867:
863:
859:
856:
853:
850:
846:
843:
839:
835:
831:
828:
825:
821:
818:
814:
811:
807:
804:
801:
798:
794:
790:
786:
782:
780:Protein NBL4.
779:
776:
773:
770:
766:
765:cell motility
762:
759:
756:protein that
755:
752:Filopodin, a
751:
748:
744:
740:
737:
734:
730:
726:
723:
719:
716:
712:
709:
705:
701:
698:
694:
690:
687:
686:
685:
679:
677:
675:
669:
667:
663:
659:
655:
651:
647:
643:
639:
636:
632:
628:
624:
620:
619:binding sites
616:
612:
608:
604:
600:
596:
592:
584:
582:
580:
576:
572:
568:
564:
560:
556:
552:
548:
544:
540:
536:
532:
528:
515:
512:
510:
506:
503:
499:
495:
492:
490:
486:
482:
478:
475:
472:
468:
463:
459:
456:
453:
451:
447:
444:
440:
436:
433:
431:
427:
424:
421:
419:
415:
412:
409:
406:
402:
399:
396:
394:
390:
386:
382:
377:
370:
365:
360:
349:
346:
344:
340:
337:
333:
329:
326:
324:
320:
316:
312:
309:
306:
302:
297:
293:
290:
287:
285:
281:
278:
274:
270:
267:
265:
261:
258:
255:
253:
249:
246:
243:
241:
237:
233:
229:
224:
217:
212:
207:
196:
193:
191:
187:
184:
180:
176:
173:
171:
167:
163:
159:
156:
153:
149:
144:
140:
137:
134:
132:
128:
125:
122:
120:
116:
113:
110:
108:
104:
101:
97:
93:
90:
88:
84:
81:
78:
76:
72:
69:
66:
63:
59:
56:
53:
51:
47:
43:
39:
34:
27:
22:
17:
1000:
996:
986:
951:
947:
937:
915:(8): 281–2.
912:
908:
878:
743:cytoskeletal
704:erythrocytes
700:cytoskeleton
683:
670:
588:
575:cytoskeleton
565:-associated
563:cytoskeletal
526:
524:
824:cytoplasmic
810:cytoplasmic
754:slime mould
747:lymphocytes
531:4.1 protein
527:FERM domain
379:Identifiers
226:Identifiers
119:OPM protein
36:Identifiers
888:References
797:congenital
769:chemotaxis
715:microvilli
654:structural
638:nucleotide
621:for other
579:N terminus
541:and M for
477:structures
311:structures
158:structures
131:Membranome
1075:IPR018979
1061:IPR019748
1047:IPR018980
838:cytokines
834:receptors
817:integrins
674:conserved
662:ubiquitin
658:structure
623:molecules
423:IPR018980
257:IPR019748
80:IPR018979
1083:Category
1071:InterPro
1057:InterPro
1043:InterPro
1027:10970839
970:10847681
842:hormones
827:tyrosine
808:(FAKs),
800:deafness
693:spectrin
689:Band 4.1
680:Examples
627:residues
607:membrane
603:sequence
573:and the
567:proteins
551:proteins
537:, R for
533:, E for
494:RCSB PDB
418:InterPro
328:RCSB PDB
252:InterPro
175:RCSB PDB
75:InterPro
978:7119092
929:9757824
866:enzymes
830:kinases
793:mutated
785:myosins
706:to the
635:guanine
615:domains
599:F-actin
595:helical
559:domains
557:. FERM
553:to the
547:protein
539:radixin
529:(F for
455:cd00836
398:PF09380
289:cd14473
245:PF00373
55:PF09379
1025:
1018:302071
1015:
997:EMBO J
976:
968:
927:
873:PTPN14
775:Merlin
648:. The
642:GTPase
543:moesin
509:PDBsum
483:
473:
443:SUPFAM
411:CL0266
387:FERM_C
384:Symbol
343:PDBsum
317:
307:
277:SUPFAM
234:FERM_M
231:Symbol
190:PDBsum
164:
154:
100:SUPFAM
68:CL0072
44:FERM_N
41:Symbol
974:S2CID
862:PTPN4
858:PTPN3
761:actin
758:binds
739:Talin
697:actin
535:ezrin
439:SCOPe
430:SCOP2
273:SCOPe
264:SCOP2
96:SCOPe
87:SCOP2
1069:and
1067:Pfam
1055:and
1053:Pfam
1041:and
1039:Pfam
1023:PMID
966:PMID
948:Cell
925:PMID
860:and
849:TYK2
836:for
789:VIIa
767:and
741:, a
502:PDBj
498:PDBe
481:ECOD
471:Pfam
435:1ef1
407:clan
405:Pfam
393:Pfam
336:PDBj
332:PDBe
315:ECOD
305:Pfam
269:1gc7
240:Pfam
183:PDBj
179:PDBe
162:ECOD
152:Pfam
124:1gc6
92:1gc7
64:clan
62:Pfam
50:Pfam
1013:PMC
1005:doi
956:doi
952:101
917:doi
795:in
787:X,
702:of
489:PDB
450:CDD
323:PDB
284:CDD
170:PDB
136:161
1085::
1073::
1059::
1045::
1021:.
1011:.
1001:19
999:.
995:.
972:.
964:.
950:.
946:.
923:.
913:23
911:.
896:^
864:,
666:PH
500:;
496:;
479:/
441:/
437:/
334:;
330:;
313:/
275:/
271:/
181:;
177:;
160:/
112:49
98:/
94:/
1029:.
1007::
980:.
958::
931:.
919::
851:.
844:.
819:.
802:.
771:.
735:.
710:.
695:-
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.