492:(IBT) when they have the same phenotypic effect. Alleles that are identical by type fall into two groups; those that are identical by descent (IBD) because they arose from the same allele in an earlier generation; and those that are non-identical by descent (NIBD) or identical by state (IBS) because they arose from separate mutations. Parent-offspring pairs share 50% of their genes IBD, and monozygotic twins share 100% IBD. What is relevant in linkage analysis is the inheritance (or coinheritance) of alleles at adjacent loci; therefore; it is critical importance to determine whether the alleles are identical by descent (i.e. copies from same parental alleles) or only identical by state (i.e. appearing same, but derived from two different copies of alleles). Therefore, there three categories of family-based linkage analysis – strongly modeled (the traditional lod score model), weakly model based (variance components methods), or model free. Variance component methods may be viewed as hybrids.
269:
TOBS /3/ SON 64/KLRE // CIAN0 /4/ SEE /n/, nth order cross BACK CROSS SYMBOL *n n number of times the back cross parent used left side simple cross symbol, back cross parent is the female, right side – male, Example: SEE/3*ANE, TOBS*6/CIAN0
290:
effectiveness in multiple genetic backgrounds. Often, by the time a QTL mapping population is developed and mapped, breeders have introgressed the new QTL using traditional breeding and selection methods. This can reduce the usefulness of MAS (marker-assisted selection) within breeding programs at the time when MAS could be most useful (i.e., shortly after new QTL are identified). Family-based QTL mapping removes this limitation by using existing plant breeding families.
563:
pattern across the genome, which is essential for association studies. Actually there is no better way to understand LD pattern than to know the haplotypes themselves. Haplotypes tell us how alleles are organized along the chromosome and reflect the pattern of inheritance over evaluations. Second, methods based on haplotypes can be more powerful than those based on single markers in association studies of mapping complex trait genes.
240:
405:, development in efficient algorithms, and computing power have enabled the large scale application of these methods. While linkage studies seek to identify loci cosegregate with the trait within families, association studies seek to identify particular variants that are associated with the phenotype at the population level. These are complementary methods that, together, provide means to probe the genome and describe etiology of
423:
273:
33:
136:
74:
409:. In linkage studies, we seek to identify the loci that cosegregate with a specific genomic region, tagged by polymorphic markers, within families. In contrast, in association studies, we seek a correlation between a specific genetic variation and trait variation in sample of individuals, implicating a causal role of the variant.
318:
technique are based on this type of populations. In plant context such population are hard to find as most of individuals are someway related. Other disadvantage of such method is that even if we can find such a population, it is difficult to find high allele frequency for allele of interest (usually
268:
SIMPLE CROSS SYMBOL Example / first order cross SON 64/KLRE //, second order cross IR 64/KLRE // CIAN0 /3/, third order cross TOBS /3/ SON 64/KLRE // CIAN0 /4/, fourth order cross
536:
The family-based, Tran-disequilibirum test (TDT) has gained wide popularity in recent years, this method also focuses on alleles transmitted to affect offispring, but it is formulated to take account of both the linkage and the disequilibrium that underlie the association. The test requires genotype
562:
refer to combinations of marker alleles which are located closely together on the same chromosome and which tend to be inherited together. With availability of high density SNP makers, haplotypes play an important role in association studies. First – haplotypes are critical to understanding the LD
514:
community for its potential to use existing genetic resources collections to fine map quantitative trait loci (QTL), validate candidate genes, and identify alleles of interest (Yu and
Buckler, 2006). The three elements of particular importance for conducting association mapping or interpreting the
334:
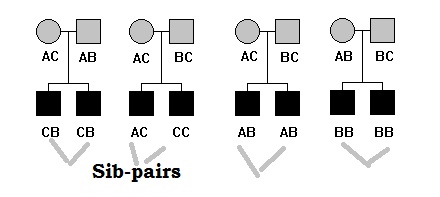
Such design include a pair of sibs from multiple independent families. The members in each sibpairs are not randomly chosen – often both siblings are chosen from one tail (upper or lower) of the distribution of the QT (concordant siblings) or one sibling is chosen from the upper tail and the other
289:
Traditional mapping populations include single family consisting of crossing between two parents or three parents often distantly related. There are some important limitations associated with traditional mapping methods. Some of which include limited polymorphism rates, and no indication of marker
572:
PEDPLOT,PEDRAW/WPEDRAW (Pedigree
Drawing/ Window Pedigree Drawing (MS-Window and X-Window version of PEDRAW)), PROGENY (Progeny Software, LLC) etc. However the pedigree drawing in plants requires some additional features such as inbreeding, selfing, mutation, polyploidy etc. which is supported in
541:
and tests the null hypothesis that the putative disease associated allele is transmitted 50% of the time from the heterogygous parents against the alternative hypothesis that the trait positive allele -associated allele will be transmitted more often. The TDT is not affected by population
571:
There are several pedigree drawing software available for human genetics context such as COPE (COllaborative
Pedigree drawing Environment), CYRILLIC, FTM (Family Tree Maker), FTREE, KINDRED, PED (PEdigree Drawing software),PEDHUNTER, PEDIGRAPH, PEDIGREE/DRAW, PEDIGREE-VISUALIZER,
305:
Broadly, there are 3 classes of study designs: study designs in which large sets of relatives from extended or nuclear families are sampled, study designs in which pairs of relatives are sampled (e.g., sibling pairs) or study designs in which unrelated individuals are sampled.
295:
483:
Genetic linkage is the phenomenon where by alleles at different loci cosegregate in families. The strength of cosegregation is measured by the recombination fraction θ, the probability of an odd number of recombination. More complex pedigree provide higher power.
557:
The TDT has been extended in context of quantitative traits and nuclear or extended pedigree families. The generalized test allows to use any family type of families in testing. QTDT has also been extended to haplotype-based association mapping.
335:
sibling is chosen from the lower tail (discordant siblings). Another sampling design could include a pair of siblings, one chosen from the upper or lower tail of the distribution and the other chosen randomly from among the remaining siblings.
633:
Rosyara, U. R.; Gonzalez-Hernandez, J. L.; Glover, K. D.; Gedye, K. R.; Stein, J. M. (2009). "Family-based mapping of quantitative trait loci in plant breeding populations with resistance to
Fusarium head blight in wheat as an illustration".
547:
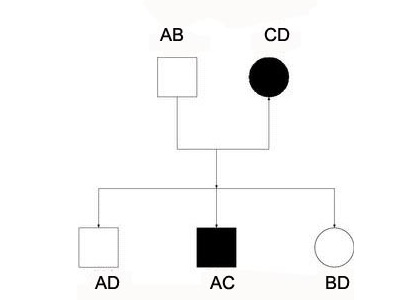
260:– an individual can be male or female and mating can be performed in random combinations, with inbreeding loops. Also plant pedigrees may contain of "selfs", i.e. offspring resulting from self-pollination of a plant.
350:
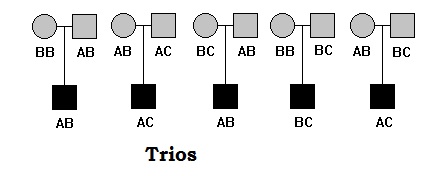
Trios include parents and one offspring (most affected). Trios are more commonly used in association studies. The concept of association mapping that each trio are unrelated, however trios are related in themselves.
294:
400:
Linkage and association analysis are primary tools for gene discovery, localization and functional analysis. While conceptual underpinning of these approaches have been long known, advances in recent decades in
280:
The idea of family-based QTL mapping comes from inheritance of marker alleles and its association with trait of interest has demonstrated how to use family-based association in plant breeding families.
537:
information on trio individuals, namely affected child and both biological parents; and at least one parent must be heterozygous for the test to be informative. The proposed test statistic is actually
256:
populations. Pedigree records are kept by plants breeders and pedigree-based selection is popular in several plant species. Plant pedigrees are different from that of humans, particularly as plant are
576:. The pedimap can be used for pedigree visualization along with phenotypic, genotypic and ibd probabilities data in every type of plant pedigrees in both diploids and tetraploids.
546:
390:
381:
In extended pedigree include multiple generation pedigree. It can be as deep or wide as the pedigree information is available. Extended pedigree are attractive for
324:
371:
236:
information include information about ancestry. Keeping pedigree records is a centuries-old tradition. Pedigrees can also be verified using gene-marker data.
610:
340:
497:
444:
99:
46:
488:(IBD) matrix estimation is a central component in mapping of Quantitative Trait Loci (QTL) using variance component models. Alleles have
356:
573:
213:
responsible for important economic, health or environmental characters. Mapping QTLs is an important activity that plant breeders and
314:
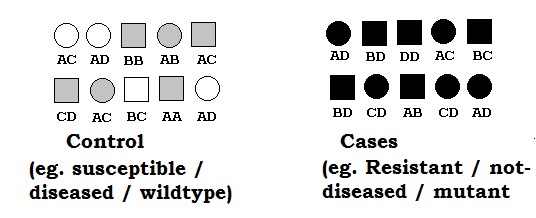
Natural collection of individuals (considered unrelated) with unknown pedigree constitutes mapping populations. The population based
470:
189:
171:
117:
60:
522:
its use to control for spurious associations and consequences in the specific case of differential selection among subgroups, and
146:
276:
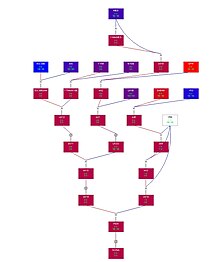
Example pedigree of
Sonalika (SONALIKA = = I53.388/AN//YT54/N10B/3/Lerma Rojo/4/B4946.A.4.18.2.IY/Y53//3*Y50 drawn using Pedimap
448:
784:
52:
542:
stratification and admixture. The concept of family-based test of association has been extended to quantitative traits.
605:
389:
595:
89:
433:
452:
437:
257:
202:
319:
mutant)in such situation. For purpose of create balance in allele frequency, usually case-control studies.
507:
533:
In contrast to population-based association, family-based association tests are becoming more popular.
370:
323:
767:
Flint-Garcia S, Thornsberry JM, Buckler ESIV (2003) "Structure of linkage disequilibrium in plants".
703:
95:
529:
and its consequences on the resolution and the application of LD mapping (Flint-Garcia et al. 2003).
590:
485:
315:
659:
402:
731:
651:
600:
538:
84:
339:
721:
711:
643:
496:
489:
585:
382:
239:
760:
Yu J, Buckler ES (2006) "Genetic association mapping and genome organization of maize".
707:
676:
Beavis W.D. (1998) "QTL analyses: power, precision, and accuracy". In: Paterson AH (ed)
17:
406:
253:
233:
748:
Glazier AM, Nadeau JH, Aitman TJ (2002) "Finding genes that underlie complex traits".
726:
691:
778:
663:
422:
272:
206:
696:
Proceedings of the
National Academy of Sciences of the United States of America
647:
559:
526:
510:(LD) and association mapping is receiving considerable attention in the plant
214:
355:
218:
716:
655:
735:
511:
209:
is the process of identifying genomic regions that potentially contain
366:
Nuclear family consists of two generation simple family pedigree.
271:
238:
210:
416:
225:
is a variant of QTL mapping where multiple-families are used.
129:
67:
26:
145:
may be in need of reorganization to comply with
Knowledge's
692:"Construction of multilocus genetic linkage maps in humans"
243:
A complex five generation plant pedigree drawn with
Pedimap
217:
routinely use to associate potential causal genes with
153:
553:
Quantitative transmission disequilibrium test (QTDT)
519:
the analysis of population structure into subgroups,
252:The method has been discussed in the context of
525:the analysis of the local structure of LD into
156:to make improvements to the overall structure.
8:
451:. Unsourced material may be challenged and
61:Learn how and when to remove these messages
611:Physiological and molecular wheat breeding
725:
715:
628:
626:
471:Learn how and when to remove this message
190:Learn how and when to remove this message
172:Learn how and when to remove this message
118:Learn how and when to remove this message
622:
7:
678:Molecular analysis of complex traits
449:adding citations to reliable sources
680:. CRC Press, Boca Raton, pp 145–161
285:Limitation of conventional methods
25:
690:Lander, E. S.; Green, P. (1987).
503:Family-based association analysis
42:This article has multiple issues.
636:Theoretical and Applied Genetics
545:
495:
421:
388:
369:
354:
338:
322:
293:
134:
98:has been specified. Please help
72:
31:
396:Linkage vs association analysis
301:Common study population mapping
50:or discuss these issues on the
539:McNemar's chi-square statistic
1:
413:Family-based linkage analysis
229:Pedigree in humans and wheat
801:
606:Nested association mapping
648:10.1007/s00122-009-1010-9
596:Marker assisted selection
567:Drawing family pedigrees
223:Family-based QTL mapping
18:Family based QTL mapping
203:Quantitative trait loci
717:10.1073/pnas.84.8.2363
508:Linkage disequilibrium
383:linkage-based analysis
277:
244:
310:Unrelated individuals
275:
242:
785:Statistical genetics
762:Curr Opin Biotechnol
445:improve this section
100:improve this article
88:to meet Knowledge's
769:Annu Rev Plant Biol
708:1987PNAS...84.2363L
591:Genetic association
486:Identity by descent
316:association mapping
264:Pedigree denotation
154:editing the article
403:molecular genetics
377:Extended pedigrees
278:
245:
601:Molecular markers
515:results include:
481:
480:
473:
200:
199:
192:
182:
181:
174:
147:layout guidelines
128:
127:
120:
90:quality standards
81:This article may
65:
16:(Redirected from
792:
753:
746:
740:
739:
729:
719:
702:(8): 2363–2367.
687:
681:
674:
668:
667:
642:(8): 1617–1631.
630:
549:
499:
490:identity by type
476:
469:
465:
462:
456:
425:
417:
392:
373:
358:
342:
326:
297:
195:
188:
177:
170:
166:
163:
157:
138:
137:
130:
123:
116:
112:
109:
103:
76:
75:
68:
57:
35:
34:
27:
21:
800:
799:
795:
794:
793:
791:
790:
789:
775:
774:
757:
756:
747:
743:
689:
688:
684:
675:
671:
632:
631:
624:
619:
586:Animal breeding
582:
569:
555:
505:
477:
466:
460:
457:
442:
426:
415:
398:
379:
364:
359:
348:
332:
312:
303:
287:
270:
266:
250:
231:
196:
185:
184:
183:
178:
167:
161:
158:
152:Please help by
151:
139:
135:
124:
113:
107:
104:
93:
77:
73:
36:
32:
23:
22:
15:
12:
11:
5:
798:
796:
788:
787:
777:
776:
773:
772:
765:
755:
754:
741:
682:
669:
621:
620:
618:
615:
614:
613:
608:
603:
598:
593:
588:
581:
578:
568:
565:
554:
551:
531:
530:
523:
520:
504:
501:
479:
478:
429:
427:
420:
414:
411:
407:complex traits
397:
394:
378:
375:
363:
362:Nuclear family
360:
353:
347:
344:
331:
328:
311:
308:
302:
299:
286:
283:
267:
265:
262:
258:hermaphroditic
254:plant breeding
249:
246:
230:
227:
198:
197:
180:
179:
142:
140:
133:
126:
125:
96:cleanup reason
80:
78:
71:
66:
40:
39:
37:
30:
24:
14:
13:
10:
9:
6:
4:
3:
2:
797:
786:
783:
782:
780:
770:
766:
763:
759:
758:
752:298:2345–2349
751:
745:
742:
737:
733:
728:
723:
718:
713:
709:
705:
701:
697:
693:
686:
683:
679:
673:
670:
665:
661:
657:
653:
649:
645:
641:
637:
629:
627:
623:
616:
612:
609:
607:
604:
602:
599:
597:
594:
592:
589:
587:
584:
583:
579:
577:
575:
566:
564:
561:
552:
550:
548:
543:
540:
534:
528:
524:
521:
518:
517:
516:
513:
509:
502:
500:
498:
493:
491:
487:
475:
472:
464:
454:
450:
446:
440:
439:
435:
430:This section
428:
424:
419:
418:
412:
410:
408:
404:
395:
393:
391:
386:
384:
376:
374:
372:
367:
361:
357:
352:
345:
343:
341:
336:
329:
327:
325:
320:
317:
309:
307:
300:
298:
296:
291:
284:
282:
274:
263:
261:
259:
255:
247:
241:
237:
235:
228:
226:
224:
221:of interest.
220:
216:
212:
208:
204:
194:
191:
176:
173:
165:
155:
149:
148:
143:This article
141:
132:
131:
122:
119:
111:
101:
97:
91:
87:
86:
79:
70:
69:
64:
62:
55:
54:
49:
48:
43:
38:
29:
28:
19:
768:
761:
749:
744:
699:
695:
685:
677:
672:
639:
635:
570:
556:
544:
535:
532:
506:
494:
482:
467:
458:
443:Please help
431:
399:
387:
380:
368:
365:
349:
337:
333:
321:
313:
304:
292:
288:
279:
251:
232:
222:
201:
186:
168:
162:January 2012
159:
144:
114:
108:January 2012
105:
82:
58:
51:
45:
44:Please help
41:
461:August 2019
215:geneticists
207:QTL mapping
205:mapping or
102:if you can.
771:54:357–374
764:17:155–160
617:References
560:Haplotypes
527:haplotypes
219:phenotypes
47:improve it
432:does not
248:In plants
53:talk page
779:Category
656:19322557
580:See also
512:genetics
330:Sibpairs
234:Pedigree
83:require
750:Science
736:3470801
704:Bibcode
664:2882803
574:Pedimap
453:removed
438:sources
85:cleanup
734:
727:304651
724:
662:
654:
660:S2CID
346:Trios
211:genes
732:PMID
652:PMID
436:any
434:cite
722:PMC
712:doi
644:doi
640:118
447:by
94:No
781::
730:.
720:.
710:.
700:84
698:.
694:.
658:.
650:.
638:.
625:^
385:.
56:.
738:.
714::
706::
666:.
646::
474:)
468:(
463:)
459:(
455:.
441:.
193:)
187:(
175:)
169:(
164:)
160:(
150:.
121:)
115:(
110:)
106:(
92:.
63:)
59:(
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.