1196:
1589:
1601:
345:
28:, an area in which molecular structure is determined from scattering data (usually of X-rays, electrons or neutrons). In fiber diffraction, the scattering pattern does not change, as the sample is rotated about a unique axis (the fiber axis). Such uniaxial symmetry is frequent with filaments or fibers consisting of biological or man-made
353:
475:
The three-dimensional sketch demonstrates that in the example experiment the collected information on the molecular structure of the polypropylene fiber is almost complete. By rotation of the plane pattern about the meridian the scattering data collected in 4 s fill an almost spherical volume of
435:
the distance between sample and detector is computed using known crystallographic data of the reference reflection, a uniformly gridded map for the representative fiber plane in reciprocal space is constructed and the diffraction data are fed into this map. The figure on the right shows the result.
652:
Saad
Mohamed (1994) "Low resolution structure and packing investigations of collagen crystalline domains in tendon using Synchrotron Radiation X-rays, Structure factors determination, evaluation of Isomorphous Replacement methods and other modeling." PhD Thesis, Université Joseph Fourier Grenoble
98:
Non-ideal fiber patterns are obtained in experiments. They only show mirror symmetry about the meridian. The reason is that the fiber axis and the incident beam (X-rays, electrons, neutrons) cannot be perfectly oriented perpendicular to each other. The corresponding geometric distortion has been
463:
there is structure information on the meridian. Of course, there is now 4-quadrant symmetry. This means that in the example pattern part of the missing information may be copied "from the lower half to the upper half" into the white areas. Thus, it frequently makes sense to tilt the fiber
119:
have carried out their own geometrical reasoning and presented an approximative equation for the fiber tilt angle β. Analysis starts by mapping the distorted 2D pattern on the representative plane of the fiber. This is the plane that contains the cylinder axis in
170:
391:
with respect to the vertical direction. This shortcoming is compensated by simple rotation of the picture. 4 straight arrows point at 4 reflection images of a chosen reference reflection. Their positions are used to determine the fiber tilt angle
136:
starts from the
Franklin approximation for the tilt angle β. It eliminates fiber tilt, unwarps the detector image, and corrects the scattering intensity. The correct equation for the determination of β has been presented by Norbert Stribeck.
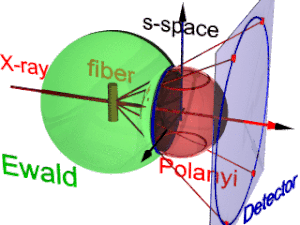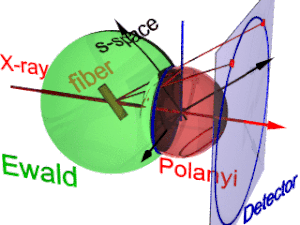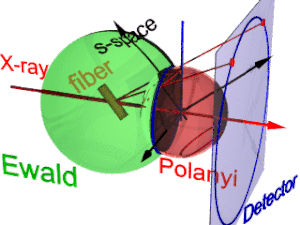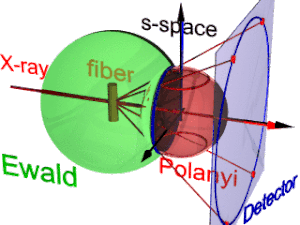
185:. Reference direction is the primary beam (label: X-ray). If the fiber is tilted away from the perpendicular direction by an angle β, as well the information about its molecular structure in reciprocal space (trihedron labelled
200:
In s-space each reflection is found on its
Polanyi-sphere. Intrinsically the ideal reflection is a point in s-space, but fiber symmetry turns it into a ring smeared out by rotation about the fiber direction.
1466:
476:
s-space. In the example the 4-quadrant symmetry has not yet been considered to fill part of the white spots. For clarity a quarter of the sphere has been cut out, but keeping the equatorial plane itself.
1461:
221:, blue ring). There up to 4 images (red spots) of the monitored reflection can show up. The position of the reflection images is a function of the orientation of the fiber in the primary beam (
1517:
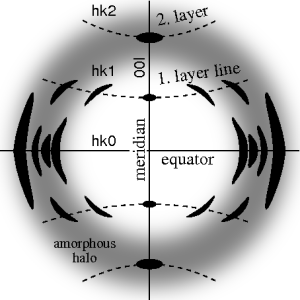
67:. In case of fiber symmetry, many more reflections than in single-crystal diffraction show up in the 2D pattern. In fiber patterns these reflections clearly appear arranged along lines (
51:
Ideal fiber diffraction pattern of a semi-crystalline material with amorphous halo and reflections on layer lines. High intensity is represented by dark color. The fiber axis is vertical
1639:
177:). Structure information is in reciprocal space (black axes), expanded on surfaces of Polanyi spheres. In the animation 1 Polanyi sphere with 1 reflection on it is monitored
304:
440:
there remain white spots at the meridian, in which structure information is missing. Only in the center of the image and at an s-value related to the scattering angle
36:, fiber symmetry is an aggravation regarding the determination of crystal structure, because reflections are smeared and may overlap in the fiber diffraction pattern.
461:
330:
433:
410:
389:
1811:
1780:
1724:
1672:
412:. The image has been recorded on a CCD detector. It shows the logarithmic intensitity in pseudo-color representation. Here bright colors represent high intensity.
252:
145:
Fibrous materials such as wool or cotton easily form aligned bundles, and were among the first biological macromolecules studied by X-ray diffraction, notably by
485:
Arnott S & Wonacott A J, The
Refinement of the Molecular & Crystal Structures of Polymers Using X-Ray Data and Stereochemical Constraints, Polymer 1966
1335:
1522:
1247:
197:
all the points of reciprocal space are found that are seen by the detector. These points are mapped on the pixels of the detector by central projection.
217:(blue ring). It does not change as the fiber is tilted. As with a slide projector the reflection circle is projected (red moving rays) on the detector (
1632:
1413:
705:
44:
diffraction pattern exposed on photographic film or on a 2D detector. 2 instead of 3 co-ordinate directions suffice to describe fiber diffraction.
1708:
1512:
1504:
1565:
1543:
1625:
1558:
1408:
1074:
939:
788:
677:
1548:
1446:
1142:
209:
with respect to the origin of s-space. Mapped onto the detector are only those points of the reflection in s-space that are both on the
795:
468:
48:
531:
Franklin RE, Gosling RG (1953) "The
Structure of Sodium Thymonucleate Fibres. II. The Cylindrically Symmetrical Patterson Function".
1570:
1428:
1398:
1327:
1734:
1280:
1827:
645:
Klug HP, Alexander LE (1974) "X-Ray
Diffraction Procedures For Polycrystalline and Amorphous Materials", 2nd ed, Wiley, New York
1682:
1553:
1476:
1350:
949:
492:
Bian W, Wang H, McCullogh I, Stubbs G (2006). "WCEN: a computer program for initial processing of fiber diffraction patterns".
1310:
613:
Rajkumar G, AL-Khayat H, Eakins F, He A, Knupp C, Squire J (2005) "FibreFix — A New
Integrated CCP13 Software Package",
332:
is valid. From the
Polanyi representation of fiber diffraction geometry the relations of the fiber mapping are established by
1388:
173:
Fiber diffraction geometry changes as the fiber is tilted (tilt-angle β is between the blue rigid axis and the axis labelled
436:
Change of scattering intensity has been considered in the unwarping process. Because of the curvature of the surface of the
667:
1770:
1403:
1393:
698:
513:
Cochran W, Crick FHC, and Vand V (1952). "The
Structure of Synthetic Polypeptides. I. The Transform of Atoms on a Helix".
1175:
1891:
1869:
1527:
800:
778:
553:
Hamilton W C, R-Factors, Statistics and Truth, Paper H5, Amer Cryst Ass
Program & Abstracts, Boulder, Colorado, 1961
193:
has its center in the sample. Its radius is 1/λ, with λ the wavelength of the incident radiation. On the surface of the
1205:
1079:
40:
considers fiber symmetry a simplification, because almost the complete obtainable structure information is in a single
833:
728:
1195:
225:). Inverted, from the positions of the reflection images the orientation of the fiber can be determined, if for the
1911:
1886:
1605:
1436:
733:
75:
becomes palpable. Bent layer lines indicate that the pattern must be straightened. Reflections are labelled by the
1832:
1451:
1380:
838:
828:
964:
1775:
1677:
1662:
1593:
1317:
1213:
1086:
1049:
843:
823:
691:
651:
1001:
680:— an introduction provided by Prof. K.C. Holmes, Max Planck Institute for Medical Research, Heidelberg.
1796:
1749:
1441:
1285:
1230:
979:
944:
1137:
954:
1059:
542:
Fraser RDB, Macrae TP, Miller A, Rowlands RJ (1976). "Digital Processing of Fibre Diffraction Patterns".
1853:
1837:
1806:
1667:
1494:
1290:
1252:
1011:
745:
524:
Donohue J, and Trueblood, K N, On the unreliability of the reliability index, Acta Crystallogr, 1956,
1218:
1091:
927:
818:
580:
Millane RP, Arnott S (1985) "Digital Processing of X-Ray Diffraction Patterns from Oriented Fibers".
1874:
1235:
1223:
1098:
1064:
1044:
257:
149:
in the early 1930s. Fiber diffraction data led to several important advances in the development of
471:
3D representation of the reciprocal space filled with scattering data from the polypropylene fiber
1801:
1729:
1484:
1295:
1240:
783:
150:
368:
before mapping it into reciprocal space. The mirror axis in the pattern is rotated by the angle
91:
artificial fiber diffraction patterns are generated by rotating a single crystal about an axis (
71:) running almost parallel to the equator. Thus, in fiber diffraction the layer line concept of
1703:
1648:
1418:
1257:
1185:
1165:
885:
755:
181:
The animation shows the geometry of fiber diffraction. It is based on the notions proposed by
112:
108:
37:
16:
Subarea of scattering, an area in which molecular structure is determined from scattering data
443:
309:
1456:
1262:
1180:
1170:
969:
902:
873:
866:
563:
James T W & Mazia D, Surface Films of Desoxyribonucleic Acid, Biochim Biophys Acta 1953
418:
395:
371:
129:
121:
1765:
1345:
1340:
1305:
1125:
1024:
959:
922:
917:
768:
714:
671:
182:
146:
125:
116:
100:
88:
72:
33:
556:
Hamilton W C, Significance Tests on the Crystallographic R Factor, Acta Crystallogr 1965
231:
1155:
1120:
1108:
1103:
1069:
1039:
1029:
988:
932:
856:
810:
206:
1905:
1300:
1113:
912:
365:
357:
29:
624:
Stribeck N (2009). "On the determination of fiber tilt angles in fiber diffraction"
1744:
1006:
996:
890:
773:
642:
Alexander LE (1979) "X-Ray Diffraction Methods in Polymer Science", Wiley, New York
437:
226:
210:
194:
190:
76:
602:
Polanyi M, Weissenberg K (1923) "Das Röntgen-Faserdiagramm (Zweite Mitteilung)".
1489:
1160:
1034:
861:
664:
154:
1739:
1617:
1054:
740:
169:
132:
is computed that is refined iteratively. The digital method frequently called
25:
205:
rings represent each reflection on the Polanyi sphere, because scattering is
763:
570:
Marvin DA (2017) "Fibre diffraction studies of biological macromolecules".
467:
344:
47:
1360:
1130:
878:
674:— Software (Linux, Mac, Windows) for the analysis of fiber patterns
333:
1370:
352:
503:
Bunn C W, Chemical Crystallography, University of Oxford, 2nd Ed, 1967
506:
Campbell Smith P J & Arnott S, LALS (etc.) Acta Crystallogr 1978
1365:
683:
466:
351:
343:
168:
46:
591:
Polanyi M (1921) "Das Röntgen-Faserdiagramm (Erste Mitteilung)".
1621:
687:
158:
360:
mapped into (the representative plane of) reciprocal space
1467:
Zeitschrift für Kristallographie – New Crystal Structures
1462:
Zeitschrift für Kristallographie – Crystalline Materials
364:
The figure on the left shows a typical fiber pattern of
1355:
87:. Reflections on the meridian are 00l-reflections. In
648:
Warren BE (1990) "X-Ray Diffraction". Dover, New York
446:
421:
398:
374:
312:
260:
234:
59:. In the ideal pattern, the fiber axis is called the
1862:
1846:
1820:
1789:
1758:
1717:
1691:
1655:
1536:
1503:
1475:
1427:
1379:
1326:
1273:
1204:
987:
978:
901:
809:
754:
721:
455:
427:
404:
383:
324:
298:
246:
213:and on the Polanyi sphere. These points form the
157:and the Watson-Crick model of double-stranded
1633:
699:
8:
128:first an approximation of the mapping into
1640:
1626:
1618:
1533:
984:
806:
751:
706:
692:
684:
445:
420:
397:
373:
311:
285:
277:
269:
261:
259:
233:
63:, the perpendicular direction is called
79:hkl, i.e. 3 digits. Reflections on the
189:) is tilted. In reciprocal space the
7:
1600:
940:Phase transformation crystallography
107:(German: "Lagenkugel") intersecting
1447:Journal of Chemical Crystallography
153:, e.g., the original models of the
14:
55:The ideal fiber pattern exhibits
1735:Dual-polarization interferometry
1599:
1588:
1587:
1194:
1766:Analytical ultracentrifugation
1389:Bilbao Crystallographic Server
286:
278:
270:
262:
1:
1771:Size exclusion chromatography
299:{\displaystyle |h|+|k|\neq 0}
1870:Protein structure prediction
1828:Hydrogen–deuterium exchange
1649:Protein structural analysis
1437:Crystal Growth & Design
729:Timeline of crystallography
103:introducing the concept of
1928:
1248:Nuclear magnetic resonance
165:Fiber diffraction geometry
1883:
1833:Site-directed mutagenesis
1583:
1452:Journal of Crystal Growth
1192:
572:Prog. Biophys. Mol. Biol.
1678:Electron crystallography
1663:Cryo-electron microscopy
1318:Single particle analysis
1176:Hermann–Mauguin notation
348:A measured fiber pattern
336:and spherical geometry.
1797:Fluorescence anisotropy
1759:Translational Diffusion
1750:Fluorescence anisotropy
1442:Crystallography Reviews
1286:Isomorphous replacement
1080:Lomer–Cottrell junction
582:J. Macromol. Sci. Phys.
456:{\displaystyle 2\beta }
415:After determination of
325:{\displaystyle l\neq 0}
99:extensively studied by
93:rotating crystal method
83:-th layer line share l=
955:Spinodal decomposition
670:July 23, 2012, at the
615:Fibre Diffraction Rev.
472:
457:
429:
428:{\displaystyle \beta }
406:
405:{\displaystyle \beta }
385:
384:{\displaystyle ~\phi }
361:
349:
326:
300:
248:
178:
52:
1892:Quaternary structure→
1854:Equilibrium unfolding
1838:Chemical modification
1807:Dielectric relaxation
1668:X-ray crystallography
1495:Gregori Aminoff Prize
1291:Molecular replacement
544:J. Appl. Crystallogr.
494:J. Appl. Crystallogr.
470:
458:
430:
407:
386:
355:
347:
327:
301:
249:
172:
50:
1790:Rotational Diffusion
801:Structure prediction
444:
419:
396:
372:
310:
258:
232:
42:two-dimensional (2D)
1887:←Tertiary structure
1065:Cottrell atmosphere
1045:Partial dislocation
789:Restriction theorem
247:{\displaystyle hkl}
57:4-quadrant symmetry
1802:Flow birefringence
1730:Circular dichroism
1485:Carl Hermann Medal
1296:Molecular dynamics
1143:Defects in diamond
1138:Stone–Wales defect
784:Reciprocal lattice
746:Biocrystallography
473:
453:
425:
402:
381:
362:
350:
340:Pattern correction
322:
296:
244:
179:
151:structural biology
53:
1912:Protein structure
1899:
1898:
1875:Molecular docking
1704:Mass spectrometry
1699:Fiber diffraction
1692:Medium resolution
1615:
1614:
1579:
1578:
1186:Thermal ellipsoid
1151:
1150:
1060:Frank–Read source
1020:
1019:
886:Aperiodic crystal
852:
851:
734:Crystallographers
678:Fiber Diffraction
626:Acta Crystallogr.
533:Acta Crystallogr.
515:Acta Crystallogr.
377:
356:Fiber pattern of
215:reflection circle
134:Fraser correction
113:Rosalind Franklin
38:Materials science
22:Fiber diffraction
1919:
1776:Light scattering
1642:
1635:
1628:
1619:
1603:
1602:
1591:
1590:
1534:
1457:Kristallografija
1311:Gerchberg–Saxton
1206:Characterisation
1198:
1181:Structure factor
985:
970:Ostwald ripening
807:
752:
708:
701:
694:
685:
462:
460:
459:
454:
434:
432:
431:
426:
411:
409:
408:
403:
390:
388:
387:
382:
375:
331:
329:
328:
323:
305:
303:
302:
297:
289:
281:
273:
265:
253:
251:
250:
245:
223:Polanyi equation
130:reciprocal space
122:reciprocal space
105:Polanyi's sphere
24:is a subarea of
1927:
1926:
1922:
1921:
1920:
1918:
1917:
1916:
1902:
1901:
1900:
1895:
1894:
1889:
1879:
1858:
1842:
1816:
1785:
1754:
1713:
1687:
1656:High resolution
1651:
1646:
1616:
1611:
1575:
1532:
1499:
1471:
1423:
1375:
1346:CrystalExplorer
1322:
1306:Phase retrieval
1269:
1200:
1199:
1190:
1147:
1126:Schottky defect
1025:Perfect crystal
1016:
1012:Abnormal growth
974:
960:Supersaturation
923:Miscibility gap
904:
897:
848:
805:
769:Bravais lattice
750:
717:
715:Crystallography
712:
672:Wayback Machine
661:
639:
482:
464:intentionally.
442:
441:
417:
416:
394:
393:
370:
369:
342:
308:
307:
256:
255:
230:
229:
219:detector circle
207:point symmetric
183:Michael Polanyi
167:
147:William Astbury
143:
141:Historical role
126:crystallography
117:Raymond Gosling
101:Michael Polanyi
89:crystallography
73:crystallography
34:crystallography
17:
12:
11:
5:
1925:
1923:
1915:
1914:
1904:
1903:
1897:
1896:
1890:
1885:
1884:
1881:
1880:
1878:
1877:
1872:
1866:
1864:
1860:
1859:
1857:
1856:
1850:
1848:
1844:
1843:
1841:
1840:
1835:
1830:
1824:
1822:
1818:
1817:
1815:
1814:
1809:
1804:
1799:
1793:
1791:
1787:
1786:
1784:
1783:
1778:
1773:
1768:
1762:
1760:
1756:
1755:
1753:
1752:
1747:
1742:
1737:
1732:
1727:
1721:
1719:
1715:
1714:
1712:
1711:
1706:
1701:
1695:
1693:
1689:
1688:
1686:
1685:
1680:
1675:
1670:
1665:
1659:
1657:
1653:
1652:
1647:
1645:
1644:
1637:
1630:
1622:
1613:
1612:
1610:
1609:
1597:
1584:
1581:
1580:
1577:
1576:
1574:
1573:
1568:
1563:
1562:
1561:
1556:
1551:
1540:
1538:
1531:
1530:
1525:
1520:
1515:
1509:
1507:
1501:
1500:
1498:
1497:
1492:
1487:
1481:
1479:
1473:
1472:
1470:
1469:
1464:
1459:
1454:
1449:
1444:
1439:
1433:
1431:
1425:
1424:
1422:
1421:
1416:
1411:
1406:
1401:
1396:
1391:
1385:
1383:
1377:
1376:
1374:
1373:
1368:
1363:
1358:
1353:
1348:
1343:
1338:
1332:
1330:
1324:
1323:
1321:
1320:
1315:
1314:
1313:
1303:
1298:
1293:
1288:
1283:
1281:Direct methods
1277:
1275:
1271:
1270:
1268:
1267:
1266:
1265:
1260:
1250:
1245:
1244:
1243:
1238:
1228:
1227:
1226:
1221:
1210:
1208:
1202:
1201:
1193:
1191:
1189:
1188:
1183:
1178:
1173:
1168:
1166:Ewald's sphere
1163:
1158:
1152:
1149:
1148:
1146:
1145:
1140:
1135:
1134:
1133:
1128:
1118:
1117:
1116:
1111:
1109:Frenkel defect
1106:
1104:Bjerrum defect
1096:
1095:
1094:
1084:
1083:
1082:
1077:
1072:
1070:Peierls stress
1067:
1062:
1057:
1052:
1047:
1042:
1040:Burgers vector
1032:
1030:Stacking fault
1027:
1021:
1018:
1017:
1015:
1014:
1009:
1004:
999:
993:
991:
989:Grain boundary
982:
976:
975:
973:
972:
967:
962:
957:
952:
947:
942:
937:
936:
935:
933:Liquid crystal
930:
925:
920:
909:
907:
899:
898:
896:
895:
894:
893:
883:
882:
881:
871:
870:
869:
864:
853:
850:
849:
847:
846:
841:
836:
831:
826:
821:
815:
813:
804:
803:
798:
796:Periodic table
793:
792:
791:
786:
781:
776:
771:
760:
758:
749:
748:
743:
738:
737:
736:
725:
723:
719:
718:
713:
711:
710:
703:
696:
688:
682:
681:
675:
660:
659:External links
657:
656:
655:
649:
646:
643:
638:
635:
634:
633:
622:
611:
600:
589:
578:
568:
561:
554:
551:
540:
529:
522:
511:
504:
501:
490:
481:
478:
452:
449:
424:
401:
380:
341:
338:
321:
318:
315:
295:
292:
288:
284:
280:
276:
272:
268:
264:
243:
240:
237:
166:
163:
142:
139:
109:Ewald's sphere
30:macromolecules
15:
13:
10:
9:
6:
4:
3:
2:
1924:
1913:
1910:
1909:
1907:
1893:
1888:
1882:
1876:
1873:
1871:
1868:
1867:
1865:
1863:Computational
1861:
1855:
1852:
1851:
1849:
1847:Thermodynamic
1845:
1839:
1836:
1834:
1831:
1829:
1826:
1825:
1823:
1819:
1813:
1810:
1808:
1805:
1803:
1800:
1798:
1795:
1794:
1792:
1788:
1782:
1779:
1777:
1774:
1772:
1769:
1767:
1764:
1763:
1761:
1757:
1751:
1748:
1746:
1743:
1741:
1738:
1736:
1733:
1731:
1728:
1726:
1723:
1722:
1720:
1718:Spectroscopic
1716:
1710:
1707:
1705:
1702:
1700:
1697:
1696:
1694:
1690:
1684:
1681:
1679:
1676:
1674:
1671:
1669:
1666:
1664:
1661:
1660:
1658:
1654:
1650:
1643:
1638:
1636:
1631:
1629:
1624:
1623:
1620:
1608:
1607:
1598:
1596:
1595:
1586:
1585:
1582:
1572:
1569:
1567:
1564:
1560:
1557:
1555:
1552:
1550:
1547:
1546:
1545:
1542:
1541:
1539:
1535:
1529:
1526:
1524:
1521:
1519:
1516:
1514:
1511:
1510:
1508:
1506:
1502:
1496:
1493:
1491:
1488:
1486:
1483:
1482:
1480:
1478:
1474:
1468:
1465:
1463:
1460:
1458:
1455:
1453:
1450:
1448:
1445:
1443:
1440:
1438:
1435:
1434:
1432:
1430:
1426:
1420:
1417:
1415:
1412:
1410:
1407:
1405:
1402:
1400:
1397:
1395:
1392:
1390:
1387:
1386:
1384:
1382:
1378:
1372:
1369:
1367:
1364:
1362:
1359:
1357:
1354:
1352:
1349:
1347:
1344:
1342:
1339:
1337:
1334:
1333:
1331:
1329:
1325:
1319:
1316:
1312:
1309:
1308:
1307:
1304:
1302:
1301:Patterson map
1299:
1297:
1294:
1292:
1289:
1287:
1284:
1282:
1279:
1278:
1276:
1272:
1264:
1261:
1259:
1256:
1255:
1254:
1251:
1249:
1246:
1242:
1239:
1237:
1234:
1233:
1232:
1229:
1225:
1222:
1220:
1217:
1216:
1215:
1212:
1211:
1209:
1207:
1203:
1197:
1187:
1184:
1182:
1179:
1177:
1174:
1172:
1171:Friedel's law
1169:
1167:
1164:
1162:
1159:
1157:
1154:
1153:
1144:
1141:
1139:
1136:
1132:
1129:
1127:
1124:
1123:
1122:
1119:
1115:
1114:Wigner effect
1112:
1110:
1107:
1105:
1102:
1101:
1100:
1099:Interstitials
1097:
1093:
1090:
1089:
1088:
1085:
1081:
1078:
1076:
1073:
1071:
1068:
1066:
1063:
1061:
1058:
1056:
1053:
1051:
1048:
1046:
1043:
1041:
1038:
1037:
1036:
1033:
1031:
1028:
1026:
1023:
1022:
1013:
1010:
1008:
1005:
1003:
1000:
998:
995:
994:
992:
990:
986:
983:
981:
977:
971:
968:
966:
963:
961:
958:
956:
953:
951:
948:
946:
945:Precipitation
943:
941:
938:
934:
931:
929:
926:
924:
921:
919:
916:
915:
914:
913:Phase diagram
911:
910:
908:
906:
900:
892:
889:
888:
887:
884:
880:
877:
876:
875:
872:
868:
865:
863:
860:
859:
858:
855:
854:
845:
842:
840:
837:
835:
832:
830:
827:
825:
822:
820:
817:
816:
814:
812:
808:
802:
799:
797:
794:
790:
787:
785:
782:
780:
777:
775:
772:
770:
767:
766:
765:
762:
761:
759:
757:
753:
747:
744:
742:
739:
735:
732:
731:
730:
727:
726:
724:
720:
716:
709:
704:
702:
697:
695:
690:
689:
686:
679:
676:
673:
669:
666:
663:
662:
658:
654:
650:
647:
644:
641:
640:
636:
631:
627:
623:
620:
616:
612:
609:
605:
601:
598:
594:
590:
587:
583:
579:
576:
573:
569:
566:
562:
559:
555:
552:
549:
545:
541:
538:
534:
530:
527:
523:
520:
516:
512:
509:
505:
502:
499:
495:
491:
488:
484:
483:
479:
477:
469:
465:
450:
447:
439:
422:
413:
399:
378:
367:
366:polypropylene
359:
358:polypropylene
354:
346:
339:
337:
335:
319:
316:
313:
293:
290:
282:
274:
266:
241:
238:
235:
228:
224:
220:
216:
212:
208:
204:
198:
196:
192:
188:
184:
176:
171:
164:
162:
160:
156:
152:
148:
140:
138:
135:
131:
127:
123:
118:
114:
110:
106:
102:
96:
94:
90:
86:
82:
78:
74:
70:
66:
62:
58:
49:
45:
43:
39:
35:
31:
27:
23:
19:
1745:Fluorescence
1698:
1604:
1592:
1537:Associations
1505:Organisation
997:Disclination
928:Polymorphism
891:Quasicrystal
834:Orthorhombic
774:Miller index
722:Key concepts
629:
625:
618:
614:
607:
603:
596:
592:
585:
581:
574:
571:
564:
557:
547:
543:
536:
532:
525:
518:
514:
507:
497:
493:
486:
474:
438:Ewald sphere
414:
363:
227:Miller index
222:
218:
214:
211:Ewald sphere
202:
199:
195:Ewald sphere
191:Ewald sphere
186:
180:
174:
144:
133:
104:
97:
92:
84:
80:
77:Miller index
68:
64:
60:
56:
54:
41:
21:
20:
18:
1490:Ewald Prize
1258:Diffraction
1236:Diffraction
1219:Diffraction
1161:Bragg plane
1156:Bragg's law
1035:Dislocation
950:Segregation
862:Crystallite
779:Point group
69:layer lines
1740:Absorbance
1274:Algorithms
1263:Scattering
1241:Scattering
1224:Scattering
1092:Slip bands
1055:Cross slip
905:transition
839:Tetragonal
829:Monoclinic
741:Metallurgy
637:Text books
521:, 581–586.
500:, 752–756.
480:References
334:elementary
26:scattering
1381:Databases
844:Triclinic
824:Hexagonal
764:Unit cell
756:Structure
610:, 123-130
604:Z. Physik
599:, 149-180
593:Z. Physik
588:, 193-227
567:367 - 370
560:502 - 510
539:, 678-685
489:157 - 166
451:β
423:β
400:β
379:ϕ
317:≠
291:≠
1906:Category
1821:Chemical
1594:Category
1429:Journals
1361:OctaDist
1356:JANA2020
1328:Software
1214:Electron
1131:F-center
918:Eutectic
879:Fiveling
874:Twinning
867:Equiaxed
668:Archived
577:, 43–87.
550:, 81–94.
111:. Later
61:meridian
1606:Commons
1554:Germany
1231:Neutron
1121:Vacancy
980:Defects
965:GP-zone
811:Systems
632:, 46-47
621:, 11-18
187:s-space
175:s-space
155:α-helix
65:equator
1549:France
1544:Europe
1477:Awards
1007:Growth
857:Growth
510:3 - 11
376:
1571:Japan
1518:IOBCr
1371:SHELX
1366:Olex2
1253:X-ray
903:Phase
819:Cubic
528:, 615
254:both
124:. In
32:. In
1709:SAXS
1513:IUCr
1414:ICDD
1409:ICSD
1394:CCDC
1341:Coot
1336:CCP4
1087:Slip
1050:Kink
665:WCEN
306:and
115:and
1812:NMR
1781:NMR
1725:NMR
1683:EPR
1673:NMR
1528:DMG
1523:RAS
1419:PDB
1404:COD
1399:CIF
1351:DSR
1075:GND
1002:CSL
630:A65
586:B24
575:127
508:A34
203:Two
159:DNA
95:).
1908::
1566:US
1559:UK
628:,
619:13
617:,
606:,
595:,
584:,
565:10
558:18
546:,
535:,
517:,
498:39
496:,
161:.
1641:e
1634:t
1627:v
707:e
700:t
693:v
653:1
608:9
597:7
548:9
537:6
526:9
519:5
487:7
448:2
320:0
314:l
294:0
287:|
283:k
279:|
275:+
271:|
267:h
263:|
242:l
239:k
236:h
85:i
81:i
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.