389:
20:
40:
32:
320:. When pRb is bound to E2F, E2F is inactive. As cyclin D is synthesized and activates Cdk4/6, the cyclin-Cdk targets Rb protein for phosphorylation. Upon phosphorylation, pRb changes conformation so that E2F is released and activated, binding to upstream regions of genes, initiating expression. Specifically, E2F drives the expression of other cyclins, including
381:
160:. Following cytokinesis, during G1 phase the cells monitor environment for the potential growth factors, grow larger and once achieve the threshold size (rRNA and overall protein content characteristic for a given cell type) they start progression through S phase. During S phase, the cell also duplicates the
419:, a protein responsible for activating cyclin-Cdk dimers. Without cyclin dimer activation, the cell cannot transition through the cycle. These two checkpoints have additional processes for regulation because replicating damaged DNA in S phase can be deleterious to the cell and more importantly, the organism.
301:
291:
to degrade the inhibitor releasing and allowing the S phase cyclin-Cdk to become activated and the cell moves into S phase. Once in S phase, cyclin-Cdks phosphorylate several factors on the replication complex promoting DNA replication by causing inhibitory proteins to fall off of replication
131:
The cell cycle is a process in which an ordered set of events leads to the growth and division into two daughter cells. The cell cycle is a cycle rather than a linear process because the two daughter cells produced repeat the cycle. This process contains two main phases,
79:, make DNA repairs, or proliferate based on environmental cues and molecular signaling inputs. The G1/S transition occurs late in G1 and the absence or improper application of this highly regulated checkpoint can lead to cellular transformation and disease states such as
123:(R-Point). If a cell passes through the G1/S transition the cell will continue through the cell cycle regardless of incoming mitogenic factors due to the positive feed-back loop of G1-S transcription. Positive feed-back loops include G1 cyclins and accumulation of E2F.
168:, which is critical for DNA separation in the M phase. After complete synthesis of its DNA, the cell enters the G2 phase where it continues to grow in preparation for mitosis. Following interphase, the cell transitions into mitosis, containing four sub stages:
279:. Therefore, an inhibitor, protein Slc-1, is present that interacts with the dimer so that the S phase cyclin-Cdk dimer remains inactive until the cell is ready to move into S phase. After the cell has grown and is ready to synthesize DNA, G
405:
so that it can bind to upstream regions of genes, inducing the expression of proteins including p21CIP. p21CIP binds to and inhibits any cyclin-cdk present in the cell cycle, halting the cycle until DNA damage can be corrected.
396:
Between G1 and S phase, three DNA damage checkpoints occur to ensure proper growth and synthesis of DNA prior to cell division. Damaged DNA during G1, before entry into S phase, and during S phase result in the expression of
414:
Of the four DNA damage checkpoints, two have an additional process for monitoring DNA damage other than activating p53. Before entry into S phase and during S phase, ATM/R also activates Chk1/2 that inhibits
71:
to ensure cell cycle integrity and the subsequent S phase can pause in response to improperly or partially replicated DNA. During this transition the cell makes decisions to become quiescent (enter
332:
either phosphorylates more pRb to further activate E2F and promote the expression of more Cyclin E, or it has the ability to increase expression of itself. Cyclin E also interacts with
344:
Retinoblastoma (Rb) is a cancer of the eye due to a mutant pRb protein. When pRb is mutated it becomes nonfunctional and is not able to inhibit the expression of transcription factor
283:
cyclin-Cdks phosphorylate the S phase cyclin inhibitor signaling ubiquitination, resulting in the addition of groups to the inhibitor. Ubiquitination of the inhibitor signals the
204:
As with most processes in the body, the cell cycle is highly regulated to prevent the synthesis of mutated cells and uncontrolled cell division that leads to
348:. Therefore, E2F is always active and driving the cell cycle to progress from G1 to S phase. As a result, cell growth and division is unregulated causing
243:, which come together at different points in the cycle to control cell progression through the cycle. When cyclin binds to Cdk, Cdk becomes activated and
247:
serine and threonine on other proteins causing the activation and degradation of other proteins allowing the cell to transition through the cell cycle.
739:
652:
140:(M) phase, during which the cell separates its DNA and divides into two new daughter cells. Interphase is further broken down into the
814:
942:
Wang X, Simpson ER, Brown KA (December 2015). "p53: Protection against Tumor Growth beyond
Effects on Cell Cycle and Apoptosis".
165:
101:, which then drives the transition from G1 to S phase. The G1/S transition is highly regulated by transcription factor
398:
915:
Fadila G, Jinho H, Vaddadi N, Abbas T (2015). "Novel regulation of cyclin D1 stability and the DNA damage response".
839:"The regulation of cyclin D1 degradation: roles in cancer development and the potential for therapeutic invention"
268:
428:
292:
complexes or through activation of components on the replication complex to induce DNA replication initiation.
217:
76:
272:
229:
220:(MPF) control the transition from one phase to the next based on a series of checkpoints. MPF is a protein
600:
Bartek J, Lukas J (February 2001). "Pathways governing G1/S transition and their response to DNA damage".
450:
Bartek J, Lukas J (February 2001). "Pathways governing G1/S transition and their response to DNA damage".
369:
365:
313:
108:
It is a "point of no return" beyond which the cell is committed to dividing; in yeast this is called the
361:
68:
23:
Depiction of regulation at the G1/S transition point in cell cycle progression. Figure taken from 2A of
388:
368:, one unreplicated DNA checkpoint at the end of G2, one spindle assembly checkpoint in mitosis, and a
364:
to monitor cell progression and halt the cycle when processes go awry. These checkpoints include four
768:
558:
95:
982:
109:
625:
582:
475:
959:
870:
796:
735:
699:
648:
617:
574:
531:
467:
120:
951:
924:
897:
860:
850:
786:
776:
689:
681:
609:
566:
521:
513:
459:
27:
Author credit: Alexis R. Barr, Frank S. Heldt, Tongli Zhang, Chris Bakal, and Be ́ la Nova ́
19:
189:
64:
901:
772:
562:
917:
Proceedings of the 106th Annual
Meeting of the American Association for Cancer Research
865:
838:
694:
669:
526:
501:
309:
244:
91:
39:
791:
756:
613:
463:
976:
221:
209:
113:
629:
479:
43:
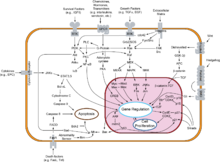
Signal transduction pathways influencing gene regulation and cellular proliferation.
586:
955:
928:
275:
components; however, the cell does not want S phase cyclins to become active in G
284:
196:
of the cell is split in two during cytokinesis resulting in two daughter cells.
185:
157:
24:
761:
Proceedings of the
National Academy of Sciences of the United States of America
304:
Crystal structure of the retinoblastoma tumour suppressor protein bound to E2F
288:
161:
133:
116:
52:
781:
325:
237:
193:
181:
177:
31:
963:
874:
855:
703:
685:
621:
578:
535:
471:
800:
757:"Subcompartments of the G1 phase of cell cycle detected by flow cytometry"
401:
protein. ATM/R protein then stabilizes and activates transcription factor
152:(GAP 2) phase and the mitotic (M) phase which in turn is broken down into
329:
321:
264:
213:
173:
169:
149:
141:
87:
72:
56:
570:
380:
670:"Proteomic snapshot of breast cancer cell cycle: G1/S transition point"
153:
145:
137:
60:
755:
Darzynkiewicz, Z; Sharpless, T; Staiano-Coico, L; Melamed, MR (1980).
416:
240:
233:
225:
192:. After duplicate DNA is separated on opposite ends of the cell, the
136:, in which the cell grows and synthesizes a copy of its DNA, and the
80:
517:
387:
379:
349:
299:
205:
38:
30:
18:
360:
To ensure proper cell division, the cell cycle utilizes numerous
549:
Massagué J (November 2004). "G1 cell-cycle control and cancer".
333:
402:
345:
317:
300:
102:
98:
890:
New DAG-dependent mechanisms modulate cell cycle progression
502:"Control of cell cycle transcription during G1 and S phases"
336:
driving the cell cycle to progress from G1 to S phase.
105:
in order to halt the cell cycle when DNA is damaged.
296:
Retinoblastoma protein (pRB) and the G1/S transition
500:Bertoli C, Skotheim JM, de Bruin RA (August 2013).
308:Another dimer present during mid G1 is composed of
734:(7th 13 ed.). Freeman, W. H. & Company.
271:, activates the expression of the S phase cyclin-
730:Lodish H, Berk A, Kaiser C, Krieger M (2012).
647:(7th ed.). Freeman, W. H. & Company.
643:Lodish H, Berk A, Kaiser C, Krieger M (2012).
495:
493:
491:
489:
410:Additional processes at DNA damage checkpoints
445:
443:
340:The role of retinoblastoma in tumor formation
25:https://dx.doi.org/10.1016/j.cels.2016.01.001
8:
208:formation. The cell cycle control system is
328:, and genes necessary for DNA replication.
188:, which are lined up and separated by the
864:
854:
790:
780:
693:
525:
832:
830:
828:
439:
725:
723:
721:
719:
717:
715:
713:
506:Nature Reviews Molecular Cell Biology
7:
668:Tenga MJ, Lazar IM (January 2013).
59:, in which the cell grows, and the
14:
384:Conceptualization of p53 pathway.
184:. In mitosis, DNA condenses into
902:10.6092/unibo/amsdottorato/6739
1:
956:10.1158/0008-5472.CAN-15-0563
929:10.1158/1538-7445.AM2015-3786
614:10.1016/S0014-5793(01)02114-7
464:10.1016/S0014-5793(01)02114-7
166:microtubule-organizing center
90:-Cdk4/6 dimer phosphorylates
55:at the boundary between the
372:checkpoint during mitosis.
316:) and transcription factor
86:During this transition, G1
999:
815:"Phases of the Cell Cycle"
429:S-phase promoting factor
218:mitosis promoting factor
923:(15 Supplement): 3786.
782:10.1073/pnas.77.11.6696
230:cyclin-dependent kinase
856:10.1186/1476-4598-6-24
837:Alao JP (April 2007).
732:Molecular Cell Biology
686:10.1002/pmic.201200188
645:Molecular Cell Biology
393:
392:p53-DNA damage complex
385:
370:chromosome segregation
366:DNA damage checkpoints
356:Cell cycle checkpoints
352:formation in the eye.
305:
69:cell cycle checkpoints
44:
36:
28:
391:
383:
303:
200:Cell cycle regulation
42:
34:
22:
96:transcription factor
67:. It is governed by
896:(Doctoral Thesis).
773:1980PNAS...77.6696D
571:10.1038/nature03094
563:2004Natur.432..298M
148:(Synthesis) phase,
127:Cell cycle overview
16:Stage in cell cycle
894:Scienze Biomediche
394:
386:
376:p53 as a regulator
306:
212:based so that the
51:is a stage in the
45:
37:
29:
741:978-1-4641-0981-2
654:978-1-4641-0981-2
557:(7015): 298–306.
121:restriction point
119:it is termed the
65:DNA is replicated
990:
968:
967:
939:
933:
932:
912:
906:
905:
885:
879:
878:
868:
858:
843:Molecular Cancer
834:
823:
822:
811:
805:
804:
794:
784:
752:
746:
745:
727:
708:
707:
697:
665:
659:
658:
640:
634:
633:
597:
591:
590:
546:
540:
539:
529:
497:
484:
483:
447:
259:In mid to late G
998:
997:
993:
992:
991:
989:
988:
987:
973:
972:
971:
944:Cancer Research
941:
940:
936:
914:
913:
909:
888:Poli A (2015).
887:
886:
882:
836:
835:
826:
813:
812:
808:
754:
753:
749:
742:
729:
728:
711:
667:
666:
662:
655:
642:
641:
637:
599:
598:
594:
548:
547:
543:
518:10.1038/nrm3629
499:
498:
487:
449:
448:
441:
437:
425:
412:
378:
358:
342:
298:
282:
278:
262:
257:
254:
202:
190:mitotic spindle
144:(GAP 1) phase,
129:
63:, during which
49:G1/S transition
17:
12:
11:
5:
996:
994:
986:
985:
975:
974:
970:
969:
950:(23): 5001–7.
934:
907:
880:
824:
806:
767:(11): 6696–9.
747:
740:
709:
660:
653:
635:
592:
541:
485:
438:
436:
433:
432:
431:
424:
421:
411:
408:
377:
374:
357:
354:
341:
338:
310:retinoblastoma
297:
294:
280:
276:
260:
256:
252:
249:
245:phosphorylates
201:
198:
128:
125:
92:retinoblastoma
15:
13:
10:
9:
6:
4:
3:
2:
995:
984:
981:
980:
978:
965:
961:
957:
953:
949:
945:
938:
935:
930:
926:
922:
918:
911:
908:
903:
899:
895:
891:
884:
881:
876:
872:
867:
862:
857:
852:
848:
844:
840:
833:
831:
829:
825:
820:
816:
810:
807:
802:
798:
793:
788:
783:
778:
774:
770:
766:
762:
758:
751:
748:
743:
737:
733:
726:
724:
722:
720:
718:
716:
714:
710:
705:
701:
696:
691:
687:
683:
679:
675:
671:
664:
661:
656:
650:
646:
639:
636:
631:
627:
623:
619:
615:
611:
608:(3): 117–22.
607:
603:
596:
593:
588:
584:
580:
576:
572:
568:
564:
560:
556:
552:
545:
542:
537:
533:
528:
523:
519:
515:
512:(8): 518–28.
511:
507:
503:
496:
494:
492:
490:
486:
481:
477:
473:
469:
465:
461:
458:(3): 117–22.
457:
453:
446:
444:
440:
434:
430:
427:
426:
422:
420:
418:
409:
407:
404:
400:
390:
382:
375:
373:
371:
367:
363:
355:
353:
351:
347:
339:
337:
335:
331:
327:
323:
319:
315:
311:
302:
295:
293:
290:
286:
274:
270:
266:
250:
248:
246:
242:
239:
235:
231:
227:
223:
219:
215:
211:
210:biochemically
207:
199:
197:
195:
191:
187:
183:
179:
175:
171:
167:
163:
159:
155:
151:
147:
143:
139:
135:
126:
124:
122:
118:
115:
114:multicellular
111:
106:
104:
100:
97:
93:
89:
84:
82:
78:
77:differentiate
74:
70:
66:
62:
58:
54:
50:
41:
33:
26:
21:
947:
943:
937:
920:
916:
910:
893:
889:
883:
846:
842:
818:
809:
764:
760:
750:
731:
680:(1): 48–60.
677:
673:
663:
644:
638:
605:
602:FEBS Letters
601:
595:
554:
550:
544:
509:
505:
455:
452:FEBS Letters
451:
413:
395:
359:
343:
307:
258:
203:
130:
107:
85:
48:
46:
819:KhanAcademy
362:checkpoints
255:/transition
224:made up of
186:chromosomes
158:cytokinesis
110:Start point
983:Cell cycle
674:Proteomics
435:References
289:proteasome
162:centrosome
134:interphase
117:eukaryotes
94:releasing
53:cell cycle
35:Cell cycle
312:protein (
267:bound to
238:threonine
232:(Cdk), a
194:cytoplasm
182:telophase
178:metaphase
112:, and in
977:Category
964:26573797
875:17407548
704:23152136
630:16090531
622:11223026
579:15549091
536:23877564
480:16090531
472:11223026
423:See also
330:Cyclin E
322:cyclin E
265:cyclin D
214:proteins
174:anaphase
170:prophase
88:cyclin D
57:G1 phase
866:1851974
801:6161370
769:Bibcode
695:4123745
587:4428026
559:Bibcode
527:4569015
263:phase,
216:of the
154:mitosis
138:mitotic
61:S phase
962:
873:
863:
849:: 24.
799:
792:350355
789:
738:
702:
692:
651:
628:
620:
585:
577:
551:Nature
534:
524:
478:
470:
417:Cdc25A
269:Cdk4/6
241:kinase
234:serine
226:cyclin
180:, and
81:cancer
626:S2CID
583:S2CID
476:S2CID
399:ATM/R
350:tumor
222:dimer
206:tumor
164:, or
960:PMID
871:PMID
797:PMID
736:ISBN
700:PMID
649:ISBN
618:PMID
575:PMID
532:PMID
468:PMID
334:Cdk2
324:and
236:and
228:and
156:and
47:The
952:doi
925:doi
898:doi
861:PMC
851:doi
787:PMC
777:doi
690:PMC
682:doi
610:doi
606:490
567:doi
555:432
522:PMC
514:doi
460:doi
456:490
403:p53
346:E2F
318:E2F
314:pRB
285:SCF
273:Cdk
103:p53
99:E2F
75:),
979::
958:.
948:75
946:.
921:75
919:.
892:.
869:.
859:.
845:.
841:.
827:^
817:.
795:.
785:.
775:.
765:77
763:.
759:.
712:^
698:.
688:.
678:13
676:.
672:.
624:.
616:.
604:.
581:.
573:.
565:.
553:.
530:.
520:.
510:14
508:.
504:.
488:^
474:.
466:.
454:.
442:^
176:,
172:,
150:G2
142:G1
83:.
73:G0
966:.
954::
931:.
927::
904:.
900::
877:.
853::
847:6
821:.
803:.
779::
771::
744:.
706:.
684::
657:.
632:.
612::
589:.
569::
561::
538:.
516::
482:.
462::
326:A
287:/
281:1
277:1
261:1
253:1
251:G
146:S
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.