240:
deviation from C corresponds to leading strand. Furthermore, the site where the deviation sign switches corresponds to the origin or terminal. The x-axis represents the chromosome locations plotted 5′ to 3′ and y-axis represents the deviation value. The major weakness of this method is its window-size dependent property. Therefore, choosing an adequate window size greatly affects the outcome of the plot. Other techniques should be combined with deviation in order to identify and locate the origin of the DNA replication with greater accuracy.
28:
20:
824:
respectively; therefore, it is expected to see a switch in GC skew sign just at the point of DNA replication origin and terminus. GC skew can also be used to study the strand biases and mechanism related to them by calculating the excess of one base over its complementary base in different milieus. Method such as GC skew, CGC skew, and Z curve are tools that can provide opportunity to better investigate the mechanism of DNA replication in different organisms.
249:
183:
Therefore, positive GC skew represents richness of G over C and the negative GC skew represents richness of C over G. As a result, one expects to see a positive GC skew and negative AT skew in the leading strand, and a negative GC skew and a positive AT skew in the lagging strand. GC or AT skew changes sign at the boundaries of the two
802:
between the leading and lagging strands. Examination of the prokaryotic genome shows a preference in third codon position for G over C and T over A. This discrimination creates an asymmetric nucleotide composition, if the coding strand is unequally distributed between the leading and lagging strands,
771:
in one strand as compared to the other. Furthermore, the time used for replication between the two strands varies and may lead to asymmetric mutational pressure between leading and lagging strand. In addition to mutations during DNA replication, transcriptional mutations can create strand specific
239:
where A, T, G, and C represent the frequency of occurrence of the equivalent base in a particular sequence in a defined length. A window sliding strategy is used to calculate deviation from C through the genome. In these plots, a positive deviation from C corresponds to lagging strand and negative
182:
In some bacterial genomes, there is an enrichment of guanine over cytosine and thymine over adenine on the leading strand and vice versa for the lagging strand. The nucleotide composition skew spectra ranges from −1, which corresponds to G = 0 or A = 0, to +1, which corresponds to T= 0 or C = 0.
170:
on each strand have equal amounts of a given base and its complement. In other words, in each DNA strand the frequency of the occurrence of T is equal to A and the frequency of the occurrence of G is equal to C because the substitution rate is presumably equal. This phenomenon is referred to as
256:
The second approach is referred to as cumulative GC skew (CGC skew). This method still uses the sliding window strategy but it takes advantage of the sum of the adjacent windows from an arbitrary start. In this scheme, the entire genome is usually plotted 5' to 3' using an arbitrary start and
191:
between the leading strand and lagging strand. DNA replication is semi-conservative and an asymmetric process itself. This asymmetry is due to the formation of the replication fork and its division into nascent leading and lagging strands. The leading strand is synthesized continuously and in
823:
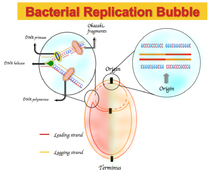
The GC skew is proven to be useful as the indicator of the DNA leading strand, lagging strand, replication origin, and replication terminal. Most bacteria and archaea contain only one DNA replication origin. The GC skew is positive and negative in the leading strand and in the lagging strand
751:
There is lack of consensus in scientific community with regard to the mechanism underlying the bias in nucleotide composition within each DNA strand. There are two major schools of thought that explain the mechanism behind the strand specific nucleotide composition in bacteria.
776:
of cytosine and ultimately mutation of cytosine to thymine in one DNA strand can increase the relative number of guanine and thymine to cytosine and adenine. In most bacteria, the majority of the genes are encoded in the leading strand. For instance, the leading strand in
178:
Any deviation from parity rule 2 will result in asymmetric base composition that discriminates the leading strand–i.e., the DNA strand that is replicated in the forward direction–from the lagging strand. This asymmetry is referred to as GC or AT skew.
1116:
Marin, A. & Xia, X. GC skew in protein-coding genes between the leading and lagging strands in bacterial genomes: new substitution models incorporating strand bias. Journal of theoretical biology 253, 508-513, doi:10.1016/j.jtbi.2008.04.004
807:, have been shown to be located mostly on the leading strand in bacteria. Therefore, a bias in the third-position codon choice of G over C can lead to GC skew. Additionally, some signal sequences are rich in guanine and thymine, such as
786:
encodes 75% of the genes. In addition, an excess of deamination and conversion of cytosine to thymine in the coding strand compared to the non-coding strand has been reported. One possible explanation is that the non-transcribed strand
145:
work in 1950 demonstrated that, in DNA, the bases guanine and cytosine were found in equal abundance, and the bases adenine and thymine were found in equal abundance. However, there was no equality between the amount of one
995:
Tillier, E. R. & Collins, R. A. The contributions of replication orientation, gene direction, and signal sequences to base-composition asymmetries in bacterial genomes. Journal of molecular evolution 50, 249-257
814:
Both mutational and selective pressure can independently introduce asymmetry in DNA strands. However, the combination and cumulative effect of both mechanisms is the most plausible explanation for GC and AT skew.
165:
A natural result of parity rule 1, at the state of equilibrium, in which there is no mutation and/or selection biases in any of the two DNA strands, is that when there is an equal substitution rate, the
957:
Necsulea, A. & Lobry, J. R. A new method for assessing the effect of replication on DNA base composition asymmetry. Molecular biology and evolution 24, 2169-2179, doi:10.1093/molbev/msm148 (2007).
795:). Another explanation is that the deamination repair activity during transcription does not occur on the coding strand. Only the transcribed strand benefits from these deamination repair events.
1107:
Eppinger, M., Baar, C., Raddatz, G., Huson, D. H. & Schuster, S. C. Comparative analysis of four
Campylobacterales. Nature Reviews. Microbiology 2, 872-885, doi:10.1038/nrmicro1024 (2004).
557:
465:
373:
278:. Unlike the previous methods, this method do not uses the sliding window strategy and is thought to perform better as to finding the origin of replication. In this method, each base's
743:
and asymmetric composition of the strands. A combination of these methods should be used for prediction of replication origin and terminal, in order to compensate for their weakness.
1133:
Charneski C.A., Honti F., Bryant J.M., Hurst L.D., Feil E.J. Atypical AT Skew in
Firmicute Genomes Results from Selection and Not from Mutation. PLoS Genetics 7(9):e1002283 (2011).
1089:
Zhang, R. & Zhang, C. T. Multiple replication origins of the archaeon
Halobacterium species NRC-1. Biochemical and biophysical research communications 302, 728-734 (2003).
266:
With the flipped definition of GC skew, the maximum value of the cumulative skew corresponds to the terminal, and the minimum value corresponds to the origin of replication.
612:
120:: the leading strand contains more guanine (G) and thymine (T), whereas the lagging strand contains more adenine (A) and cytosine (C). This phenomenon is referred to as
212:
The first approach is GC and AT asymmetry. Jean R. Lobry was the first to report, in 1996, the presence of compositional asymmetry in the genomes of three bacteria:
693:
666:
639:
737:
717:
282:
with respect to the base at the beginning of the sequence is investigated. The Z curve uses a three-dimensional representation with the following parameters:
187:, which correspond to DNA replication origin or terminus. Originally, this asymmetric nucleotide composition was explained as a different mechanism used in
1140:
Mewes, H. W. et al. MIPS: analysis and annotation of proteins from whole genomes in 2005. Nucleic Acids Res 34, D169-172, doi:10.1093/nar/gkj148 (2006).
842:"Understanding the Adaptation of Halobacterium Species NRC-1 to Its Extreme Environment through Computational Analysis of Its Genome Sequence"
791:) is single-stranded during the transcription process; therefore, it is more vulnerable to deamination compared to the transcribed strand (
1008:
Rocha, E. P. The replication-related organization of bacterial genomes. Microbiology 150, 1609-1627, doi:10.1099/mic.0.26974-0 (2004).
913:
Lobry, J. R. Asymmetric substitution patterns in the two DNA strands of bacteria. Molecular biology and evolution 13, 660-665 (1996).
925:
Chargaff, E. Chemical specificity of nucleic acids and mechanism of their enzymatic degradation. Experientia 6, 201-209 (1950).
1066:
260:
In contrast to Lobry's earlier paper, recent implementations of GC skew flips the original definition, redefining it to be:
1098:
Lobry, J. R. & Sueoka, N. Asymmetric directional mutation pressures in bacteria. Genome biology 3, RESEARCH0058 (2002).
279:
471:
379:
287:
257:
arbitrary strand. In the cumulative GC skew plot, the peaks corresponds to the switch points (terminus or origin).
159:
192:
juxtapose to the leading strand; the lagging strand is replicated through short fragments of polynucleotide (
158:. Three years later, Watson and Crick used this fact during their derivation of the structure of DNA, their
1153:
760:
108:
27:
1077:
Grigoriev, A. Analyzing genomes with cumulative skew diagrams. Nucleic Acids
Research 26, 2286-2290 (1998).
840:
Kennedy, Sean P.; Ng, Wailap Victor; Salzberg, Steven L.; Hood, Leroy; DasSarma, Shiladitya (2001-10-01).
226:
798:
The second school of thought describes the mechanism of GC and AT skew as resulting from a difference in
811:, and these sequences might have a higher frequency of occurrence in one strand compared to the other.
204:
There are three major approaches to calculate and graphically demonstrate GC skew and its properties.
1163:
151:
89:
31:
Richness of G over T in the leading strand, resulting in a GC skew sign at the origin and terminus.
564:
23:
Indication of the origin and terminus of DNA replication on a GC skew and cumulative GC skew plot.
799:
740:
65:
57:. GC skew is also a statistical method for measuring strand-specific guanine overrepresentation.
230:. The original formulas at the time were not called skew, but rather deviation from = or = :
763:. Due to the asymmetric nature of the replication process, an unequal mutational frequency and
19:
1049:
1041:
879:
861:
804:
792:
778:
756:
220:
193:
167:
1031:
869:
853:
671:
644:
617:
1036:
1019:
188:
722:
702:
175:. Hence, the second parity rule only exists when there is no mutation or substitution.
142:
117:
113:
874:
841:
1147:
788:
696:
16:
Over- or under-abundance of guanine and cytosine in a particular region of DNA or RNA
1158:
803:
as in the case for bacteria. In addition, the highly transcribed genes, such as
773:
61:
764:
184:
39:
1045:
865:
147:
883:
1053:
808:
768:
93:
85:
46:
248:
275:
214:
103:
98:
81:
77:
73:
42:
857:
1020:"Asymmetric substitution patterns in the two DNA strands of bacteria"
69:
247:
26:
18:
54:
50:
767:
efficiency during the replication process can introduce more
150:
versus the other. Chargaff's finding is referred to as
252:
Cumulative CG and AT skew for 49 bacterial chromosomes
112:), nucleotide compositions are asymmetric between the
49:
are over- or under-abundant in a particular region of
725:
705:
674:
647:
620:
567:
474:
382:
290:
72:) there is an equal frequency of the four DNA bases (
68:
and with nucleotides randomly distributed within the
695:shows the relationship between the weak and strong
731:
711:
687:
660:
633:
606:
551:
459:
367:
128:and the corresponding statistics were defined as:
755:The first one describes a bias and an asymmetric
641:represents the excess of purine over pyrimidine,
552:{\displaystyle z_{n}=(A_{n}+T_{n})-(C_{n}+G_{n})}
460:{\displaystyle y_{n}=(A_{n}+C_{n})-(G_{n}+T_{n})}
368:{\displaystyle x_{n}=(A_{n}+G_{n})-(C_{n}+T_{n})}
8:
1129:
1127:
1125:
1123:
1085:
1083:
991:
989:
987:
985:
983:
953:
951:
1004:
1002:
981:
979:
977:
975:
973:
971:
969:
967:
965:
963:
949:
947:
945:
943:
941:
939:
937:
935:
933:
931:
921:
919:
909:
907:
905:
903:
901:
899:
897:
895:
893:
759:on each DNA strand during replication and
1035:
873:
724:
704:
679:
673:
652:
646:
625:
619:
566:
540:
527:
508:
495:
479:
473:
448:
435:
416:
403:
387:
381:
356:
343:
324:
311:
295:
289:
832:
668:denotes excess of keto over amino, and
236:deviation from = as (C − G)/(C + G);
233:deviation from = as (A − T)/(A + T);
1037:10.1093/oxfordjournals.molbev.a025626
7:
92:of a DNA molecule. However, in most
1067:"Explanation of Lobry 1996 article"
60:In equilibrium conditions (without
14:
138:Asymmetric nucleotide composition
739:components can alone detect the
1024:Molecular Biology and Evolution
546:
520:
514:
488:
454:
428:
422:
396:
362:
336:
330:
304:
1:
772:nucleotide composition skew.
200:Calculation and GC skew plots
607:{\displaystyle n=0,1,2,...N}
263:GC skew = (G − C)/(G + C).
196:) in a 5' to 3' direction.
131:GC skew = (G - C)/(G + C)
1180:
274:The final approach is the
134:AT skew = (A − T)/(A + T)
1018:Lobry, J. R. (May 1996).
168:complementary nucleotides
109:Sulfolobus solfataricus
733:
713:
689:
662:
635:
608:
553:
461:
369:
253:
227:Haemophilus influenzae
32:
24:
734:
714:
690:
688:{\displaystyle z_{n}}
663:
661:{\displaystyle y_{n}}
636:
634:{\displaystyle x_{n}}
609:
554:
462:
370:
251:
30:
22:
723:
703:
672:
645:
618:
565:
472:
380:
288:
280:cumulative frequency
757:mutational pressure
805:ribosomal proteins
800:selective pressure
741:replication origin
729:
709:
685:
658:
631:
604:
549:
457:
365:
254:
160:double helix model
66:selective pressure
33:
25:
858:10.1101/gr.190201
852:(10): 1641–1650.
793:non-coding strand
732:{\displaystyle y}
712:{\displaystyle x}
221:Bacillus subtilis
194:Okazaki fragments
1171:
1134:
1131:
1118:
1114:
1108:
1105:
1099:
1096:
1090:
1087:
1078:
1075:
1069:
1064:
1058:
1057:
1039:
1015:
1009:
1006:
997:
993:
958:
955:
926:
923:
914:
911:
888:
887:
877:
837:
738:
736:
735:
730:
718:
716:
715:
710:
694:
692:
691:
686:
684:
683:
667:
665:
664:
659:
657:
656:
640:
638:
637:
632:
630:
629:
613:
611:
610:
605:
558:
556:
555:
550:
545:
544:
532:
531:
513:
512:
500:
499:
484:
483:
466:
464:
463:
458:
453:
452:
440:
439:
421:
420:
408:
407:
392:
391:
374:
372:
371:
366:
361:
360:
348:
347:
329:
328:
316:
315:
300:
299:
143:Erwin Chargaff's
1179:
1178:
1174:
1173:
1172:
1170:
1169:
1168:
1144:
1143:
1137:
1132:
1121:
1115:
1111:
1106:
1102:
1097:
1093:
1088:
1081:
1076:
1072:
1065:
1061:
1017:
1016:
1012:
1007:
1000:
994:
961:
956:
929:
924:
917:
912:
891:
846:Genome Research
839:
838:
834:
830:
821:
749:
721:
720:
701:
700:
675:
670:
669:
648:
643:
642:
621:
616:
615:
563:
562:
536:
523:
504:
491:
475:
470:
469:
444:
431:
412:
399:
383:
378:
377:
352:
339:
320:
307:
291:
286:
285:
272:
246:
210:
202:
189:DNA replication
152:Chargaff's rule
140:
17:
12:
11:
5:
1177:
1175:
1167:
1166:
1161:
1156:
1154:Bioinformatics
1146:
1145:
1142:
1141:
1136:
1135:
1119:
1109:
1100:
1091:
1079:
1070:
1059:
1030:(5): 660–665.
1010:
998:
959:
927:
915:
889:
831:
829:
826:
820:
817:
748:
745:
728:
708:
697:hydrogen bonds
682:
678:
655:
651:
628:
624:
603:
600:
597:
594:
591:
588:
585:
582:
579:
576:
573:
570:
548:
543:
539:
535:
530:
526:
522:
519:
516:
511:
507:
503:
498:
494:
490:
487:
482:
478:
456:
451:
447:
443:
438:
434:
430:
427:
424:
419:
415:
411:
406:
402:
398:
395:
390:
386:
364:
359:
355:
351:
346:
342:
338:
335:
332:
327:
323:
319:
314:
310:
306:
303:
298:
294:
271:
268:
245:
242:
209:
206:
201:
198:
139:
136:
118:lagging strand
114:leading strand
90:single strands
15:
13:
10:
9:
6:
4:
3:
2:
1176:
1165:
1162:
1160:
1157:
1155:
1152:
1151:
1149:
1139:
1138:
1130:
1128:
1126:
1124:
1120:
1113:
1110:
1104:
1101:
1095:
1092:
1086:
1084:
1080:
1074:
1071:
1068:
1063:
1060:
1055:
1051:
1047:
1043:
1038:
1033:
1029:
1025:
1021:
1014:
1011:
1005:
1003:
999:
992:
990:
988:
986:
984:
982:
980:
978:
976:
974:
972:
970:
968:
966:
964:
960:
954:
952:
950:
948:
946:
944:
942:
940:
938:
936:
934:
932:
928:
922:
920:
916:
910:
908:
906:
904:
902:
900:
898:
896:
894:
890:
885:
881:
876:
871:
867:
863:
859:
855:
851:
847:
843:
836:
833:
827:
825:
818:
816:
812:
810:
809:chi sequences
806:
801:
796:
794:
790:
789:coding strand
785:
784:
781:
775:
770:
766:
762:
761:transcription
758:
753:
746:
744:
742:
726:
706:
698:
680:
676:
653:
649:
626:
622:
601:
598:
595:
592:
589:
586:
583:
580:
577:
574:
571:
568:
559:
541:
537:
533:
528:
524:
517:
509:
505:
501:
496:
492:
485:
480:
476:
467:
449:
445:
441:
436:
432:
425:
417:
413:
409:
404:
400:
393:
388:
384:
375:
357:
353:
349:
344:
340:
333:
325:
321:
317:
312:
308:
301:
296:
292:
283:
281:
277:
269:
267:
264:
261:
258:
250:
243:
241:
237:
234:
231:
229:
228:
223:
222:
217:
216:
207:
205:
199:
197:
195:
190:
186:
180:
176:
174:
173:parity rule 2
169:
163:
161:
157:
156:parity rule 2
153:
149:
144:
137:
135:
132:
129:
127:
123:
119:
115:
111:
110:
105:
101:
100:
95:
91:
87:
83:
79:
75:
71:
67:
63:
58:
56:
52:
48:
44:
41:
37:
29:
21:
1112:
1103:
1094:
1073:
1062:
1027:
1023:
1013:
849:
845:
835:
822:
813:
797:
782:
779:
754:
750:
560:
468:
376:
284:
273:
265:
262:
259:
255:
238:
235:
232:
225:
219:
213:
211:
208:GC asymmetry
203:
181:
177:
172:
164:
155:
141:
133:
130:
125:
121:
107:
97:
59:
38:is when the
35:
34:
1164:Nucleotides
774:Deamination
185:replichores
102:) and some
40:nucleotides
1148:Categories
828:References
765:DNA repair
88:) on both
62:mutational
1046:0737-4038
866:1088-9051
769:mutations
747:Mechanism
518:−
426:−
334:−
884:11591641
783:subtilis
780:Bacillus
244:CGC skew
116:and the
94:bacteria
86:cytosine
47:cytosine
1117:(2008).
1054:8676740
996:(2000).
276:Z curve
270:Z curve
215:E. coli
126:AT skew
104:archaea
99:E. coli
82:thymine
78:guanine
74:adenine
43:guanine
36:GC skew
1052:
1044:
882:
875:311145
872:
864:
561:Where
224:, and
106:(e.g.
96:(e.g.
84:, and
70:genome
1050:PMID
1042:ISSN
880:PMID
862:ISSN
819:Uses
719:and
148:pair
124:and
45:and
1159:DNA
1032:doi
870:PMC
854:doi
154:or
64:or
55:RNA
53:or
51:DNA
1150::
1122:^
1082:^
1048:.
1040:.
1028:13
1026:.
1022:.
1001:^
962:^
930:^
918:^
892:^
878:.
868:.
860:.
850:11
848:.
844:.
699:.
614:,
218:,
162:.
122:GC
80:,
76:,
1056:.
1034::
886:.
856::
787:(
727:y
707:x
681:n
677:z
654:n
650:y
627:n
623:x
602:N
599:.
596:.
593:.
590:,
587:2
584:,
581:1
578:,
575:0
572:=
569:n
547:)
542:n
538:G
534:+
529:n
525:C
521:(
515:)
510:n
506:T
502:+
497:n
493:A
489:(
486:=
481:n
477:z
455:)
450:n
446:T
442:+
437:n
433:G
429:(
423:)
418:n
414:C
410:+
405:n
401:A
397:(
394:=
389:n
385:y
363:)
358:n
354:T
350:+
345:n
341:C
337:(
331:)
326:n
322:G
318:+
313:n
309:A
305:(
302:=
297:n
293:x
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.