544:
395:
135:
208:
197:
307:
523:. On average, there is approximately 1 protein-altering mutation every 7 coding bases, but some CCRs can have over 100 bases in sequence with no observed protein-altering mutations, some without even synonymous mutations. These patterns of constraint between genomes may provide clues to the sources of rare
514:
While it is well known that the genome of one individual can have extensive differences when compared to the genome of another, recent research has found that some coding regions are highly constrained, or resistant to mutation, between individuals of the same species. This is similar to the concept
211:
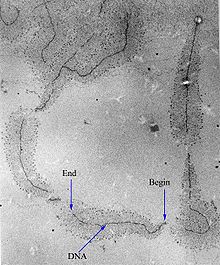
An electron-micrograph of DNA strands decorated by hundreds of RNAP molecules too small to be resolved. Each RNAP is transcribing an RNA strand, which can be seen branching off from the DNA. "Begin" indicates the 3' end of the DNA, where RNAP initiates transcription; "End" indicates the 5' end, where
582:
within a DNA sequence is straightforward, identifying coding sequences is not, because the cell translates only a subset of all open reading frames to proteins. Currently CDS prediction uses sampling and sequencing of mRNA from cells, although there is still the problem of determining which parts of
611:
in viruses, but would require a known coding strand to compare the potential overlapping coding strand with. An alternative method using single genome sequences would not require multiple genome sequences to execute comparisons but would require at least 50 nucleotides overlapping in order to be
385:
in the coding region can have very diverse effects on the phenotype of the organism. While some mutations in this region of DNA/RNA can result in advantageous changes, others can be harmful and sometimes even lethal to an organism's survival. In contrast, changes in the non-coding region may not
203:: RNA Polymerase (RNAP) uses a template DNA strand and begins coding at the promoter sequence (green) and ends at the terminator sequence (red) in order to encompass the entire coding region into the pre-mRNA (teal). The pre-mRNA is polymerised 5' to 3' and the template DNA read 3' to 5'
58:. Studying the length, composition, regulation, splicing, structures, and functions of coding regions compared to non-coding regions over different species and time periods can provide a significant amount of important information regarding gene organization and evolution of
125:
strand is not coded continuously but is interrupted by "silent" non-coding regions. This was the first indication that there needed to be a distinction between the parts of the genome that code for protein, now called coding regions, and those that do not.
149:
than non-coding regions. There is further research that discovered that the longer the coding strand, the higher the GC-content. Short coding strands are comparatively still GC-poor, similar to the low GC-content of the base composition translational
602:
occurs relatively often in both DNA and RNA viruses as an evolutionary advantage to reduce genome size while retaining the ability to produce various proteins from the available coding regions. For both DNA and RNA,
370:, in particular the 5' splicing site, which is one of the substrates for the first step in splicing. The coding regions are within the exons, which become covalently joined together to form the
329:
is one form of regulation of the coding region. The gene that would have been transcribed can be silenced by targeting a specific sequence. The bases in this sequence would be blocked using
402:
that may exist within coding regions. Such alterations may or may not have phenotypic changes, depending on whether or not they code for different amino acids during translation.
145:
The evidence suggests that there is a general interdependence between base composition patterns and coding region availability. The coding region is thought to contain a higher
184:
compared to accessory and non-essential regions (gene-poor). However, it is still unclear whether this came about through neutral and random mutation or through a pattern of
587:, the latter also including prediction of DNA sequences that code not only for protein but also for other functional elements such as RNA genes and regulatory sequences.
362:
ultimately determines what part of the sequence becomes translated and expressed, and this process involves cutting out introns and putting together exons. Where the RNA
188:. There is also debate on whether the methods used, such as gene windows, to ascertain the relationship between GC-content and coding region are accurate and unbiased.
1172:
109:
refers to all exons within a genome, the coding region refers to a singular section of the DNA or RNA which specifically codes for a certain kind of protein.
1395:
450:), or passed on from a parent to its offspring. Such mutated coding regions are present in all cells within the organism. Other forms of mutations are acquired (
519:. Researchers termed these highly constrained sequences constrained coding regions (CCRs), and have also discovered that such regions may be involved in high
486:
There exist multiple transcription and translation mechanisms to prevent lethality due to deleterious mutations in the coding region. Such measures include
1443:
169:, which are changes from purine to pyrimidine or pyrimidine to purine. The transitions are less likely to change the encoded amino acid and remain a
1207:
English: The structure of a mature eukaryotic mRNA. A fully processed mRNA includes the 5' cap, 5' UTR, coding region, 3' UTR, and poly(A) tail.
1478:
Havrilla, J. M., Pedersen, B. S., Layer, R. M., & Quinlan, A. R. (2018). A map of constrained coding regions in the human genome.
532:
1466:
Peretó J. (2011) Wobble
Hypothesis (Genetics). In: Gargaud M. et al. (eds) Encyclopedia of Astrobiology. Springer, Berlin, Heidelberg
410:, in which a change in nucleotides does not result in any change in amino acid after transcription and translation. There also exist
813:
1173:
https://www.khanacademy.org/science/biology/gene-expression-central-dogma/transcription-of-dna-into-rna/a/overview-of-transcription
268:
688:
264:
1079:"The nucleotide composition of microbial genomes indicates differential patterns of selection on core and accessory genomes"
341:
418:, or single base pair changes in the coding region, that code for different amino acids during translation, are called
310:
The coding region (teal) is flanked by untranslated regions, the 5' cap, and the poly(A) tail which together form the
1186:
1130:"Relationship between gene expression and GC-content in mammals: statistical significance and biological relevance"
180:
This indicates that essential coding regions (gene-rich) are higher in GC-content and more stable and resistant to
414:, where base alterations in the coding region code for a premature stop codon, producing a shorter final protein.
344:
manages the abundance of RNA or protein made in a cell, the regulation of these mechanisms can be controlled by a
714:
Höglund M, Säll T, Röhme D (February 1990). "On the origin of coding sequences from random open reading frames".
543:
121:
published "Why Genes in Pieces" which first began to explore the idea that the gene is a mosaic—that each full
478:, resulting in a mutation present in the offspring's DNA while being absent in both the sperm and egg cells.
1429:
What is a gene mutation and how do mutations occur? - Genetics Home
Reference - NIH. (n.d.). Retrieved from
236:
binds to the promoter sequence and moves along the template strand to the coding region. RNAP then adds RNA
229:
524:
487:
1788:
1412:
793:
280:
162:
1600:
454:) during an organism's lifetime, and may not be constant cell-to-cell. These changes can be caused by
970:
961:
Oliver JL, Marín A (September 1996). "A relationship between GC content and coding-sequence length".
876:
765:
Sakharkar MK, Chow VT, Kangueane P (2004). "Distributions of exons and introns in the human genome".
723:
633:
503:
371:
67:
1430:
579:
520:
495:
431:
423:
353:
345:
221:
91:
87:
1688:
356:
will then determine the location and time that expression will occur for a protein coding region.
177:
of a codon) which is usually beneficial to the organism during translation and protein formation.
90:
of the RNA, and so therefore, an exon would be partially made up of coding regions. The 3' and 5'
902:
837:
747:
608:
604:
516:
427:
349:
1730:"A Simple Method to Detect Candidate Overlapping Genes in Viruses Using Single Genome Sequences"
1275:
394:
1759:
1710:
1669:
1620:
1581:
1532:
1389:
1355:
1257:
1151:
1110:
1059:
986:
943:
894:
819:
809:
774:
739:
499:
447:
419:
411:
185:
95:
1599:
Rogozin IB, Spiridonov AN, Sorokin AV, Wolf YI, Jordan IK, Tatusov RL, Koonin EV (May 2002).
1749:
1741:
1700:
1659:
1651:
1612:
1571:
1563:
1522:
1512:
1483:
1345:
1312:
1247:
1239:
1141:
1100:
1090:
1049:
1041:
978:
933:
884:
801:
731:
599:
528:
471:
451:
134:
1550:
Furuno M, Kasukawa T, Saito R, Adachi J, Suzuki H, Baldarelli R, et al. (June 2003).
849:
584:
467:
407:
225:
170:
141:
transitions (blue) are elevated compared to transversions (red) in GC-rich coding regions.
974:
880:
727:
1754:
1729:
1664:
1639:
1527:
1500:
1374:
1252:
1227:
1105:
1078:
805:
657:
651:
639:
491:
415:
334:
272:
233:
207:
158:
118:
1616:
1576:
1551:
1054:
1029:
1777:
621:
475:
1705:
1004:
196:
1783:
906:
751:
560:
552:
406:
There are various forms of mutations that can occur in coding regions. One form is
359:
288:
166:
165:, which are changes from purine to purine or pyrimidine to pyrimidine, compared to
122:
692:
303:
chain, eventually forming the protein defined in the initial DNA coding region.
1228:"Alkylation of template strand of coding region causes effective gene silencing"
645:
463:
367:
363:
300:
276:
256:
583:
a given mRNA are actually translated to protein. CDS prediction is a subset of
470:
and are not passed down to offspring. Changes in the coding region can also be
1095:
591:
564:
459:
326:
296:
259:
formed encompasses multiple parts important for its eventual translation into
237:
174:
151:
146:
59:
1517:
1077:
Bohlin J, Eldholm V, Pettersson JH, Brynildsrud O, Snipen L (February 2017).
743:
1745:
1317:
1300:
595:
571:
556:
548:
63:
1763:
1714:
1673:
1655:
1624:
1585:
1536:
1359:
1350:
1333:
1261:
1155:
1114:
1063:
947:
823:
778:
527:
or potentially even embryonic lethality. Clinically validated variants and
17:
990:
1243:
1146:
1129:
1045:
938:
921:
898:
663:
443:
382:
284:
181:
323:
The coding region can be modified in order to regulate gene expression.
306:
1567:
982:
735:
648:
Entire coding sequence lies within the bounds of a larger external gene
455:
260:
249:
55:
1601:"Purifying and directional selection in overlapping prokaryotic genes"
1226:
Shinohara K, Sasaki S, Minoshima M, Bando T, Sugiyama H (2006-02-13).
1205:
1501:"Analytical Biases Associated with GC-Content in Molecular Evolution"
1413:
https://www.singerinstruments.com/resource/what-are-genetic-mutation/
1411:
Yang, J. (2016, March 23). What are
Genetic Mutation? Retrieved from
889:
864:
292:
245:
563:), wherein coding DNA regions occur to a greater extent in lighter (
1487:
542:
330:
252:. This continues until the RNAP reaches the termination sequence.
206:
106:
102:
1728:
Schlub TE, Buchmann JP, Holmes EC (October 2018). Malik H (ed.).
1689:"Detecting overlapping coding sequences with pairwise alignments"
1431:
https://ghr.nlm.nih.gov/primer/mutationsanddisorders/genemutation
466:). Acquired mutations can also be a result of copy-errors during
636:
The portion of the mRNA transcription product that is translated
627:
241:
105:
and there is a clear distinction between these terms. While the
83:
79:
35:
78:
Although this term is also sometimes used interchangeably with
217:
51:
47:
920:
Lercher MJ, Urrutia AO, Pavlícek A, Hurst LD (October 2003).
86:
is composed of the coding region as well as the 3' and 5'
531:
in CCRs have been previously linked to disorders such as
922:"A unification of mosaic structures in the human genome"
654:
Parts of genomes that do not encode protein-coding genes
240:
complementary to the coding region in order to form the
607:
can detect overlapping coding regions, including short
474:(new); such changes are thought to occur shortly after
1334:"Recognition of the 5' splice site by the spliceosome"
94:
of the RNA, which do not code for protein, are termed
666:
Parts of genomes with no relevant biological function
660:
Molecules that do not encode proteins, so have no CDS
1562:(6B). Cold Spring Harbor Laboratory Press: 1478–87.
630:
The entire portion of the strand that is transcribed
212:
the longer RNA molecules are completely transcribed.
101:
There is often confusion between coding regions and
228:and the termination sequence on the 3' end. During
1638:Chirico N, Vianelli A, Belshaw R (December 2010).
1171:Overview of transcription. (n.d.). Retrieved from
798:Recent Advances in Nutrigenetics and Nutrigenomics
386:always result in detectable changes in phenotype.
27:Portion of gene's sequence which codes for protein
800:. Vol. 108. Academic Press. pp. 17–50.
263:. The coding region in an mRNA is flanked by the
1276:"DNA alkylation Gene Ontology Term (GO:0006305)"
1128:Sémon M, Mouchiroud D, Duret L (February 2005).
535:, developmental delay and severe heart disease.
366:cuts, however, is guided by the recognition of
291:to the coding region, 3 nucleotides at a time (
1552:"CDS annotation in full-length cDNA sequence"
8:
1030:"DNA helix: the importance of being GC-rich"
98:regions and are not discussed on this page.
1394:: CS1 maint: numeric names: authors list (
1301:"Eukaryotic and prokaryotic gene structure"
794:"Advances in Technologies and Study Design"
1005:"ROSALIND | Glossary | Gene coding region"
796:. In Bouchard, C.; Ordovas, J. M. (eds.).
1753:
1704:
1663:
1575:
1526:
1516:
1349:
1316:
1251:
1145:
1104:
1094:
1053:
937:
888:
161:type is altered slightly: there are more
66:. This can further assist in mapping the
506:of the third base within an mRNA codon.
393:
305:
255:After transcription and maturation, the
195:
133:
1444:"DNA proofreading and repair (article)"
676:
624:The DNA strand that codes for a protein
551:of a human, showing an overview of the
295:). The tRNAs transfer their associated
173:(especially if they occur in the third
157:GC-rich areas are also where the ratio
1387:
845:
835:
642:The other elements that make up a gene
220:, the coding region is flanked by the
82:, it is not the exact same thing: the
1474:
1472:
1425:
1423:
1421:
1407:
1405:
1187:"Translation: DNA to mRNA to Protein"
1167:
1165:
682:
680:
462:, or other environmental agents (ex.
7:
1687:Firth AE, Brown CM (February 2005).
691:. The Wellcome Trust. Archived from
1376:English: Example of silent mutation
792:Parnell, Laurence D. (2012-01-01).
422:. Other types of mutations include
806:10.1016/B978-0-12-398397-8.00002-2
533:infantile epileptic encephalopathy
287:facilitates the attachment of the
25:
687:Twyman, Richard (1 August 2003).
510:Constrained coding regions (CCRs)
398:Examples of the various forms of
1644:Proceedings. Biological Sciences
498:following replication, and the '
1734:Molecular Biology and Evolution
352:begins in a strand of DNA. The
1640:"Why genes overlap in viruses"
963:Journal of Molecular Evolution
716:Journal of Molecular Evolution
515:of interspecies constraint in
46:), is the portion of a gene's
1:
1706:10.1093/bioinformatics/bti007
1617:10.1016/S0168-9525(02)02649-5
1191:Scitable: By Nature Education
342:regulation of gene expression
70:and developing gene therapy.
1499:Romiguier J, Roux C (2017).
1028:Vinogradov AE (April 2003).
442:Some forms of mutations are
863:Gilbert W (February 1978).
1805:
569:
1299:Shafee T, Lowe R (2017).
1096:10.1186/s12864-017-3543-7
539:Coding sequence detection
1518:10.3389/fgene.2017.00016
1373:Jonsta247 (2013-05-10),
1338:Acta Biochimica Polonica
1185:Clancy, Suzanne (2008).
1134:Human Molecular Genetics
926:Human Molecular Genetics
578:While identification of
154:like TAG, TAA, and TGA.
1305:WikiJournal of Medicine
1280:www.informatics.jax.org
1656:10.1098/rspb.2010.1052
1351:10.18388/abp.1998_4346
1232:Nucleic Acids Research
1204:Plociam (2005-08-08),
1034:Nucleic Acids Research
865:"Why genes in pieces?"
575:
525:developmental diseases
502:' which describes the
403:
315:
269:3' untranslated region
265:5' untranslated region
213:
204:
192:Structure and function
142:
1746:10.1093/molbev/msy155
1318:10.15347/wjm/2017.002
570:Further information:
546:
397:
309:
234:RNA Polymerase (RNAP)
224:on the 5' end of the
210:
199:
139:Point mutation types:
137:
1332:Konarska MM (1998).
494:during replication,
424:frameshift mutations
372:mature messenger RNA
92:untranslated regions
88:untranslated regions
38:, also known as the
975:1996JMolE..43..216O
881:1978Natur.271..501G
728:1990JMolE..30..104H
609:open reading frames
605:pairwise alignments
580:open reading frames
521:purifying selection
517:conserved sequences
354:regulatory sequence
346:regulatory sequence
333:, which create the
1605:Trends in Genetics
1568:10.1101/gr.1060303
1244:10.1093/nar/gkl005
1147:10.1093/hmg/ddi038
1046:10.1093/nar/gkg296
983:10.1007/pl00006080
939:10.1093/hmg/ddg251
736:10.1007/bf02099936
664:Non-functional DNA
576:
448:germline mutations
420:missense mutations
412:nonsense mutations
404:
350:open reading frame
316:
214:
205:
143:
1740:(10): 2572–2581.
1650:(1701): 3809–17.
767:In Silico Biology
529:de novo mutations
500:Wobble Hypothesis
452:somatic mutations
348:found before the
222:promoter sequence
54:that codes for a
16:(Redirected from
1796:
1768:
1767:
1757:
1725:
1719:
1718:
1708:
1684:
1678:
1677:
1667:
1635:
1629:
1628:
1596:
1590:
1589:
1579:
1547:
1541:
1540:
1530:
1520:
1496:
1490:
1476:
1467:
1464:
1458:
1457:
1455:
1454:
1440:
1434:
1427:
1416:
1409:
1400:
1399:
1393:
1385:
1384:
1383:
1370:
1364:
1363:
1353:
1329:
1323:
1322:
1320:
1296:
1290:
1289:
1287:
1286:
1272:
1266:
1265:
1255:
1223:
1217:
1216:
1215:
1214:
1201:
1195:
1194:
1182:
1176:
1169:
1160:
1159:
1149:
1125:
1119:
1118:
1108:
1098:
1074:
1068:
1067:
1057:
1025:
1019:
1018:
1016:
1015:
1001:
995:
994:
958:
952:
951:
941:
917:
911:
910:
892:
890:10.1038/271501a0
860:
854:
853:
847:
843:
841:
833:
831:
830:
789:
783:
782:
762:
756:
755:
711:
705:
704:
702:
700:
695:on 28 March 2007
689:"Gene Structure"
684:
600:gene overlapping
559:(which includes
408:silent mutations
21:
1804:
1803:
1799:
1798:
1797:
1795:
1794:
1793:
1774:
1773:
1772:
1771:
1727:
1726:
1722:
1686:
1685:
1681:
1637:
1636:
1632:
1598:
1597:
1593:
1556:Genome Research
1549:
1548:
1544:
1498:
1497:
1493:
1480:Nature Genetics
1477:
1470:
1465:
1461:
1452:
1450:
1442:
1441:
1437:
1428:
1419:
1410:
1403:
1386:
1381:
1379:
1372:
1371:
1367:
1331:
1330:
1326:
1298:
1297:
1293:
1284:
1282:
1274:
1273:
1269:
1225:
1224:
1220:
1212:
1210:
1203:
1202:
1198:
1184:
1183:
1179:
1170:
1163:
1127:
1126:
1122:
1076:
1075:
1071:
1027:
1026:
1022:
1013:
1011:
1003:
1002:
998:
960:
959:
955:
919:
918:
914:
862:
861:
857:
844:
834:
828:
826:
816:
791:
790:
786:
764:
763:
759:
713:
712:
708:
698:
696:
686:
685:
678:
673:
618:
585:gene prediction
574:
568:
561:Giemsa-staining
541:
512:
496:mismatch repair
492:DNA Polymerases
484:
468:DNA replication
440:
416:Point mutations
400:point mutations
392:
380:
321:
299:to the growing
244:, substituting
226:template strand
194:
171:silent mutation
132:
115:
76:
40:coding sequence
28:
23:
22:
15:
12:
11:
5:
1802:
1800:
1792:
1791:
1786:
1776:
1775:
1770:
1769:
1720:
1693:Bioinformatics
1679:
1630:
1591:
1542:
1491:
1488:10.1101/220814
1468:
1459:
1435:
1417:
1401:
1365:
1324:
1291:
1267:
1238:(4): 1189–95.
1218:
1196:
1177:
1161:
1120:
1069:
1040:(7): 1838–44.
1020:
996:
953:
932:(19): 2411–5.
912:
855:
846:|journal=
814:
784:
757:
722:(2): 104–108.
706:
675:
674:
672:
669:
668:
667:
661:
658:Non-coding RNA
655:
652:Non-coding DNA
649:
643:
640:Gene structure
637:
631:
625:
617:
614:
540:
537:
511:
508:
483:
480:
439:
436:
391:
390:Mutation types
388:
379:
376:
320:
317:
271:(3'-UTR), the
193:
190:
159:point mutation
131:
128:
119:Walter Gilbert
114:
111:
75:
72:
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
1801:
1790:
1787:
1785:
1782:
1781:
1779:
1765:
1761:
1756:
1751:
1747:
1743:
1739:
1735:
1731:
1724:
1721:
1716:
1712:
1707:
1702:
1699:(3): 282–92.
1698:
1694:
1690:
1683:
1680:
1675:
1671:
1666:
1661:
1657:
1653:
1649:
1645:
1641:
1634:
1631:
1626:
1622:
1618:
1614:
1611:(5): 228–32.
1610:
1606:
1602:
1595:
1592:
1587:
1583:
1578:
1573:
1569:
1565:
1561:
1557:
1553:
1546:
1543:
1538:
1534:
1529:
1524:
1519:
1514:
1510:
1506:
1502:
1495:
1492:
1489:
1485:
1481:
1475:
1473:
1469:
1463:
1460:
1449:
1445:
1439:
1436:
1432:
1426:
1424:
1422:
1418:
1414:
1408:
1406:
1402:
1397:
1391:
1378:
1377:
1369:
1366:
1361:
1357:
1352:
1347:
1344:(4): 869–81.
1343:
1339:
1335:
1328:
1325:
1319:
1314:
1310:
1306:
1302:
1295:
1292:
1281:
1277:
1271:
1268:
1263:
1259:
1254:
1249:
1245:
1241:
1237:
1233:
1229:
1222:
1219:
1209:
1208:
1200:
1197:
1192:
1188:
1181:
1178:
1174:
1168:
1166:
1162:
1157:
1153:
1148:
1143:
1139:
1135:
1131:
1124:
1121:
1116:
1112:
1107:
1102:
1097:
1092:
1088:
1084:
1080:
1073:
1070:
1065:
1061:
1056:
1051:
1047:
1043:
1039:
1035:
1031:
1024:
1021:
1010:
1009:rosalind.info
1006:
1000:
997:
992:
988:
984:
980:
976:
972:
969:(3): 216–23.
968:
964:
957:
954:
949:
945:
940:
935:
931:
927:
923:
916:
913:
908:
904:
900:
896:
891:
886:
882:
878:
875:(5645): 501.
874:
870:
866:
859:
856:
851:
839:
825:
821:
817:
815:9780123983978
811:
807:
803:
799:
795:
788:
785:
780:
776:
773:(4): 387–93.
772:
768:
761:
758:
753:
749:
745:
741:
737:
733:
729:
725:
721:
717:
710:
707:
694:
690:
683:
681:
677:
670:
665:
662:
659:
656:
653:
650:
647:
644:
641:
638:
635:
632:
629:
626:
623:
622:Coding strand
620:
619:
615:
613:
610:
606:
601:
597:
593:
588:
586:
581:
573:
566:
562:
558:
554:
550:
545:
538:
536:
534:
530:
526:
522:
518:
509:
507:
505:
501:
497:
493:
489:
481:
479:
477:
476:fertilization
473:
469:
465:
461:
457:
453:
449:
445:
437:
435:
433:
429:
425:
421:
417:
413:
409:
401:
396:
389:
387:
384:
377:
375:
373:
369:
365:
361:
357:
355:
351:
347:
343:
338:
336:
332:
328:
324:
318:
313:
308:
304:
302:
298:
294:
290:
286:
282:
278:
274:
270:
267:(5'-UTR) and
266:
262:
258:
253:
251:
247:
243:
239:
235:
231:
230:transcription
227:
223:
219:
209:
202:
201:Transcription
198:
191:
189:
187:
183:
178:
176:
172:
168:
167:transversions
164:
160:
155:
153:
148:
140:
136:
129:
127:
124:
120:
112:
110:
108:
104:
99:
97:
93:
89:
85:
81:
73:
71:
69:
65:
61:
57:
53:
49:
45:
41:
37:
33:
32:coding region
19:
1789:Biochemistry
1737:
1733:
1723:
1696:
1692:
1682:
1647:
1643:
1633:
1608:
1604:
1594:
1559:
1555:
1545:
1508:
1504:
1494:
1479:
1462:
1451:. Retrieved
1448:Khan Academy
1447:
1438:
1380:, retrieved
1375:
1368:
1341:
1337:
1327:
1308:
1304:
1294:
1283:. Retrieved
1279:
1270:
1235:
1231:
1221:
1211:, retrieved
1206:
1199:
1190:
1180:
1140:(3): 421–7.
1137:
1133:
1123:
1086:
1083:BMC Genomics
1082:
1072:
1037:
1033:
1023:
1012:. Retrieved
1008:
999:
966:
962:
956:
929:
925:
915:
872:
868:
858:
827:. Retrieved
797:
787:
770:
766:
760:
719:
715:
709:
697:. Retrieved
693:the original
589:
577:
553:human genome
513:
488:proofreading
485:
441:
405:
399:
381:
368:splice sites
360:RNA splicing
358:
339:
331:alkyl groups
325:
322:
311:
254:
248:in place of
215:
200:
179:
156:
144:
138:
123:nucleic acid
116:
100:
77:
68:human genome
43:
39:
31:
29:
1505:Front Genet
646:Nested gene
634:Mature mRNA
612:sensitive.
592:prokaryotes
460:carcinogens
364:spliceosome
312:mature mRNA
301:polypeptide
297:amino acids
281:translation
277:Poly-A tail
257:mature mRNA
238:nucleotides
163:transitions
152:stop codons
130:Composition
60:prokaryotes
18:Gene coding
1778:Categories
1453:2023-05-22
1382:2019-11-19
1285:2019-10-30
1213:2019-11-19
1089:(1): 151.
1014:2019-10-31
829:2019-11-07
671:References
596:eukaryotes
567:) regions.
547:Schematic
504:degeneracy
482:Prevention
444:hereditary
428:insertions
340:While the
327:Alkylation
319:Regulation
175:nucleotide
147:GC-content
96:non-coding
74:Definition
64:eukaryotes
1482:, 88–95.
848:ignored (
838:cite book
744:0022-2844
572:Karyotype
557:G banding
549:karyogram
438:Formation
432:deletions
383:Mutations
378:Mutations
335:silencing
279:. During
186:selection
117:In 1978,
1764:30099499
1715:15347574
1674:20610432
1625:12047938
1586:12819146
1537:28261263
1390:citation
1360:10397335
1262:16500890
1156:15590696
1115:28187704
1064:12654999
948:12915446
824:22656372
779:15217358
616:See also
590:In both
490:by some
456:mutagens
426:such as
337:effect.
285:ribosome
182:mutation
1755:6188560
1665:2992710
1528:5309256
1253:1383623
1106:5303225
991:8703087
971:Bibcode
907:4216649
877:Bibcode
752:5978109
724:Bibcode
699:6 April
565:GC rich
472:de novo
261:protein
250:thymine
113:History
56:protein
1762:
1752:
1713:
1672:
1662:
1623:
1584:
1577:403693
1574:
1535:
1525:
1511:: 16.
1358:
1260:
1250:
1154:
1113:
1103:
1062:
1055:152811
1052:
989:
946:
905:
899:622185
897:
869:Nature
822:
812:
777:
750:
742:
293:codons
283:, the
275:, and
273:5' cap
246:uracil
232:, the
103:exomes
1311:(1).
903:S2CID
748:S2CID
289:tRNAs
107:exome
34:of a
1760:PMID
1711:PMID
1670:PMID
1621:PMID
1582:PMID
1533:PMID
1396:link
1356:PMID
1258:PMID
1152:PMID
1111:PMID
1060:PMID
987:PMID
944:PMID
895:PMID
850:help
820:PMID
810:ISBN
775:PMID
740:ISSN
701:2003
628:Exon
594:and
242:mRNA
84:exon
80:exon
62:and
36:gene
30:The
1784:DNA
1750:PMC
1742:doi
1701:doi
1660:PMC
1652:doi
1648:277
1613:doi
1572:PMC
1564:doi
1523:PMC
1513:doi
1484:doi
1346:doi
1313:doi
1248:PMC
1240:doi
1142:doi
1101:PMC
1091:doi
1050:PMC
1042:doi
979:doi
934:doi
885:doi
873:271
802:doi
732:doi
555:on
430:or
218:DNA
216:In
52:RNA
50:or
48:DNA
44:CDS
1780::
1758:.
1748:.
1738:35
1736:.
1732:.
1709:.
1697:21
1695:.
1691:.
1668:.
1658:.
1646:.
1642:.
1619:.
1609:18
1607:.
1603:.
1580:.
1570:.
1560:13
1558:.
1554:.
1531:.
1521:.
1507:.
1503:.
1471:^
1446:.
1420:^
1404:^
1392:}}
1388:{{
1354:.
1342:45
1340:.
1336:.
1307:.
1303:.
1278:.
1256:.
1246:.
1236:34
1234:.
1230:.
1189:.
1164:^
1150:.
1138:14
1136:.
1132:.
1109:.
1099:.
1087:18
1085:.
1081:.
1058:.
1048:.
1038:31
1036:.
1032:.
1007:.
985:.
977:.
967:43
965:.
942:.
930:12
928:.
924:.
901:.
893:.
883:.
871:.
867:.
842::
840:}}
836:{{
818:.
808:.
769:.
746:.
738:.
730:.
720:30
718:.
679:^
598:,
464:UV
458:,
434:.
374:.
1766:.
1744::
1717:.
1703::
1676:.
1654::
1627:.
1615::
1588:.
1566::
1539:.
1515::
1509:8
1486::
1456:.
1433:.
1415:.
1398:)
1362:.
1348::
1321:.
1315::
1309:4
1288:.
1264:.
1242::
1193:.
1175:.
1158:.
1144::
1117:.
1093::
1066:.
1044::
1017:.
993:.
981::
973::
950:.
936::
909:.
887::
879::
852:)
832:.
804::
781:.
771:4
754:.
734::
726::
703:.
446:(
314:.
42:(
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.