111:
22:
333:
Takeshima, Hirohiko; Iguchi, Kei-ichiro & Nishida, Mutsumi (2005): Unexpected
Ceiling of Genetic Differentiation in the Control Region of the Mitochondrial DNA between Different Subspecies of the Ayu
130:
of the human mitogenome, the most variable sites of HVR1 are numbered 16024-16383 (this subsequence is called HVR-I), and the most variable sites of HVR2 are numbered 57-372 (
397:
Antibodies are remarkably specific, thanks to hypervariable regions in both light and heavy chains. The hyperbariable regions for the antigen-binding site.
122:. HVR1 is considered a "low resolution" region and HVR2 is considered a "high resolution" region. Getting HVR1 and HVR2 DNA tests can help determine one's
98:
repeat (in the case of nuclear DNA) or have substitutions (in the case of mitochondrial DNA). Changes or repeats in the hypervariable region are highly
369:
252:
227:
123:
383:
161:
accumulate mutations faster and more freely. It is not known whether such hypovariable control regions are more widespread. In the
114:
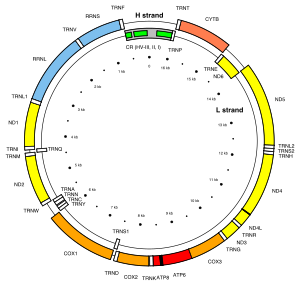
Human mitochondrial genome showing hypervariable regions I to III (green boxes) located in the control region (CR; grey box).
37:
442:
242:
127:
43:
437:
208:
204:
48:
416:
181:
are far lower in the control region than elsewhere. This phenomenon completely defies explanation at present.
110:
99:
31:
247:
194:
119:
375:
257:
154:
447:
190:
146:
340:
412:
422:
379:
300:
87:
348:
290:
231:
212:
321:
431:
279:"Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation"
162:
322:"Annotated mtDNA reference sequences: revised Cambridge Reference Sequence (rCRS)"
79:
220:
200:
178:
118:
There are two mitochondrial hypervariable regions used in human mitochondrial
95:
203:, hypervariable regions form the antigen-binding site and are found on both
170:
142:
91:
304:
423:
DNA: Forensic and Legal
Applications, Explanation of Hypervariable Regions
174:
352:
295:
278:
150:
83:
211:. They also contribute to the specificity of each antibody. In a
158:
368:
Michael Stein; Paul Zei; Gloria Hwang; Radhika
Breaden (2000).
177:
rate is not markedly lowered, but sequence differences between
15:
219:
segments of each heavy or light chain fold together at the
157:
evolves remarkably slowly. Even functional mitochondrial
67:
A highly polymorphic nuclear or mitochondrial DNA region
29:It has been suggested that this article should be
8:
363:
361:
415:at the U.S. National Library of Medicine
294:
109:
269:
277:van Oven M, Kayser M (February 2009).
173:protacanthopterygian, control region
7:
223:to form an antigen binding pocket.
38:Hypervariable region (nucleic acid)
253:Human mitochondrial DNA haplogroup
228:Complementarity-determining region
14:
371:Cracking The Boards: USMLE Step 1
20:
324:. Retrieved on 4 February 2016.
44:Hypervariable region (antibody)
1:
243:Cambridge Reference Sequence
155:mitochondrial control region
128:Cambridge Reference Sequence
464:
225:
188:
417:Medical Subject Headings
120:genealogical DNA testing
78:) is a location within
336:Plecoglossus altivelis
167:Plecoglossus altivelis
145:, for example certain
115:
248:Genealogical DNA test
195:Somatic hypermutation
134:HVR-II) and 438-574 (
113:
35:into articles titled
413:Hypervariable+region
376:The Princeton Review
258:mtDNA control region
72:hypervariable region
443:Genetic engineering
191:V(D)J recombination
147:Protacanthopterygii
353:10.2108/zsj.22.401
296:10.1002/humu.20921
116:
438:Genetic genealogy
126:. In the revised
88:mitochondrial DNA
65:
64:
455:
400:
399:
394:
392:
365:
356:
331:
325:
315:
309:
308:
298:
274:
232:Framework region
60:
57:
24:
23:
16:
463:
462:
458:
457:
456:
454:
453:
452:
428:
427:
409:
404:
403:
390:
388:
386:
367:
366:
359:
355:(HTML abstract)
332:
328:
316:
312:
276:
275:
271:
266:
239:
234:
218:
213:variable domain
197:
189:Main articles:
187:
108:
68:
61:
55:
52:
25:
21:
12:
11:
5:
461:
459:
451:
450:
445:
440:
430:
429:
426:
425:
420:
408:
407:External links
405:
402:
401:
384:
357:
347:(4): 401–410.
326:
310:
289:(2): E386–94.
283:Human Mutation
268:
267:
265:
262:
261:
260:
255:
250:
245:
238:
235:
216:
186:
183:
107:
104:
66:
63:
62:
28:
26:
19:
13:
10:
9:
6:
4:
3:
2:
460:
449:
446:
444:
441:
439:
436:
435:
433:
424:
421:
418:
414:
411:
410:
406:
398:
387:
385:9780375761638
381:
377:
373:
372:
364:
362:
358:
354:
350:
346:
343:
342:
337:
330:
327:
323:
319:
314:
311:
306:
302:
297:
292:
288:
284:
280:
273:
270:
263:
259:
256:
254:
251:
249:
246:
244:
241:
240:
236:
233:
229:
224:
222:
214:
210:
206:
202:
196:
192:
184:
182:
180:
176:
172:
168:
164:
160:
156:
152:
148:
144:
139:
137:
133:
129:
125:
121:
112:
106:Mitochondrial
105:
103:
101:
97:
93:
89:
85:
81:
77:
73:
59:
50:
46:
45:
40:
39:
34:
33:
27:
18:
17:
396:
389:. Retrieved
370:
344:
339:
335:
329:
318:PhyloTree mt
317:
313:
286:
282:
272:
209:heavy chains
198:
166:
140:
135:
131:
117:
75:
71:
69:
53:
42:
36:
30:
391:5 September
143:bony fishes
100:polymorphic
96:nucleotides
80:nuclear DNA
448:Antibodies
432:Categories
341:Zool. Sci.
264:References
226:See also:
221:N-terminus
201:antibodies
185:Antibodies
179:subspecies
171:East Asian
138:HVR-III).
124:haplogroup
92:base pairs
215:, the 3 H
90:in which
56:June 2022
305:18853457
237:See also
175:mutation
141:In some
151:Gadidae
82:or the
49:discuss
419:(MeSH)
382:
303:
169:), an
153:, the
84:D-loop
205:light
159:genes
136:i.e.,
132:i.e.,
32:split
393:2011
380:ISBN
301:PMID
230:and
207:and
193:and
149:and
41:and
349:doi
291:doi
199:In
163:Ayu
94:of
86:of
76:HVR
51:)
47:. (
434::
395:.
378:.
374:.
360:^
345:22
338:.
320:.
299:.
287:30
285:.
281:.
102:.
70:A
351::
307:.
293::
217:V
165:(
74:(
58:)
54:(
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.