649:
335:
181:
40:
690:
Following synthesis of HIV's doubled stranded DNA genome, integrase binds to the long tandem repeats flanking the genome on both ends. Using its endonucleolytic activity, integrase cleaves a di or trinucleotide from both 3' ends of the genome in a processing known as 3'-processing. The specificity of
698:
through SN2-type nucleophilic attack. The 3' ends are covalently linked to the target DNA. The 5' over hangs of the viral genome are then cleaved using host repair enzymes, those same enzymes are believed to be responsible for the integration of the 5' end into the host genome forming the provirus.
674:
The N-terminus is composed of 50 amino acid residues which contain a conserved histidine, histidine, cytosine, cytosine sequence which chelates zinc ions, furthermore enhancing the enzymatic activity of the catalytic core domain. As metal chelation is vital in integrase efficacy, it is a target for
678:
The catalytic core domain, like the N-terminus, contains highly conserved amino acid residues -Asp64, Asp116, Glu152- as the conserved DDE (Asp-Asp-Glu) motif contributes to the endonuclease and polynucleotide transferase functions of integrase. Mutations in these regions inactivates integrase and
613:
replication. Integration is a "point of no return" for the cell, which becomes a permanent carrier of the viral genome (provirus). Integration is in part responsible for the persistence of retroviral infections. After integration, the viral gene expression and particle production may take place
584:(dimer-of-dimers), with all three domains being important for multimerization and viral DNA binding. In addition, several host cellular proteins have been shown to interact with IN to facilitate the integration process: e.g., the host factor, human chromatin-associated protein
2175:
594:(HFV), an agent harmless to humans, has an integrase similar to HIV IN and is therefore a model of HIV IN function; a 2010 crystal structure of the HFV integrase assembled on viral DNA ends has been determined.
2185:
2180:
691:
cleavage is improved through the use of cofactors such as Mn and Mg which interact with the DDE motif of the catalytic core domain, acting as cofactors to integrase function.
2300:
2278:
2622:
2617:
2096:
434:
293:
139:
2674:
962:
Lodi PJ, Ernst JA, Kuszewski J, Hickman AB, Engelman A, Craigie R, et al. (August 1995). "Solution structure of the DNA binding domain of HIV-1 integrase".
719:, demonstrated that the compound has potent antiviral activity. On October 12, 2007, the Food and Drug Administration (U.S.) approved the integrase inhibitor
2206:
3202:
3197:
3192:
3187:
3149:
3144:
580:(RSV) have been reported, with the first structures determined in 1994. Biochemical data and structural data suggest that retroviral IN functions as a
3524:
3182:
3177:
2101:
2653:
2648:
2643:
2367:
2268:
3905:
621:
3'-processing, in which two or three nucleotides are removed from one or both 3' ends of the viral DNA to expose an invariant CA dinucleotide.
3668:
1907:
382:
241:
87:
506:
671:, catalytic core domain, and C-terminus, which each have distinct properties and functions contributing to the efficacy of HIV integrase.
760:
Beck BJ, Freudenreich O, Worth JL (2010). "Patients with Human
Immunodeficiency Virus Infection and Acquired Immunodeficiency Syndrome".
2480:
2041:
3638:
3572:
2799:
1611:
1405:
777:
3266:
2934:
1752:
1258:"HIV-1 Reverse Transcriptase/Integrase Dual Inhibitors: A Review of Recent Advances and Structure-activity Relationship Studies"
614:
immediately or at some point in the future, the timing depends on the activity of the chromosomal locus hosting the provirus.
602:
Integration occurs following production of the double-stranded linear viral DNA by the viral RNA/DNA-dependent DNA polymerase
624:
the strand transfer reaction, in which the processed 3' ends of the viral DNA are covalently ligated to host chromosomal DNA.
530:
454:
313:
159:
3785:
2325:
2320:
1880:
1698:
573:
3259:
3254:
2360:
2190:
2078:
2046:
2031:
3887:
3822:
3494:
3470:
3277:
3162:
3068:
2994:
2919:
2814:
2698:
2633:
2553:
2519:
2470:
1713:
637:
514:
652:
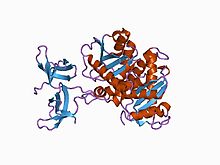
Structural depiction of the HIV catalytic core domain based on the works of Feng, L. and
Kvaratskhelia, M. from the
2563:
2490:
2455:
588:, tightly binds HIV IN and directs the HIV pre-integration complex towards highly expressed genes for integration.
3445:
3249:
2450:
2311:
2289:
2264:
2239:
2159:
1767:
1727:
1672:
1649:
1621:
1589:
1455:
1542:
3944:
3832:
3758:
3696:
3648:
3600:
3552:
3504:
3383:
3232:
3127:
2888:
2794:
2774:
2757:
2607:
2495:
2430:
1776:
1757:
1688:
1422:
1377:
572:
Crystal and NMR structures of the individual domains and 2-domain constructs of integrases from HIV-1, HIV-2,
442:
301:
147:
2353:
1634:
535:
2597:
4049:
4007:
3843:
3728:
2568:
2088:
1861:
1771:
695:
486:
3977:
3868:
3748:
3620:
3480:
1942:
1897:
1599:
1584:
1520:
1500:
1398:
609:
The main function of IN is to insert the viral DNA into the host chromosomal DNA, an essential step for
603:
438:
297:
143:
1383:
795:"Non-Enzymatic Functions of Retroviral Integrase: The Next Target for Novel Anti-HIV Drug Development"
44:
solution structure of the n-terminal zn binding domain of hiv-1 integrase (e form), nmr, 38 structures
3779:
3717:
3404:
2848:
2839:
2761:
2708:
2529:
2420:
1856:
1851:
1731:
1594:
1574:
1483:
521:
395:
254:
100:
3450:
2844:
2834:
2753:
2119:
2111:
1892:
1559:
1495:
1459:
736:
712:
629:
3972:
3849:
1982:
1920:
1871:
1606:
1433:
1348:
1204:"The role of HIV integration in viral persistence: no more whistling past the proviral graveyard"
682:
The C-terminus domain binds to host DNA non-specifically and stabilizes the integration complex.
581:
577:
554:
HH-CC zinc-binding domain (a three-helical bundle stabilized by coordination of a Zn(II) cation),
4039:
3424:
3172:
3111:
2876:
1997:
1987:
1977:
1902:
1886:
1845:
1831:
1826:
1564:
1527:
1340:
1287:
1233:
1184:
1135:
1086:
1035:
979:
944:
886:
826:
773:
591:
429:
288:
134:
4044:
3686:
3292:
3029:
3024:
3019:
3014:
3009:
3004:
2983:
2330:
1992:
1972:
1914:
1810:
1708:
1693:
1639:
1537:
1532:
1391:
1330:
1322:
1277:
1269:
1223:
1215:
1174:
1166:
1125:
1117:
1076:
1066:
1053:
Hare S, Di Nunzio F, Labeja A, Wang J, Engelman A, Cherepanov P (July 2009). Luban J (ed.).
1025:
1017:
971:
934:
924:
876:
868:
816:
806:
765:
546:
All retroviral IN proteins contain three canonical domains, connected by flexible linkers:
421:
280:
126:
3916:
3373:
3363:
3116:
2253:
2243:
2065:
2036:
1662:
1569:
1554:
648:
498:
3990:
3873:
3418:
3243:
3138:
2744:
2664:
2221:
2060:
1967:
1959:
1836:
1799:
1335:
1310:
1282:
1257:
1228:
1203:
1179:
1154:
1130:
1105:
1081:
1054:
1030:
1005:
939:
912:
881:
856:
821:
794:
769:
708:
494:
1373:
1155:"Efficient HIV-1 in vitro reverse transcription: optimal capsid stability is required"
4033:
3939:
3590:
3051:
2688:
2405:
2380:
2376:
1490:
1352:
525:
490:
387:
246:
92:
1273:
363:
209:
68:
3659:
3611:
3563:
3515:
3436:
3221:
3057:
2734:
2543:
2410:
2384:
1579:
1549:
1478:
724:
510:
502:
417:
276:
122:
2345:
1006:"Recent advances in the discovery of small-molecule inhibitors of HIV-1 integrase"
222:
1071:
375:
234:
80:
3999:
3542:
2862:
2825:
2145:
2135:
1736:
1473:
1414:
1121:
720:
1326:
1170:
872:
3881:
3813:
3410:
3216:
3047:
3043:
1947:
1790:
1703:
1468:
1418:
668:
565:
561:
551:
478:
811:
17:
3927:
3922:
3857:
3101:
2978:
2968:
2956:
2729:
2393:
2226:
1676:
1442:
929:
1344:
1291:
1237:
1188:
1139:
1090:
1039:
1021:
948:
890:
830:
983:
391:
334:
250:
180:
96:
39:
2509:
2073:
1653:
1055:"Structural basis for functional tetramerization of lentiviral integrase"
741:
633:
370:
229:
75:
975:
2024:
2009:
1625:
716:
913:"Integrase and integration: biochemical activities of HIV-1 integrase"
911:
Delelis O, Carayon K, Saïb A, Deprez E, Mouscadet JF (December 2008).
762:
Massachusetts
General Hospital Handbook of General Hospital Psychiatry
653:
3769:
3707:
3455:
3394:
3287:
2785:
2441:
2002:
1437:
1219:
474:
449:
308:
154:
528:
bound to the ends of the viral DNA ends has been referred to as the
1368:
3340:
3330:
3325:
3315:
3310:
3305:
3300:
2907:
2902:
2897:
2019:
2014:
1866:
647:
628:
Both reactions are catalyzed in the same active site, and involve
585:
2176:
CDP-diacylglycerol—glycerol-3-phosphate 3-phosphatidyltransferase
723:(MK-0518, brand name Isentress). The second integrase inhibitor,
3892:
3723:
3335:
3320:
2749:
2485:
1930:
1925:
1821:
1816:
1515:
1510:
1505:
857:"Structural Biology of HIV Integrase Strand Transfer Inhibitors"
411:
358:
270:
216:
204:
116:
63:
2349:
1387:
3953:
3949:
3932:
3078:
2929:
660:
610:
482:
493:
links between) its genetic information into that of the host
1256:
Mahboubi-Rabbani M, Abbasi M, Hajimahdi Z, Zarghi A (2021).
1309:
Maertens GN, Engelman AN, Cherepanov P (January 2022).
1004:
Choi E, Mallareddy JR, Lu D, Kolluru S (October 2018).
694:
The newly generated 3'OH groups disrupt the host DNA's
497:
it infects. Retroviral INs are not to be confused with
667:
is a 32kDa viral protein consisting of three domains-
2186:
CDP-diacylglycerol—inositol 3-phosphatidyltransferase
2191:
CDP-diacylglycerol—choline O-phosphatidyltransferase
3998:
3989:
3966:
3903:
3830:
3821:
3812:
3768:
3756:
3747:
3706:
3694:
3685:
3658:
3646:
3637:
3610:
3598:
3589:
3562:
3550:
3541:
3514:
3502:
3493:
3468:
3435:
3393:
3381:
3372:
3357:
3275:
3230:
3215:
3160:
3125:
3110:
3095:
3066:
3042:
2992:
2977:
2964:
2955:
2917:
2886:
2875:
2824:
2812:
2784:
2772:
2743:
2725:
2696:
2687:
2662:
2631:
2605:
2596:
2585:
2551:
2542:
2517:
2508:
2468:
2440:
2428:
2419:
2401:
2392:
2310:
2288:
2263:
2238:
2219:
2199:
2181:
CDP-diacylglycerol—serine O-phosphatidyltransferase
2168:
2158:
2128:
2110:
2087:
2059:
1958:
1798:
1789:
1766:
1726:
1671:
1648:
1620:
1454:
1430:
534:; IN is a key component in this and the retroviral
448:
428:
410:
405:
381:
369:
357:
349:
344:
327:
307:
287:
269:
264:
240:
228:
215:
203:
195:
190:
173:
153:
133:
115:
110:
86:
74:
62:
54:
49:
32:
855:Jóźwik IK, Passos DO, Lyumkis D (September 2020).
339:Crystal structure of the RSV two-domain integrase.
185:Crystal structure of the RSV two-domain integrase.
1311:"Structure and function of retroviral integrase"
2361:
1399:
8:
2207:N-acetylglucosamine-1-phosphate transferase
2097:UTP—glucose-1-phosphate uridylyltransferase
1110:Cold Spring Harbor Perspectives in Medicine
727:, was approved in the U.S. in August 2012.
3995:
3986:
3827:
3818:
3765:
3753:
3703:
3691:
3655:
3643:
3607:
3595:
3559:
3547:
3511:
3499:
3390:
3378:
3369:
3227:
3122:
3107:
3063:
2989:
2974:
2961:
2883:
2821:
2781:
2769:
2740:
2693:
2602:
2593:
2548:
2514:
2437:
2425:
2416:
2398:
2368:
2354:
2346:
2235:
2165:
1795:
1786:
1451:
1406:
1392:
1384:
1262:Iranian Journal of Pharmaceutical Research
402:
333:
261:
179:
107:
38:
3525:Parainfluenza hemagglutinin-neuraminidase
2301:serine/threonine-specific protein kinases
2279:serine/threonine-specific protein kinases
2102:Galactose-1-phosphate uridylyltransferase
1376:at the U.S. National Library of Medicine
1334:
1281:
1227:
1178:
1129:
1080:
1070:
1029:
938:
928:
880:
820:
810:
675:the development of retroviral therapies.
636:protein-DNA intermediate (in contrast to
1159:Signal Transduction and Targeted Therapy
617:Retroviral INs catalyzes two reactions:
752:
638:Ser/Tyr recombinase-catalyzed reactions
1153:Burdick RC, Pathak VK (January 2021).
557:a catalytic core domain (RNaseH fold),
324:
170:
29:
3669:Respiratory syncytial virus G protein
1251:
1249:
1247:
1208:The Journal of Clinical Investigation
7:
999:
997:
995:
993:
906:
904:
902:
900:
850:
848:
846:
844:
842:
840:
1104:Craigie R, Bushman FD (July 2012).
861:Trends in Pharmacological Sciences
770:10.1016/b978-1-4377-1927-7.00026-1
25:
3573:Mumps hemagglutinin-neuraminidase
1612:Glucose-1,6-bisphosphate synthase
2935:Hepatitis B virus DNA polymerase
1753:Ribose-phosphate diphosphokinase
2766:(non-enveloped circular ds-DNA)
1367:PDB-101 Molecule of the Month:
1274:10.22037/ijpr.2021.115446.15370
1202:Maldarelli F (February 2016).
764:. Elsevier. pp. 353–370.
707:In November 2005, data from a
1:
3786:Vesiculovirus matrix proteins
2326:Protein-histidine tele-kinase
2321:Protein-histidine pros-kinase
2200:Glycosyl-1-phosphotransferase
679:prevents genome integration.
406:Available protein structures:
265:Available protein structures:
111:Available protein structures:
33:Integrase Zinc binding domain
2032:RNA-dependent RNA polymerase
1315:Nature Reviews. Microbiology
1072:10.1371/journal.ppat.1000515
328:Integrase DNA binding domain
3823:Structure and genome of HIV
2880:(circular partially ds-DNA)
1939:RNA-directed DNA polymerase
1807:DNA-directed DNA polymerase
1122:10.1101/cshperspect.a006890
515:site-specific recombination
513:integrase, as discussed in
4066:
2456:Herpesvirus glycoprotein B
2292:: protein-dual-specificity
1327:10.1038/s41579-021-00586-9
1171:10.1038/s41392-020-00458-3
873:10.1016/j.tips.2020.06.003
711:of an investigational HIV
2451:HHV capsid portal protein
799:Frontiers in Microbiology
401:
332:
260:
178:
106:
37:
2269:protein-serine/threonine
2169:Phosphatidyltransferases
1758:Thiamine diphosphokinase
1378:Medical Subject Headings
812:10.3389/fmicb.2011.00210
3446:Influenza hemagglutinin
930:10.1186/1742-4690-5-114
696:phosphodiester linkages
536:pre-integration complex
4008:Gag-onc fusion protein
2089:Nucleotidyltransferase
1772:nucleotidyltransferase
1699:Nucleoside-diphosphate
1022:10.4155/fsoa-2018-0060
703:Antiretroviral therapy
656:
598:Function and mechanism
522:macromolecular complex
3978:murine leukemia virus
3869:Reverse transcriptase
3749:Indiana vesiculovirus
3621:Measles hemagglutinin
1943:Reverse transcriptase
1106:"HIV DNA integration"
686:Integration mechanism
651:
604:reverse transcriptase
507:used in biotechnology
174:Integrase core domain
2709:Early 35 kDa protein
1732:diphosphotransferase
1714:Thiamine-diphosphate
1421:-containing groups (
632:without involving a
564:DNA-binding domain (
467:Retroviral integrase
2314:: protein-histidine
2232:; protein acceptor)
2120:mRNA capping enzyme
2112:Guanylyltransferase
976:10.1021/bi00031a002
737:Integrase inhibitor
713:integrase inhibitor
630:transesterification
3973:Rous sarcoma virus
1590:Phosphoinositide 3
1434:phosphotransferase
657:
578:Rous Sarcoma Virus
4027:
4026:
4023:
4022:
4019:
4018:
4015:
4014:
3962:
3961:
3808:
3807:
3804:
3803:
3800:
3799:
3796:
3795:
3743:
3742:
3739:
3738:
3681:
3680:
3677:
3676:
3633:
3632:
3629:
3628:
3585:
3584:
3581:
3580:
3537:
3536:
3533:
3532:
3489:
3488:
3464:
3463:
3353:
3352:
3349:
3348:
3211:
3210:
3091:
3090:
3087:
3086:
3038:
3037:
2951:
2950:
2947:
2946:
2943:
2942:
2871:
2870:
2858:
2857:
2808:
2807:
2721:
2720:
2717:
2716:
2683:
2682:
2581:
2580:
2577:
2576:
2538:
2537:
2504:
2503:
2464:
2463:
2343:
2342:
2339:
2338:
2215:
2214:
2154:
2153:
2055:
2054:
1968:Template-directed
1722:
1721:
1689:Phosphomevalonate
1369:135 HIV Integrase
1010:Future Science OA
970:(31): 9826–9833.
793:Masuda T (2011).
592:Human foamy virus
464:
463:
460:
459:
455:structure summary
323:
322:
319:
318:
314:structure summary
169:
168:
165:
164:
160:structure summary
16:(Redirected from
4057:
3996:
3987:
3828:
3819:
3766:
3754:
3704:
3692:
3687:Zaire ebolavirus
3656:
3644:
3608:
3596:
3560:
3548:
3512:
3500:
3391:
3379:
3370:
3293:3C-like protease
3228:
3123:
3108:
3064:
2990:
2984:Duplornaviricota
2975:
2962:
2884:
2822:
2782:
2770:
2741:
2694:
2603:
2594:
2549:
2515:
2438:
2426:
2417:
2399:
2370:
2363:
2356:
2347:
2331:Histidine kinase
2254:tyrosine kinases
2244:protein-tyrosine
2236:
2166:
1973:RNA polymerase I
1796:
1787:
1640:Aspartate kinase
1635:Phosphoglycerate
1452:
1408:
1401:
1394:
1385:
1356:
1338:
1296:
1295:
1285:
1253:
1242:
1241:
1231:
1220:10.1172/JCI80564
1199:
1193:
1192:
1182:
1150:
1144:
1143:
1133:
1101:
1095:
1094:
1084:
1074:
1050:
1044:
1043:
1033:
1001:
988:
987:
959:
953:
952:
942:
932:
908:
895:
894:
884:
852:
835:
834:
824:
814:
790:
784:
783:
757:
654:protein database
499:phage integrases
403:
337:
325:
262:
183:
171:
108:
42:
30:
27:Class of enzymes
21:
4065:
4064:
4060:
4059:
4058:
4056:
4055:
4054:
4030:
4029:
4028:
4011:
3958:
3917:transactivators
3899:
3792:
3735:
3673:
3625:
3577:
3529:
3485:
3460:
3431:
3374:Influenza virus
3364:Negarnaviricota
3361:
3359:
3345:
3271:
3219:
3207:
3156:
3117:Kitrinoviricota
3114:
3099:
3097:
3083:
3055:
3034:
2981:
2966:
2939:
2913:
2879:
2867:
2854:
2804:
2765:
2747:
2733:
2727:
2713:
2679:
2658:
2627:
2589:
2588:(Duplodnaviria,
2587:
2586:circular ds-DNA
2573:
2534:
2500:
2460:
2409:
2403:
2388:
2374:
2344:
2335:
2306:
2284:
2259:
2230:
2224:
2220:2.7.10-2.7.13:
2211:
2195:
2162:: miscellaneous
2150:
2124:
2106:
2083:
2066:exoribonuclease
2063:
2051:
2037:Polyadenylation
1954:
1780:
1774:
1762:
1744:
1740:
1734:
1718:
1680:
1667:
1644:
1616:
1446:
1440:
1432:
1426:
1412:
1364:
1359:
1308:
1304:
1302:Further reading
1299:
1255:
1254:
1245:
1201:
1200:
1196:
1152:
1151:
1147:
1103:
1102:
1098:
1065:(7): e1000515.
1052:
1051:
1047:
1003:
1002:
991:
961:
960:
956:
910:
909:
898:
854:
853:
838:
792:
791:
787:
780:
759:
758:
754:
750:
733:
705:
688:
646:
600:
544:
340:
186:
45:
28:
23:
22:
15:
12:
11:
5:
4063:
4061:
4053:
4052:
4047:
4042:
4032:
4031:
4025:
4024:
4021:
4020:
4017:
4016:
4013:
4012:
4010:
4005:
4003:
3993:
3991:Fusion protein
3984:
3983:
3982:
3981:
3980:
3975:
3964:
3963:
3960:
3959:
3957:
3956:
3947:
3942:
3937:
3936:
3935:
3930:
3925:
3912:
3910:
3901:
3900:
3898:
3897:
3896:
3895:
3890:
3878:
3877:
3876:
3874:HIV-1 protease
3871:
3866:
3854:
3853:
3852:
3839:
3837:
3825:
3816:
3810:
3809:
3806:
3805:
3802:
3801:
3798:
3797:
3794:
3793:
3791:
3790:
3789:
3788:
3780:matrix protein
3775:
3773:
3763:
3751:
3745:
3744:
3741:
3740:
3737:
3736:
3734:
3733:
3732:
3731:
3726:
3718:matrix protein
3713:
3711:
3701:
3689:
3683:
3682:
3679:
3678:
3675:
3674:
3672:
3671:
3665:
3663:
3653:
3641:
3635:
3634:
3631:
3630:
3627:
3626:
3624:
3623:
3617:
3615:
3605:
3593:
3587:
3586:
3583:
3582:
3579:
3578:
3576:
3575:
3569:
3567:
3557:
3545:
3539:
3538:
3535:
3534:
3531:
3530:
3528:
3527:
3521:
3519:
3509:
3497:
3491:
3490:
3487:
3486:
3484:
3483:
3477:
3475:
3466:
3465:
3462:
3461:
3459:
3458:
3453:
3448:
3442:
3440:
3433:
3432:
3430:
3429:
3428:
3427:
3419:viral envelope
3415:
3414:
3413:
3405:matrix protein
3400:
3398:
3388:
3376:
3367:
3360:negative-sense
3355:
3354:
3351:
3350:
3347:
3346:
3344:
3343:
3338:
3333:
3328:
3323:
3318:
3313:
3308:
3303:
3298:
3297:
3296:
3284:
3282:
3273:
3272:
3270:
3269:
3264:
3263:
3262:
3257:
3252:
3244:viral envelope
3239:
3237:
3225:
3213:
3212:
3209:
3208:
3206:
3205:
3200:
3195:
3190:
3185:
3180:
3175:
3169:
3167:
3158:
3157:
3155:
3154:
3153:
3152:
3147:
3139:viral envelope
3134:
3132:
3120:
3105:
3098:positive-sense
3093:
3092:
3089:
3088:
3085:
3084:
3082:
3081:
3075:
3073:
3061:
3040:
3039:
3036:
3035:
3033:
3032:
3027:
3022:
3017:
3012:
3007:
3001:
2999:
2987:
2972:
2959:
2953:
2952:
2949:
2948:
2945:
2944:
2941:
2940:
2938:
2937:
2932:
2926:
2924:
2915:
2914:
2912:
2911:
2910:
2900:
2895:
2893:
2881:
2873:
2872:
2869:
2868:
2866:
2865:
2859:
2856:
2855:
2853:
2852:
2842:
2837:
2831:
2829:
2819:
2810:
2809:
2806:
2805:
2803:
2802:
2797:
2791:
2789:
2779:
2767:
2745:Polyomaviridae
2738:
2723:
2722:
2719:
2718:
2715:
2714:
2712:
2711:
2705:
2703:
2691:
2685:
2684:
2681:
2680:
2678:
2677:
2671:
2669:
2660:
2659:
2657:
2656:
2651:
2646:
2640:
2638:
2629:
2628:
2626:
2625:
2620:
2614:
2612:
2600:
2591:
2590:Varidnaviria?)
2583:
2582:
2579:
2578:
2575:
2574:
2572:
2571:
2566:
2560:
2558:
2546:
2540:
2539:
2536:
2535:
2533:
2532:
2526:
2524:
2512:
2506:
2505:
2502:
2501:
2499:
2498:
2493:
2488:
2483:
2477:
2475:
2466:
2465:
2462:
2461:
2459:
2458:
2453:
2447:
2445:
2435:
2423:
2421:Herpes simplex
2414:
2396:
2390:
2389:
2377:Viral proteins
2375:
2373:
2372:
2365:
2358:
2350:
2341:
2340:
2337:
2336:
2334:
2333:
2328:
2323:
2317:
2315:
2308:
2307:
2305:
2304:
2295:
2293:
2286:
2285:
2283:
2282:
2273:
2271:
2261:
2260:
2258:
2257:
2248:
2246:
2233:
2228:
2222:protein kinase
2217:
2216:
2213:
2212:
2210:
2209:
2203:
2201:
2197:
2196:
2194:
2193:
2188:
2183:
2178:
2172:
2170:
2163:
2156:
2155:
2152:
2151:
2149:
2148:
2143:
2132:
2130:
2126:
2125:
2123:
2122:
2116:
2114:
2108:
2107:
2105:
2104:
2099:
2093:
2091:
2085:
2084:
2082:
2081:
2076:
2070:
2068:
2061:Phosphorolytic
2057:
2056:
2053:
2052:
2050:
2049:
2044:
2039:
2034:
2029:
2028:
2027:
2022:
2017:
2007:
2006:
2005:
1995:
1990:
1985:
1980:
1975:
1970:
1964:
1962:
1960:RNA polymerase
1956:
1955:
1953:
1952:
1951:
1950:
1940:
1936:
1935:
1934:
1933:
1928:
1923:
1912:
1911:
1910:
1905:
1900:
1895:
1884:
1878:
1877:
1876:
1869:
1864:
1859:
1854:
1843:
1842:
1841:
1834:
1829:
1824:
1819:
1808:
1804:
1802:
1800:DNA polymerase
1793:
1784:
1778:
1764:
1763:
1761:
1760:
1755:
1749:
1747:
1742:
1738:
1724:
1723:
1720:
1719:
1717:
1716:
1711:
1706:
1701:
1696:
1691:
1685:
1683:
1678:
1669:
1668:
1666:
1665:
1659:
1657:
1646:
1645:
1643:
1642:
1637:
1631:
1629:
1618:
1617:
1615:
1614:
1609:
1604:
1603:
1602:
1597:
1587:
1585:Diacylglycerol
1582:
1577:
1572:
1567:
1562:
1557:
1552:
1547:
1546:
1545:
1535:
1530:
1525:
1524:
1523:
1518:
1513:
1508:
1503:
1496:Phosphofructo-
1493:
1488:
1487:
1486:
1476:
1471:
1465:
1463:
1449:
1444:
1428:
1427:
1413:
1411:
1410:
1403:
1396:
1388:
1382:
1381:
1371:
1363:
1362:External links
1360:
1358:
1357:
1305:
1303:
1300:
1298:
1297:
1268:(2): 333–369.
1243:
1214:(2): 438–447.
1194:
1145:
1116:(7): a006890.
1096:
1059:PLOS Pathogens
1045:
989:
954:
896:
867:(9): 611–626.
836:
785:
778:
751:
749:
746:
745:
744:
739:
732:
729:
704:
701:
687:
684:
645:
642:
626:
625:
622:
599:
596:
570:
569:
558:
555:
543:
540:
477:produced by a
462:
461:
458:
457:
452:
446:
445:
432:
426:
425:
415:
408:
407:
399:
398:
385:
379:
378:
373:
367:
366:
361:
355:
354:
351:
347:
346:
342:
341:
338:
330:
329:
321:
320:
317:
316:
311:
305:
304:
291:
285:
284:
274:
267:
266:
258:
257:
244:
238:
237:
232:
226:
225:
220:
213:
212:
207:
201:
200:
197:
193:
192:
188:
187:
184:
176:
175:
167:
166:
163:
162:
157:
151:
150:
137:
131:
130:
120:
113:
112:
104:
103:
90:
84:
83:
78:
72:
71:
66:
60:
59:
56:
52:
51:
47:
46:
43:
35:
34:
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
4062:
4051:
4050:Viral enzymes
4048:
4046:
4043:
4041:
4038:
4037:
4035:
4009:
4006:
4004:
4001:
3997:
3994:
3992:
3988:
3985:
3979:
3976:
3974:
3971:
3970:
3969:
3968:
3965:
3955:
3951:
3948:
3946:
3943:
3941:
3938:
3934:
3931:
3929:
3926:
3924:
3921:
3920:
3919:
3918:
3914:
3913:
3911:
3909:
3907:
3902:
3894:
3891:
3889:
3886:
3885:
3884:
3883:
3879:
3875:
3872:
3870:
3867:
3865:
3862:
3861:
3860:
3859:
3855:
3851:
3848:
3847:
3846:
3845:
3841:
3840:
3838:
3836:
3834:
3829:
3826:
3824:
3820:
3817:
3815:
3811:
3787:
3784:
3783:
3782:
3781:
3777:
3776:
3774:
3771:
3767:
3764:
3762:
3760:
3755:
3752:
3750:
3746:
3730:
3727:
3725:
3722:
3721:
3720:
3719:
3715:
3714:
3712:
3709:
3705:
3702:
3700:
3698:
3693:
3690:
3688:
3684:
3670:
3667:
3666:
3664:
3661:
3657:
3654:
3652:
3650:
3645:
3642:
3640:
3636:
3622:
3619:
3618:
3616:
3613:
3609:
3606:
3604:
3602:
3597:
3594:
3592:
3588:
3574:
3571:
3570:
3568:
3565:
3561:
3558:
3556:
3554:
3549:
3546:
3544:
3540:
3526:
3523:
3522:
3520:
3517:
3513:
3510:
3508:
3506:
3501:
3498:
3496:
3495:Parainfluenza
3492:
3482:
3479:
3478:
3476:
3474:
3472:
3467:
3457:
3454:
3452:
3451:Neuraminidase
3449:
3447:
3444:
3443:
3441:
3438:
3434:
3426:
3423:
3422:
3421:
3420:
3416:
3412:
3409:
3408:
3407:
3406:
3402:
3401:
3399:
3396:
3392:
3389:
3387:
3385:
3380:
3377:
3375:
3371:
3368:
3365:
3356:
3342:
3339:
3337:
3334:
3332:
3329:
3327:
3324:
3322:
3319:
3317:
3314:
3312:
3309:
3307:
3304:
3302:
3299:
3294:
3291:
3290:
3289:
3286:
3285:
3283:
3281:
3279:
3274:
3268:
3265:
3261:
3258:
3256:
3253:
3251:
3248:
3247:
3246:
3245:
3241:
3240:
3238:
3236:
3234:
3229:
3226:
3223:
3218:
3214:
3204:
3201:
3199:
3196:
3194:
3191:
3189:
3186:
3184:
3181:
3179:
3176:
3174:
3171:
3170:
3168:
3166:
3164:
3159:
3151:
3148:
3146:
3143:
3142:
3141:
3140:
3136:
3135:
3133:
3131:
3129:
3124:
3121:
3118:
3113:
3109:
3106:
3103:
3094:
3080:
3077:
3076:
3074:
3072:
3070:
3065:
3062:
3059:
3053:
3049:
3045:
3041:
3031:
3028:
3026:
3023:
3021:
3018:
3016:
3013:
3011:
3008:
3006:
3003:
3002:
3000:
2998:
2996:
2991:
2988:
2985:
2980:
2976:
2973:
2970:
2963:
2960:
2958:
2954:
2936:
2933:
2931:
2928:
2927:
2925:
2923:
2921:
2916:
2909:
2906:
2905:
2904:
2901:
2899:
2896:
2894:
2892:
2890:
2885:
2882:
2878:
2874:
2864:
2861:
2860:
2850:
2846:
2843:
2841:
2838:
2836:
2833:
2832:
2830:
2827:
2823:
2820:
2818:
2816:
2811:
2801:
2798:
2796:
2793:
2792:
2790:
2787:
2783:
2780:
2778:
2776:
2771:
2768:
2763:
2759:
2755:
2751:
2746:
2742:
2739:
2736:
2731:
2724:
2710:
2707:
2706:
2704:
2702:
2700:
2695:
2692:
2690:
2689:Baculoviridae
2686:
2676:
2673:
2672:
2670:
2668:
2666:
2661:
2655:
2652:
2650:
2647:
2645:
2642:
2641:
2639:
2637:
2635:
2630:
2624:
2621:
2619:
2616:
2615:
2613:
2611:
2609:
2604:
2601:
2599:
2595:
2592:
2584:
2570:
2567:
2565:
2562:
2561:
2559:
2557:
2555:
2550:
2547:
2545:
2541:
2531:
2528:
2527:
2525:
2523:
2521:
2516:
2513:
2511:
2507:
2497:
2494:
2492:
2489:
2487:
2484:
2482:
2479:
2478:
2476:
2474:
2472:
2467:
2457:
2454:
2452:
2449:
2448:
2446:
2443:
2439:
2436:
2434:
2432:
2427:
2424:
2422:
2418:
2415:
2412:
2407:
2406:Duplodnaviria
2402:linear ds-DNA
2400:
2397:
2395:
2391:
2386:
2382:
2378:
2371:
2366:
2364:
2359:
2357:
2352:
2351:
2348:
2332:
2329:
2327:
2324:
2322:
2319:
2318:
2316:
2313:
2309:
2303:
2302:
2297:
2296:
2294:
2291:
2287:
2281:
2280:
2275:
2274:
2272:
2270:
2266:
2262:
2256:
2255:
2250:
2249:
2247:
2245:
2241:
2237:
2234:
2231:
2223:
2218:
2208:
2205:
2204:
2202:
2198:
2192:
2189:
2187:
2184:
2182:
2179:
2177:
2174:
2173:
2171:
2167:
2164:
2161:
2157:
2147:
2144:
2141:
2137:
2134:
2133:
2131:
2127:
2121:
2118:
2117:
2115:
2113:
2109:
2103:
2100:
2098:
2095:
2094:
2092:
2090:
2086:
2080:
2077:
2075:
2072:
2071:
2069:
2067:
2062:
2058:
2048:
2045:
2043:
2040:
2038:
2035:
2033:
2030:
2026:
2023:
2021:
2018:
2016:
2013:
2012:
2011:
2008:
2004:
2001:
2000:
1999:
1996:
1994:
1991:
1989:
1986:
1984:
1981:
1979:
1976:
1974:
1971:
1969:
1966:
1965:
1963:
1961:
1957:
1949:
1946:
1945:
1944:
1941:
1938:
1937:
1932:
1929:
1927:
1924:
1922:
1919:
1918:
1916:
1913:
1909:
1906:
1904:
1901:
1899:
1896:
1894:
1891:
1890:
1888:
1885:
1882:
1879:
1875:
1874:
1870:
1868:
1865:
1863:
1860:
1858:
1855:
1853:
1850:
1849:
1847:
1844:
1840:
1839:
1835:
1833:
1830:
1828:
1825:
1823:
1820:
1818:
1815:
1814:
1812:
1809:
1806:
1805:
1803:
1801:
1797:
1794:
1792:
1788:
1785:
1782:
1773:
1769:
1765:
1759:
1756:
1754:
1751:
1750:
1748:
1745:
1733:
1729:
1725:
1715:
1712:
1710:
1707:
1705:
1702:
1700:
1697:
1695:
1692:
1690:
1687:
1686:
1684:
1681:
1674:
1670:
1664:
1661:
1660:
1658:
1655:
1651:
1647:
1641:
1638:
1636:
1633:
1632:
1630:
1627:
1623:
1619:
1613:
1610:
1608:
1605:
1601:
1600:Class II PI 3
1598:
1596:
1593:
1592:
1591:
1588:
1586:
1583:
1581:
1578:
1576:
1575:Deoxycytidine
1573:
1571:
1568:
1566:
1563:
1561:
1558:
1556:
1553:
1551:
1548:
1544:
1543:ADP-thymidine
1541:
1540:
1539:
1536:
1534:
1531:
1529:
1526:
1522:
1519:
1517:
1514:
1512:
1509:
1507:
1504:
1502:
1499:
1498:
1497:
1494:
1492:
1489:
1485:
1482:
1481:
1480:
1477:
1475:
1472:
1470:
1467:
1466:
1464:
1461:
1457:
1453:
1450:
1447:
1439:
1435:
1429:
1424:
1420:
1416:
1409:
1404:
1402:
1397:
1395:
1390:
1389:
1386:
1379:
1375:
1372:
1370:
1366:
1365:
1361:
1354:
1350:
1346:
1342:
1337:
1332:
1328:
1324:
1320:
1316:
1312:
1307:
1306:
1301:
1293:
1289:
1284:
1279:
1275:
1271:
1267:
1263:
1259:
1252:
1250:
1248:
1244:
1239:
1235:
1230:
1225:
1221:
1217:
1213:
1209:
1205:
1198:
1195:
1190:
1186:
1181:
1176:
1172:
1168:
1164:
1160:
1156:
1149:
1146:
1141:
1137:
1132:
1127:
1123:
1119:
1115:
1111:
1107:
1100:
1097:
1092:
1088:
1083:
1078:
1073:
1068:
1064:
1060:
1056:
1049:
1046:
1041:
1037:
1032:
1027:
1023:
1019:
1016:(9): FSO338.
1015:
1011:
1007:
1000:
998:
996:
994:
990:
985:
981:
977:
973:
969:
965:
958:
955:
950:
946:
941:
936:
931:
926:
922:
918:
917:Retrovirology
914:
907:
905:
903:
901:
897:
892:
888:
883:
878:
874:
870:
866:
862:
858:
851:
849:
847:
845:
843:
841:
837:
832:
828:
823:
818:
813:
808:
804:
800:
796:
789:
786:
781:
779:9781437719277
775:
771:
767:
763:
756:
753:
747:
743:
740:
738:
735:
734:
730:
728:
726:
722:
718:
714:
710:
709:phase 2 study
702:
700:
697:
692:
685:
683:
680:
676:
672:
670:
666:
663:
662:
655:
650:
643:
641:
639:
635:
631:
623:
620:
619:
618:
615:
612:
607:
605:
597:
595:
593:
589:
587:
583:
579:
575:
567:
563:
559:
556:
553:
549:
548:
547:
541:
539:
537:
533:
532:
527:
526:macromolecule
523:
518:
516:
512:
508:
504:
500:
496:
492:
488:
484:
480:
476:
472:
468:
456:
453:
451:
447:
444:
440:
436:
433:
431:
427:
423:
419:
416:
413:
409:
404:
400:
397:
393:
389:
386:
384:
380:
377:
374:
372:
368:
365:
362:
360:
356:
352:
348:
343:
336:
331:
326:
315:
312:
310:
306:
303:
299:
295:
292:
290:
286:
282:
278:
275:
272:
268:
263:
259:
256:
252:
248:
245:
243:
239:
236:
233:
231:
227:
224:
221:
218:
214:
211:
208:
206:
202:
198:
194:
189:
182:
177:
172:
161:
158:
156:
152:
149:
145:
141:
138:
136:
132:
128:
124:
121:
118:
114:
109:
105:
102:
98:
94:
91:
89:
85:
82:
79:
77:
73:
70:
67:
65:
61:
57:
53:
48:
41:
36:
31:
19:
18:HIV integrase
3915:
3904:
3880:
3863:
3856:
3842:
3831:
3778:
3757:
3716:
3695:
3660:glycoprotein
3647:
3612:glycoprotein
3599:
3564:glycoprotein
3551:
3516:glycoprotein
3503:
3469:
3437:glycoprotein
3417:
3403:
3382:
3276:
3267:Nucleocapsid
3242:
3231:
3222:Pisuviricota
3161:
3137:
3126:
3067:
3058:Pisuviricota
2993:
2918:
2887:
2813:
2773:
2735:Monodnaviria
2697:
2663:
2632:
2606:
2598:Epstein–Barr
2552:
2544:Adenoviridae
2518:
2469:
2429:
2411:Varidnaviria
2298:
2276:
2251:
2139:
1872:
1837:
1595:Class I PI 3
1560:Pantothenate
1431:2.7.1-2.7.4:
1415:Transferases
1321:(1): 20–34.
1318:
1314:
1265:
1261:
1211:
1207:
1197:
1162:
1158:
1148:
1113:
1109:
1099:
1062:
1058:
1048:
1013:
1009:
967:
964:Biochemistry
963:
957:
920:
916:
864:
860:
802:
798:
788:
761:
755:
725:elvitegravir
706:
693:
689:
681:
677:
673:
664:
659:
658:
627:
616:
608:
601:
590:
571:
545:
529:
519:
503:recombinases
470:
466:
465:
58:Integrase_Zn
4000:oncoprotein
3112:Hepatitis C
2877:Hepatitis B
2863:Agnoprotein
2826:oncoprotein
2800:VP2 and VP3
2146:Transposase
2136:Recombinase
1781:-nucleoside
1607:Sphingosine
721:Raltegravir
345:Identifiers
191:Identifiers
50:Identifiers
4034:Categories
3425:M2 protein
3411:M1 protein
3217:SARS-CoV-2
1948:Telomerase
1791:Polymerase
1565:Mevalonate
1528:Riboflavin
1419:phosphorus
1374:Integrases
923:(1): 114.
748:References
669:N-terminus
562:C-terminal
552:N-terminal
509:, such as
487:integrates
479:retrovirus
418:structures
277:structures
123:structures
3967:Multiple
3864:Integrase
3102:Riboviria
2979:Rotavirus
2969:Riboviria
2730:Riboviria
2140:Integrase
2064:3' to 5'
1709:Guanylate
1704:Uridylate
1694:Adenylate
1538:Thymidine
1533:Shikimate
1353:235787691
1165:(1): 13.
665:integrase
542:Structure
524:of an IN
481:(such as
376:IPR001037
235:IPR001584
81:IPR003308
4040:Virology
3260:Membrane
3255:Envelope
2849:SV40 Tag
2510:Vaccinia
2074:RNase PH
1682:acceptor
1663:Creatine
1656:acceptor
1628:acceptor
1570:Pyruvate
1555:Glycerol
1516:Platelet
1491:Galacto-
1462:acceptor
1345:34244677
1292:34567166
1238:26829624
1189:33436564
1140:22762018
1091:19609359
1040:30416746
949:19091057
891:32624197
831:22016749
742:Integron
731:See also
634:covalent
582:tetramer
566:SH3 fold
531:intasome
491:covalent
473:) is an
435:RCSB PDB
371:InterPro
353:IN_DBD_C
294:RCSB PDB
230:InterPro
140:RCSB PDB
76:InterPro
4045:Enzymes
3591:Measles
3044:Rhinov.
2491:ICP34.5
2025:PrimPol
2010:Primase
1484:Hepatic
1479:Fructo-
1336:8671357
1283:8457747
1229:4731194
1180:7804106
1131:3385939
1082:2705190
1031:6222271
984:7632683
940:2615046
882:7429322
822:3192317
805:: 210.
717:MK-0518
511:λ phage
489:(forms
485:) that
364:PF00552
210:PF00665
69:PF02022
3770:capsid
3708:capsid
3456:HA-tag
3395:capsid
3358:ss-RNA
3288:ORF1ab
3096:ss-RNA
3056:etc. (
2965:ds-RNA
2786:capsid
2654:EBNA-3
2649:EBNA-2
2644:EBNA-1
2442:capsid
2312:2.7.13
2290:2.7.12
2265:2.7.11
2240:2.7.10
2079:PNPase
2047:PNPase
2003:POLRMT
1998:ssRNAP
1511:Muscle
1474:Gluco-
1438:kinase
1380:(MeSH)
1351:
1343:
1333:
1290:
1280:
1236:
1226:
1187:
1177:
1138:
1128:
1089:
1079:
1038:
1028:
982:
947:
937:
889:
879:
829:
819:
776:
644:In HIV
576:, and
475:enzyme
450:PDBsum
424:
414:
396:SUPFAM
350:Symbol
309:PDBsum
283:
273:
255:SUPFAM
223:CL0219
196:Symbol
155:PDBsum
129:
119:
101:SUPFAM
55:Symbol
3906:VRAPs
3888:gp120
3543:Mumps
3341:ORF9b
3331:ORF7b
3326:ORF7a
3316:ORF3d
3311:ORF3c
3306:ORF3b
3301:ORF3a
3295:(NS5)
3250:Spike
3052:Hep A
3048:Polio
2908:HBeAg
2903:HBcAg
2898:HBsAg
2762:HaPyV
2758:MCPyV
2726:other
2665:ncRNA
2623:LMP-2
2618:LMP-1
2496:ICP47
2481:vmw65
2381:early
2160:2.7.8
2129:Other
1768:2.7.7
1728:2.7.6
1673:2.7.4
1650:2.7.3
1622:2.7.2
1506:Liver
1469:Hexo-
1456:2.7.1
1349:S2CID
586:LEDGF
392:SCOPe
383:SCOP2
251:SCOPe
242:SCOP2
97:SCOPe
88:SCOP2
3893:gp41
3833:VSPs
3759:VSPs
3729:VP24
3724:VP40
3697:VSPs
3649:VSPs
3601:VSPs
3553:VSPs
3505:VSPs
3471:VNPs
3384:VSPs
3336:ORF8
3321:ORF6
3278:VNPs
3233:VSPs
3203:NS5B
3198:NS5A
3193:NS4B
3188:NS4A
3163:VNPs
3128:VSPs
3069:VNPs
3030:NSP6
3025:NSP5
3020:NSP4
3015:NSP3
3010:NSP2
3005:NSP1
2995:VNPs
2920:VNPs
2889:VSPs
2845:LTag
2840:MTag
2835:STag
2815:VNPs
2775:VSPs
2754:MPyV
2750:SV40
2699:VNPs
2675:EBER
2634:VNPs
2608:VSPs
2554:VNPs
2530:B13R
2520:VNPs
2486:ICP8
2471:VNPs
2431:VSPs
2385:late
2383:and
2299:see
2277:see
2252:see
1626:COOH
1425:2.7)
1341:PMID
1288:PMID
1234:PMID
1185:PMID
1136:PMID
1087:PMID
1036:PMID
980:PMID
945:PMID
887:PMID
827:PMID
774:ISBN
520:The
495:cell
443:PDBj
439:PDBe
422:ECOD
412:Pfam
388:1ihw
359:Pfam
302:PDBj
298:PDBe
281:ECOD
271:Pfam
247:2itg
219:clan
217:Pfam
205:Pfam
148:PDBj
144:PDBe
127:ECOD
117:Pfam
93:1wjb
64:Pfam
3954:Vpx
3952:or
3950:Vpu
3945:Vif
3940:Nef
3933:Vpr
3928:Rev
3923:Tat
3882:env
3858:pol
3850:p24
3844:gag
3639:RSV
3481:NS1
3183:NS3
3178:NS2
3079:VPg
2957:RNA
2930:HBx
2795:VP1
2569:E1B
2564:E1A
2394:DNA
2042:PAP
1983:III
1917:/Y
1908:TDT
1889:/X
1881:III
1873:Pfu
1848:/B
1838:Taq
1813:/A
1580:PFP
1550:NAD
1331:PMC
1323:doi
1278:PMC
1270:doi
1224:PMC
1216:doi
1212:126
1175:PMC
1167:doi
1126:PMC
1118:doi
1077:PMC
1067:doi
1026:PMC
1018:doi
972:doi
935:PMC
925:doi
877:PMC
869:doi
817:PMC
807:doi
766:doi
661:HIV
611:HIV
574:SIV
550:an
483:HIV
430:PDB
289:PDB
199:rve
135:PDB
4036::
3814:RT
3173:P7
3150:E2
3145:E1
3050:,
3046:,
2760:,
2756:,
2752:,
2267::
2242::
2227:PO
1988:IV
1978:II
1887:IV
1883:/C
1846:II
1832:T7
1777:PO
1770::
1730::
1677:PO
1675::
1652::
1624::
1460:OH
1458::
1443:PO
1423:EC
1417::
1347:.
1339:.
1329:.
1319:20
1317:.
1313:.
1286:.
1276:.
1266:20
1264:.
1260:.
1246:^
1232:.
1222:.
1210:.
1206:.
1183:.
1173:.
1161:.
1157:.
1134:.
1124:.
1112:.
1108:.
1085:.
1075:.
1061:.
1057:.
1034:.
1024:.
1012:.
1008:.
992:^
978:.
968:34
966:.
943:.
933:.
919:.
915:.
899:^
885:.
875:.
865:41
863:.
859:.
839:^
825:.
815:.
801:.
797:.
772:.
715:,
640:.
606:.
568:).
560:a
538:.
517:.
505:)
471:IN
441:;
437:;
420:/
394:/
390:/
300:;
296:;
279:/
253:/
249:/
146:;
142:;
125:/
99:/
95:/
4002::
3908::
3835::
3772::
3761::
3710::
3699::
3662::
3651::
3614::
3603::
3566::
3555::
3518::
3507::
3473::
3439::
3397::
3386::
3366:)
3362:(
3280::
3235::
3224:)
3220:(
3165::
3130::
3119:)
3115:(
3104:)
3100:(
3071::
3060:)
3054:,
2997::
2986:)
2982:(
2971:)
2967:(
2922::
2891::
2851:)
2847:(
2828::
2817::
2788::
2777::
2764:)
2748:(
2737:)
2732:,
2728:(
2701::
2667::
2636::
2610::
2556::
2522::
2473::
2444::
2433::
2413:)
2408:,
2404:(
2387:)
2379:(
2369:e
2362:t
2355:v
2229:4
2225:(
2142:)
2138:(
2020:2
2015:1
1993:V
1931:κ
1926:ι
1921:η
1915:V
1903:μ
1898:λ
1893:β
1867:ζ
1862:ε
1857:δ
1852:α
1827:ν
1822:θ
1817:γ
1811:I
1783:)
1779:4
1775:(
1746:)
1743:7
1741:O
1739:2
1737:P
1735:(
1679:4
1654:N
1521:2
1501:1
1448:)
1445:4
1441:(
1436:/
1407:e
1400:t
1393:v
1355:.
1325::
1294:.
1272::
1240:.
1218::
1191:.
1169::
1163:6
1142:.
1120::
1114:2
1093:.
1069::
1063:5
1042:.
1020::
1014:4
986:.
974::
951:.
927::
921:5
893:.
871::
833:.
809::
803:2
782:.
768::
501:(
469:(
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.