493:
glycines and threonine coordinate with the K ion, while the side-chains of valine and tyrosine are directed into the protein core to impose geometric constraint on the filter. As a result, the KcsA tetramer harbors four equal spaced K binding sites, with each side composed of a cage formed by eight oxygen atoms that sit on the vertices of a cube. The oxygen atoms that surround K ions in the filter are arranged like the water molecules that encircle hydrated K ions in the cavity of the channel; this suggests that oxygen coordination and binding sites in the selectivity filter are paying for the energetic cost of K dehydration. Because the Na+ ion is too small for these K-sized binding sites, dehydration energy is not compensated and thus, the filter selects against other extraneous ions. Additionally, the KcsA channel is blocked by
514:
residues in the selectivity filter. There is evidence to suggest that the main pH sensor is in the cytoplasmic domain. Exchanging negatively charged amino acids for neutral ones made the KcsA channel insensitive to pH even though there were no amino-acid changes at the transmembrane region. In addition, between the pH of 6 and 7, histidine is one of the few titratable side chains of histidines; they are absent in the transmembrane and extracellular segments of TM2 but present at KcsA's C-terminus. This highlights a possible mechanism for the slow opening of KcsA which is particularly pH sensitive, especially as the conformational propagation of channel opening signal from the C-terminus to the selectivity filter could be important in coordinating the structural changes needed for conductance along the entire pore.
527:
initially stabilizes the selectivity filter. The collapse of the filter region prevents entry into or facilitate exit from the inactivated state. Glu71, a key part of the selectivity filter signature sequence that is conserved among K ion channels, plays a pivotal role in gating as its ability to reorient itself in the direction of the transmembrane voltage field is able to provide an explanation for voltage gating events in KcsA. The orientation of amino acids in the filter region might play significant physiological role in modulating potassium fluxes in eukaryotes and prokaryotes under steady-state conditions.
424:
588:. However, the dynamic behavior of KcsA makes analysis of the channel difficult as a crystal structure inevitably provides a static, spatially and temporally averaged image of a channel. To bridge the gap between molecular structure and physiological behavior, an understanding of the atomic resolution dynamics of potassium channels is required.
215:
structural and mechanistic insight on the molecular basis for K ion selection and conduction. As one of the most studied ion channels to this day, KcsA is a template for research on K channel function and its elucidated structure underlies computational modeling of channel dynamics for both prokaryotic and eukaryotic species.
313:
29:
475:
junction to block the passage of any potassium ions. At pH 4 however, KcsA undergoes millisecond-timescale conformational exchanges filter permeating and nonpermeating states and between the open and closed conformations of the M2 helices. While these distinct conformational changes occur in separate
299:
while also gating electrical conductance. In 2011, the crystal structure of full length KcsA was resolved to reveal that hindrance by the previously truncated residues permits only straightforward expansion of the intercellular ion passage region of the protein. This research provides a more detailed
269:
cytoplasmic domain of the native protein (residues 126–158) increases the stability of crystallized samples. A model of KcsA at the 3.2A resolution was produced that confirmed the tetrameric arrangement of the protein around a center pore, with one helix of each subunit facing the inside axis and the
513:
The pH-dependent conductance of KcsA indicates that the opening of the ion channel occurs when the protein is exposed to a more acidic environment. NMR studies performed by the Riek group show that pH sensitivity occurs in both the C-terminal TM2 region of the protein as well as with Tyr78 and Gly79
517:
NMR studies also suggest that a complex hydrogen bond network between Tyr78, Gly79, Glu71 and Asp80 exists in the KcsA filter region, and further acts as a pH-sensitive trigger for conductance. The mutation of key residues in the region, including E71A, results in a large energy cost of 4 kcal mol,
376:
begins with a gate region formed by M2 helices at 18 Å in diameter, and then opens into a wide cavity (~10 Å across) near the middle of the membrane. In these regions, K ions are in contact with surrounding water molecules but when they enter the channel from the selectivity filter at the top, the
214:
and a highly selective pore region, responsible for the gating and shuttling of K ions out of the cell. The amino acid sequence found in the selectivity filter of KcsA is highly conserved among both prokaryotic and eukaryotic K voltage channels; as a result, research on KcsA has provided important
629:
of new medications. In addition, homology models based on the closed state KcsA crystal structure have been generated computationally to construct a multiple state representation of the hERG cardiac K channel. Such models reveal the flexibility of the hERG channel and can consistently predict the
492:
The sequence TVGYG is especially important for maintaining the potassium specificity of KcsA. The glycines in this selectivity filter sequence have dihedral angles that allow carbonyl oxygen atoms in the protein backbone of the filter to point in one direction, toward the ions along the pore. The
459:
helices at the end of the pore. At low pH, the M2 helix is protonated, shifting the ion channel from closed to open conformation. As ions flow through the channel, voltage gating mechanisms are thought to induce interactions between Glu71 and Asp80 in the selectivity filter, which destabilize the
409:
sequence, (Threonine, Valine, Glycine, Tyrosine, Glycine), characteristic of potassium channels. Within this region, coordination between the TVGYG amino acids and incoming K ions allows for conduction of ions through the channel. The selectivity filter of KcsA contains four ion binding sites,
526:
In 2006, the Perozo group proposed a mechanistic explanation for the effects of voltage fields on KcsA gating. After adding a depolarizing current to the channel, the reorientation of Glu71 towards the intracellular pore occurs, thereby disrupting the Glu71-Asp80 carboxyl-carboxylate pair that
316:
The crystal structure of KcsA. Only two of the four subunits are shown here. The protein is shown in green, backbone carbonyl groups (oxygen = red, carbon = green) and potassium ions (occupying the S2 and S4 sites) and oxygen atoms of water molecules (S1 and S3) are purple and red spheres
274:
fragments were attached to KcsA crystals to further stabilize the channel. In the early 2000s, evidence for the occupation of the selectivity filter by two K atom during the transport process emerged, based on energy and electrostatic calculations made to model the pore region. Continued
231:
and his colleagues in 1998. In the years leading up to this, research on the structure of K channels was centered on the use of small toxin binding to reveal the location of the pore and selectivity filter among channel residues. MacKinnon's group theorized the
630:
binding affinity of a set of diverse ion channel-interacting ligands. Analysis of the complex ligand-hERG structures can be used to guide the synthesis of drug analogs with reduced hERG liability, based on drug structure and docking potential.
33:
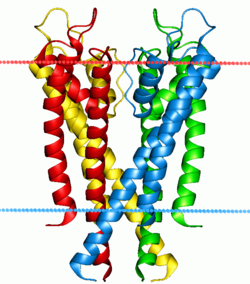
The four subunits forming the channel are drawn in different colors. They surround a central pore, guarded by the selectivity filter made up of the P-loops from each of the subunits. The blue and red dots indicate the boundaries of the
414:
simulations suggest the filter is flexible. The presence of TVGYG in the filter region of KcsA is conserved even in more complex eukaryotic channels, thus making KcsA an optimal system for studying K channel conductance across species.
300:
look into the motion of separate channel regions during ion conduction. In the present day, KcsA studies are focused on using the prokaryotic channel as a model for the channel dynamics of larger eukaryotic K channels, including
427:
KcsA transitions from a closed to open conformation upon protonation of the M2 helix at low pH. Voltage gating results in the collapse of the selectivity filter and subsequent inactivation. Image is adapted from
Thompson et al.
518:
equivalent to the loss of the hydrogen bond between Glu71 and Tyr78 and the water-mediated hydrogen bond between Glu71 and Asp80 in KcsA(E71A). These studies further highlight the role of pH gating in KcsA channel function.
596:
Due to the high sequence similarity between the pore of KcsA and other eukaryotic K ion channel proteins, KcsA has provided important insight into the behavior of other important voltage conducting proteins such as the
579:
has yet to be discussed: the best resolved and most applied crystal structure of KcsA appears to be that of the ‘closed' form of the channel. This is reasonable as the closed state of the channel is favored at
345:
and are slightly kinked, opening up to face the outside of the cell like a flower. These two TM helices are linked by a reentrant loop, dispersed symmetrically around a common axis corresponding to the central
290:
the ion channel from pH 7 to pH 4, corresponds to conformational changes in two regions: transition to the ion-exchanging state of the selectivity filter, and the opening of the arrangement of TM2 at the
896:
Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R (Apr 1998). "The structure of the potassium channel: molecular basis of K conduction and selectivity".
262:
ion channels, also provided a method to understand the mechanisms of K channels conduction at a more rudimentary level, thereby providing even great impetus for the study of KcsA.
544:
and multiple models are used to describe different aspects of the selectivity. Models explaining selectivity based on field strength concept developed by George
Eisenman based on
350:. The pore region spans approximately 30 amino acid residues and can be divided into three parts: a selectivity filter near the extracellular side, a dilated water-filled
1393:
Cordero-Morales JF, Cuello LG, Zhao Y, Jogini V, Cortes DM, Roux B, Perozo E (Apr 2006). "Molecular determinants of gating at the potassium-channel selectivity filter".
337:
composed of four identical, single-domain subunits (each with two α-helices) arranged so that one M2 helix faces the central pore, while the other M1 helix faces the
157:
939:
Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R (Nov 2001). "Chemistry of ion coordination and hydration revealed by a K channel-Fab complex at 2.0 A resolution".
548:
have been applied to KcsA. An alternative explanation for the selectivity of KcsA is based on the close-fit model (also known as the snug-fit model) developed by
113:
101:
354:
at the center, and a closed gate near the cytoplasmic side formed by four packed M2 helices. This architecture is found to be highly conserved in the potassium
1591:
Noskov SY, Bernèche S, Roux B (Oct 2004). "Control of ion selectivity in potassium channels by electrostatic and dynamic properties of carbonyl ligands".
572:
ion. Further work has studied thermodynamic differences in ion binding, topological considerations, and the number of continuous ion binding sites.
560:
carbonyl oxygen atoms that make up the selectivity filter are held at a precise position that allows them to substitute for water molecules in the
484:. The dynamics of this exchange stereochemical configurations in the filter provides the physical basis for simultaneous K conductance and gating.
368:
The overall length of the pore is 45 Å, and its diameter varies considerably within the distinct regions of the inner tunnel. Travelling from the
783:
Yu FH, Yarov-Yarovoy V, Gutman GA, Catterall WA (Dec 2005). "Overview of molecular relationships in the voltage-gated ion channel superfamily".
410:
although it is proposed that only two of these four positions are occupied at one time. The selectivity filter is about 3 Å in diameter. though
276:
240:
segments, and even suggested presence of pore-forming “loops” in the filter region made of short segments of amino acids that interacted with
81:
467:
In the nonconducting conformation of KcsA at pH 7, K is bound tightly to coordinating oxygens of the selectivity filter and the four TM2
1968:
Rajamani R, Tounge BA, Li J, Reynolds CH (Mar 2005). "A two-state homology model of the hERG K channel: application to ligand binding".
283:
have since provided even more insight into channel structure and the forces gating the switch from channel inactivation to conduction.
254:
protein, attracted the attention of the scientific community especially as the K channel signature sequence began to appear in other
280:
460:
conductive conformation and facilitate entry into a long-lived nonconducting state that resembles the C-type–inactivation of
177:
1933:
Sanguinetti MC, Mitcheson JS (Mar 2005). "Predicting drug-hERG channel interactions that cause acquired long QT syndrome".
381:
composition of the pore-lining residues within KcsA, the side chains lining the internal pore and cavity are predominantly
275:
investigation of the various opened and closed, inactive and active conformations of KcsA by other imaging methods such as
869:
2013:
498:
614:
461:
270:
other facing outwards. Three years later, a higher resolution model was produced by Morais-Cabral and Zhou after
436:
because the KcsA structure provides a framework for understanding K channel conduction, which has three parts:
165:
377:
cavity is so narrow that K ions must shed any hydrating waters in order to enter the cell. In regards to the
598:
265:
The crystal structure of KcsA was solved by the MacKinnon group in 1998 after discovery that removal of the
1644:"Negative conductance caused by entry of sodium and cesium ions into the potassium channels of squid axons"
675:"A prokaryotic potassium ion channel with two predicted transmembrane segments from Streptomyces lividans"
2008:
585:
481:
456:
237:
224:
211:
161:
1876:"Tuning the ion selectivity of tetrameric cation channels by changing the number of ion binding sites"
1887:
1828:
1771:
1714:
1600:
1547:
1449:
1342:
1114:
1059:
948:
905:
739:
673:
Schrempf H, Schmidt O, Kümmerlen R, Hinnah S, Müller D, Betzler M, Steinkamp T, Wagner R (Nov 1995).
382:
94:
549:
440:
271:
1817:"Selectivity in K channels is due to topological control of the permeant ion's coordinated state"
1704:
1624:
1418:
1224:
1181:
1138:
972:
808:
765:
411:
355:
228:
621:
and various drug compounds. Such tests can screen for drug-hERG channel interactions that cause
1105:
Lu Z, Klem AM, Ramu Y (Oct 2001). "Ion conduction pore is conserved among potassium channels".
1985:
1950:
1915:
1856:
1797:
1740:
1673:
1616:
1573:
1516:
1475:
1410:
1370:
1311:
1273:
1216:
1173:
1130:
1087:
1028:
964:
921:
843:
800:
757:
704:
644:
541:
406:
402:
351:
258:
genes. The simplicity of the two transmembrane helices in KcsA, as opposed to the six in many
245:
193:
152:
839:
1977:
1942:
1905:
1895:
1846:
1836:
1787:
1779:
1730:
1722:
1663:
1655:
1608:
1563:
1555:
1506:
1465:
1457:
1402:
1360:
1350:
1303:
1263:
1255:
1208:
1165:
1122:
1077:
1067:
1018:
1010:
956:
913:
835:
792:
747:
694:
686:
622:
545:
373:
347:
144:
639:
561:
322:
259:
1891:
1832:
1775:
1718:
1604:
1551:
1453:
1346:
1118:
1063:
952:
909:
743:
1910:
1875:
1851:
1816:
1792:
1759:
1735:
1692:
1668:
1643:
1568:
1535:
1470:
1437:
1365:
1330:
1268:
1243:
1082:
1047:
1023:
998:
690:
649:
557:
553:
477:
386:
1559:
752:
723:
699:
674:
2002:
1511:
1494:
1199:
Hille B, Armstrong CM, MacKinnon R (Oct 1999). "Ion channels: from idea to reality".
976:
565:
398:
369:
342:
338:
330:
296:
241:
106:
86:
35:
1422:
870:"Nobel Lecture: Potassium Channels and the Atomic Basis of Selective Ion Conduction"
423:
62:
1628:
1228:
1142:
812:
769:
197:
140:
1185:
443:, channel gating by pH sensitivity, and voltage-gated channel inactivation. K ion
118:
1244:"Potassium channels in myelinated nerve. Selective permeability to small cations"
999:"Conformational dynamics of the KcsA potassium channel governs gating properties"
74:
1783:
1726:
917:
626:
603:
476:
regions of the channel, the molecular behavior of each region is linked by both
468:
452:
433:
363:
255:
250:
203:
1981:
1946:
1880:
Proceedings of the
National Academy of Sciences of the United States of America
1821:
Proceedings of the
National Academy of Sciences of the United States of America
1335:
Proceedings of the
National Academy of Sciences of the United States of America
1052:
Proceedings of the
National Academy of Sciences of the United States of America
1760:"The predominant role of coordination number in potassium channel selectivity"
1461:
1307:
576:
569:
444:
378:
359:
292:
266:
1900:
1841:
1355:
1072:
1046:
Uysal S, Cuello LG, Cortes DM, Koide S, Kossiakoff AA, Perozo E (Jul 2011).
502:
472:
437:
287:
1989:
1954:
1919:
1860:
1801:
1744:
1620:
1577:
1520:
1479:
1414:
1374:
1315:
1220:
1177:
1134:
1091:
1032:
968:
847:
804:
761:
325:, with a central pore running down the center made up of two transmembrane
1677:
1495:"GCN4 enhances the stability of the pore domain of potassium channel KcsA"
1277:
925:
708:
90:
1659:
1259:
334:
233:
69:
1709:
1693:"Tuning ion coordination architectures to enable selective partitioning"
1612:
1294:
Noskov SY, Roux B (Dec 2006). "Ion selectivity in potassium channels".
796:
540:
The precise mechanism of potassium channel selectivity continues to be
494:
326:
286:
In 2007, Riek et al. showed that the channel opening that results from
1406:
1126:
1014:
960:
295:. This model explains the ability of KcsA to simultaneous select for
172:
1438:"Role of the KcsA channel cytoplasmic domain in pH-dependent gating"
1169:
997:
Baker KA, Tzitzilonis C, Kwiatkowski W, Choe S, Riek R (Nov 2007).
223:
KcsA was the first potassium ion channel to be characterized using
1212:
1048:"Mechanism of activation gating in the full-length KcsA K channel"
826:
Roux B (2005). "Ion conduction and selectivity in K(+) channels".
311:
447:
occurs at the upper selectivity filter region of the pore, while
1874:
Derebe MG, Sauer DB, Zeng W, Alam A, Shi N, Jiang Y (Jan 2011).
618:
609:
301:
248:
between KcsA and other channels in the Kv family, including the
134:
57:
1536:"Cation selective glass electrodes and their mode of operation"
575:
In addition, a major limitation of crystal structure study and
341:. The inner helices are tilted by about 25° in relation to the
312:
28:
1331:"Molecular mechanism of pH sensing in KcsA potassium channels"
863:
861:
859:
857:
244:
passing through the channel The discovery of strong sequence
389:
amino acids are present that contact the dehydrated K ions.
329:(the outer-helix M1 and the inner-helix M2), which span the
581:
448:
207:
1329:
Thompson AN, Posson DJ, Parsa PV, Nimigean CM (May 2008).
1388:
1386:
1384:
724:"Exploring the open pore of the potassium channel from
828:
Annual Review of
Biophysics and Biomolecular Structure
401:
mouth of the channel made up of pore helices, plus a
1156:Choe S (Feb 2002). "Potassium channel structures".
372:region outwards (bottom to top in the picture) the
171:
151:
133:
128:
112:
100:
80:
68:
56:
48:
43:
21:
1436:Hirano M, Onishi Y, Yanagida T, Ide T (Nov 2011).
722:Meuser D, Splitt H, Wagner R, Schrempf H (1999).
584:, at which the crystal structure was solved by
192:(K channel of streptomyces A) is a prokaryotic
1289:
1287:
891:
889:
887:
885:
883:
668:
666:
664:
397:The wider end of the cone corresponds to the
321:The structure of KcsA is that of an inverted
8:
1970:Bioorganic & Medicinal Chemistry Letters
1758:Thomas M, Jayatilaka D, Corry B (Oct 2007).
992:
990:
988:
986:
625:, are essential for determining the cardiac
617:studies to model the interactions between
125:
1909:
1899:
1850:
1840:
1791:
1734:
1708:
1667:
1567:
1510:
1469:
1395:Nature Structural & Molecular Biology
1364:
1354:
1267:
1081:
1071:
1022:
1003:Nature Structural & Molecular Biology
751:
698:
613:potassium channel. KcsA has been used in
840:10.1146/annurev.biophys.34.040204.144655
422:
660:
432:The KcsA channel is considered a model
1642:Bezanilla F, Armstrong CM (Nov 1972).
18:
1493:Yuchi Z, Pau VP, Yang DS (Dec 2008).
202:that has been studied extensively in
7:
385:, but within the selectivity filter
1935:Trends in Pharmacological Sciences
1815:Bostick DL, Brooks CL (May 2007).
691:10.1002/j.1460-2075.1995.tb00201.x
14:
1648:The Journal of General Physiology
1248:The Journal of General Physiology
568:ion, but they are too far from a
16:Prokaryotic potassium ion channel
1512:10.1111/j.1742-4658.2008.06747.x
210:activated protein possesses two
27:
1691:Varma S, Rempe SB (Aug 2007).
1:
1560:10.1016/S0006-3495(62)86959-8
753:10.1016/S0014-5793(99)01579-3
129:Available protein structures:
1158:Nature Reviews. Neuroscience
1784:10.1529/biophysj.107.108167
1727:10.1529/biophysj.107.107482
918:10.1126/science.280.5360.69
2030:
1982:10.1016/j.bmcl.2005.01.008
1947:10.1016/j.tips.2005.01.003
478:electrostatic interactions
462:voltage-dependent channels
333:. The channel itself is a
1462:10.1016/j.bpj.2011.09.024
1308:10.1016/j.bpc.2006.05.033
623:acquired long QT syndrome
501:requires the presence of
124:
26:
196:from the soil bacterium
1901:10.1073/pnas.1013636108
1842:10.1073/pnas.0700554104
1534:Eisenman G (Mar 1962).
1356:10.1073/pnas.0800873105
1073:10.1073/pnas.1105112108
785:Pharmacological Reviews
429:
318:
212:transmembrane segments
22:KcsA potassium channel
1296:Biophysical Chemistry
726:Streptomyces lividans
586:X-ray crystallography
426:
315:
225:x-ray crystallography
199:Streptomyces lividans
1660:10.1085/jgp.60.5.588
1260:10.1085/jgp.61.6.669
1242:Hille B (Jun 1973).
868:Roderick MacKinnon.
405:that is formed by a
1892:2011PNAS..108..598D
1833:2007PNAS..104.9260B
1776:2007BpJ....93.2635T
1764:Biophysical Journal
1719:2007BpJ....93.1093V
1697:Biophysical Journal
1613:10.1038/nature02943
1605:2004Natur.431..830N
1552:1962BpJ.....2..259E
1546:(2 Pt 2): 259–323.
1540:Biophysical Journal
1454:2011BpJ...101.2157H
1442:Biophysical Journal
1347:2008PNAS..105.6900T
1119:2001Natur.413..809L
1064:2011PNAS..10811896U
953:2001Natur.414...43Z
910:1998Sci...280...69D
744:1999FEBSL.462..447M
550:Francisco Bezanilla
542:studied and debated
236:arrangement of the
2014:Bacterial proteins
797:10.1124/pr.57.4.13
471:converge near the
430:
412:molecular dynamics
403:selectivity filter
393:Selectivity filter
319:
229:Roderick MacKinnon
876:. Nobel Media AB.
645:Potassium channel
194:potassium channel
187:
186:
183:
182:
178:structure summary
2021:
1994:
1993:
1965:
1959:
1958:
1930:
1924:
1923:
1913:
1903:
1871:
1865:
1864:
1854:
1844:
1812:
1806:
1805:
1795:
1755:
1749:
1748:
1738:
1712:
1688:
1682:
1681:
1671:
1639:
1633:
1632:
1588:
1582:
1581:
1571:
1531:
1525:
1524:
1514:
1499:The FEBS Journal
1490:
1484:
1483:
1473:
1433:
1427:
1426:
1407:10.1038/nsmb1069
1390:
1379:
1378:
1368:
1358:
1326:
1320:
1319:
1291:
1282:
1281:
1271:
1239:
1233:
1232:
1196:
1190:
1189:
1153:
1147:
1146:
1127:10.1038/35101535
1113:(6858): 809–13.
1102:
1096:
1095:
1085:
1075:
1043:
1037:
1036:
1026:
1015:10.1038/nsmb1311
994:
981:
980:
961:10.1038/35102009
936:
930:
929:
893:
878:
877:
865:
852:
851:
823:
817:
816:
780:
774:
773:
755:
719:
713:
712:
702:
679:The EMBO Journal
670:
126:
31:
19:
2029:
2028:
2024:
2023:
2022:
2020:
2019:
2018:
1999:
1998:
1997:
1967:
1966:
1962:
1932:
1931:
1927:
1873:
1872:
1868:
1814:
1813:
1809:
1757:
1756:
1752:
1710:physics/0608180
1690:
1689:
1685:
1641:
1640:
1636:
1599:(7010): 830–4.
1590:
1589:
1585:
1533:
1532:
1528:
1505:(24): 6228–36.
1492:
1491:
1487:
1435:
1434:
1430:
1392:
1391:
1382:
1328:
1327:
1323:
1293:
1292:
1285:
1241:
1240:
1236:
1201:Nature Medicine
1198:
1197:
1193:
1155:
1154:
1150:
1104:
1103:
1099:
1058:(29): 11896–9.
1045:
1044:
1040:
1009:(11): 1089–95.
996:
995:
984:
938:
937:
933:
904:(5360): 69–77.
895:
894:
881:
867:
866:
855:
825:
824:
820:
782:
781:
777:
721:
720:
716:
672:
671:
662:
658:
640:Calcium channel
636:
594:
538:
533:
524:
511:
490:
451:rises from the
421:
395:
310:
221:
102:OPM superfamily
39:
17:
12:
11:
5:
2027:
2025:
2017:
2016:
2011:
2001:
2000:
1996:
1995:
1976:(6): 1737–41.
1960:
1925:
1886:(2): 598–602.
1866:
1827:(22): 9260–5.
1807:
1770:(8): 2635–43.
1750:
1683:
1654:(5): 588–608.
1634:
1583:
1526:
1485:
1448:(9): 2157–62.
1428:
1380:
1341:(19): 6900–5.
1321:
1283:
1234:
1207:(10): 1105–9.
1191:
1170:10.1038/nrn727
1148:
1097:
1038:
982:
947:(6859): 43–8.
931:
879:
874:Nobelprize.org
853:
818:
775:
738:(3): 447–452.
714:
685:(21): 5170–8.
659:
657:
654:
653:
652:
650:Sodium channel
647:
642:
635:
632:
607:and the human
593:
590:
562:hydrated shell
537:
534:
532:
529:
523:
522:Voltage Gating
520:
510:
509:pH Sensitivity
507:
489:
486:
420:
417:
394:
391:
356:channel family
343:lipid membrane
339:lipid membrane
309:
306:
272:monoclonal Fab
220:
217:
206:research. The
185:
184:
181:
180:
175:
169:
168:
155:
149:
148:
138:
131:
130:
122:
121:
116:
110:
109:
104:
98:
97:
84:
78:
77:
72:
66:
65:
60:
54:
53:
50:
46:
45:
41:
40:
32:
24:
23:
15:
13:
10:
9:
6:
4:
3:
2:
2026:
2015:
2012:
2010:
2007:
2006:
2004:
1991:
1987:
1983:
1979:
1975:
1971:
1964:
1961:
1956:
1952:
1948:
1944:
1941:(3): 119–24.
1940:
1936:
1929:
1926:
1921:
1917:
1912:
1907:
1902:
1897:
1893:
1889:
1885:
1881:
1877:
1870:
1867:
1862:
1858:
1853:
1848:
1843:
1838:
1834:
1830:
1826:
1822:
1818:
1811:
1808:
1803:
1799:
1794:
1789:
1785:
1781:
1777:
1773:
1769:
1765:
1761:
1754:
1751:
1746:
1742:
1737:
1732:
1728:
1724:
1720:
1716:
1711:
1706:
1703:(4): 1093–9.
1702:
1698:
1694:
1687:
1684:
1679:
1675:
1670:
1665:
1661:
1657:
1653:
1649:
1645:
1638:
1635:
1630:
1626:
1622:
1618:
1614:
1610:
1606:
1602:
1598:
1594:
1587:
1584:
1579:
1575:
1570:
1565:
1561:
1557:
1553:
1549:
1545:
1541:
1537:
1530:
1527:
1522:
1518:
1513:
1508:
1504:
1500:
1496:
1489:
1486:
1481:
1477:
1472:
1467:
1463:
1459:
1455:
1451:
1447:
1443:
1439:
1432:
1429:
1424:
1420:
1416:
1412:
1408:
1404:
1400:
1396:
1389:
1387:
1385:
1381:
1376:
1372:
1367:
1362:
1357:
1352:
1348:
1344:
1340:
1336:
1332:
1325:
1322:
1317:
1313:
1309:
1305:
1302:(3): 279–91.
1301:
1297:
1290:
1288:
1284:
1279:
1275:
1270:
1265:
1261:
1257:
1254:(6): 669–86.
1253:
1249:
1245:
1238:
1235:
1230:
1226:
1222:
1218:
1214:
1213:10.1038/13415
1210:
1206:
1202:
1195:
1192:
1187:
1183:
1179:
1175:
1171:
1167:
1164:(2): 115–21.
1163:
1159:
1152:
1149:
1144:
1140:
1136:
1132:
1128:
1124:
1120:
1116:
1112:
1108:
1101:
1098:
1093:
1089:
1084:
1079:
1074:
1069:
1065:
1061:
1057:
1053:
1049:
1042:
1039:
1034:
1030:
1025:
1020:
1016:
1012:
1008:
1004:
1000:
993:
991:
989:
987:
983:
978:
974:
970:
966:
962:
958:
954:
950:
946:
942:
935:
932:
927:
923:
919:
915:
911:
907:
903:
899:
892:
890:
888:
886:
884:
880:
875:
871:
864:
862:
860:
858:
854:
849:
845:
841:
837:
833:
829:
822:
819:
814:
810:
806:
802:
798:
794:
791:(4): 387–95.
790:
786:
779:
776:
771:
767:
763:
759:
754:
749:
745:
741:
737:
733:
729:
727:
718:
715:
710:
706:
701:
696:
692:
688:
684:
680:
676:
669:
667:
665:
661:
655:
651:
648:
646:
643:
641:
638:
637:
633:
631:
628:
624:
620:
616:
612:
611:
606:
605:
600:
591:
589:
587:
583:
578:
573:
571:
567:
563:
559:
555:
551:
547:
546:Coulomb's law
543:
535:
530:
528:
521:
519:
515:
508:
506:
504:
500:
496:
487:
485:
483:
479:
474:
470:
465:
463:
458:
457:transmembrane
454:
450:
446:
442:
439:
435:
425:
418:
416:
413:
408:
404:
400:
399:extracellular
392:
390:
388:
384:
380:
375:
371:
370:intracellular
366:
365:
361:
357:
353:
349:
344:
340:
336:
332:
331:lipid bilayer
328:
324:
317:respectively.
314:
307:
305:
303:
298:
294:
289:
284:
282:
278:
273:
268:
263:
261:
257:
253:
252:
247:
243:
239:
238:transmembrane
235:
230:
226:
218:
216:
213:
209:
205:
201:
200:
195:
191:
179:
176:
174:
170:
167:
163:
159:
156:
154:
150:
146:
142:
139:
136:
132:
127:
123:
120:
117:
115:
111:
108:
105:
103:
99:
96:
92:
88:
85:
83:
79:
76:
73:
71:
67:
64:
61:
59:
55:
51:
47:
42:
37:
36:lipid bilayer
30:
25:
20:
2009:Ion channels
1973:
1969:
1963:
1938:
1934:
1928:
1883:
1879:
1869:
1824:
1820:
1810:
1767:
1763:
1753:
1700:
1696:
1686:
1651:
1647:
1637:
1596:
1592:
1586:
1543:
1539:
1529:
1502:
1498:
1488:
1445:
1441:
1431:
1401:(4): 311–8.
1398:
1394:
1338:
1334:
1324:
1299:
1295:
1251:
1247:
1237:
1204:
1200:
1194:
1161:
1157:
1151:
1110:
1106:
1100:
1055:
1051:
1041:
1006:
1002:
944:
940:
934:
901:
897:
873:
831:
827:
821:
788:
784:
778:
735:
732:FEBS Letters
731:
725:
717:
682:
678:
608:
602:
595:
592:Applications
574:
539:
525:
516:
512:
491:
488:Kselectivity
466:
431:
396:
367:
364:prokaryotes.
320:
285:
264:
249:
222:
198:
189:
188:
615:mutagenesis
599:drosophilla
577:simulations
473:cytoplasmic
453:protonation
441:selectivity
383:hydrophobic
256:prokaryotic
204:ion channel
114:OPM protein
44:Identifiers
2003:Categories
834:: 153–71.
656:References
582:neutral pH
558:main chain
445:permeation
379:amino acid
360:eukaryotes
293:C-terminus
267:C-terminus
260:eukaryotic
234:tetrameric
141:structures
977:205022645
601:-derived
566:potassium
554:Armstrong
497:ions and
482:allostery
449:pH gating
438:Potassium
308:Structure
288:titrating
75:IPR013099
1990:15745831
1955:15749156
1920:21187421
1861:17519335
1802:17573427
1745:17513348
1621:15483608
1578:13889686
1521:19016844
1480:22067153
1423:20765018
1415:16532009
1375:18443286
1316:16843584
1221:10502800
1178:11836519
1135:11677598
1092:21730186
1033:17922011
969:11689936
848:15869387
805:16382097
762:10622743
634:See also
536:Function
531:Research
419:Function
358:in both
335:tetramer
246:homology
158:RCSB PDB
70:InterPro
1911:3021048
1888:Bibcode
1852:1890482
1829:Bibcode
1793:1989715
1772:Bibcode
1736:1929028
1715:Bibcode
1678:4644327
1669:2226091
1629:4414885
1601:Bibcode
1569:1366487
1548:Bibcode
1471:3207171
1450:Bibcode
1366:2383984
1343:Bibcode
1278:4541077
1269:2203488
1229:5216271
1143:4364245
1115:Bibcode
1083:3141920
1060:Bibcode
1024:3525321
949:Bibcode
926:9525859
906:Bibcode
898:Science
813:2643413
770:6231397
740:Bibcode
709:7489706
564:of the
469:helices
434:channel
327:helices
219:History
63:PF07885
1988:
1953:
1918:
1908:
1859:
1849:
1800:
1790:
1743:
1733:
1676:
1666:
1627:
1619:
1593:Nature
1576:
1566:
1519:
1478:
1468:
1421:
1413:
1373:
1363:
1314:
1276:
1266:
1227:
1219:
1186:825973
1184:
1176:
1141:
1133:
1107:Nature
1090:
1080:
1031:
1021:
975:
967:
941:Nature
924:
846:
811:
803:
768:
760:
707:
700:394625
697:
627:safety
604:Shaker
570:sodium
556:. The
505:ions.
499:gating
352:cavity
297:K ions
251:Shaker
242:K ions
173:PDBsum
147:
137:
95:SUPFAM
49:Symbol
1705:arXiv
1625:S2CID
1419:S2CID
1225:S2CID
1182:S2CID
1139:S2CID
973:S2CID
809:S2CID
766:S2CID
428:2008.
407:TVGYG
387:polar
277:ssNMR
91:SCOPe
82:SCOP2
1986:PMID
1951:PMID
1916:PMID
1857:PMID
1798:PMID
1741:PMID
1674:PMID
1617:PMID
1574:PMID
1517:PMID
1476:PMID
1411:PMID
1371:PMID
1312:PMID
1274:PMID
1217:PMID
1174:PMID
1131:PMID
1088:PMID
1029:PMID
965:PMID
922:PMID
844:PMID
801:PMID
758:PMID
705:PMID
619:hERG
610:hERG
552:and
480:and
374:pore
362:and
348:pore
323:cone
302:hERG
279:and
190:KcsA
166:PDBj
162:PDBe
145:ECOD
135:Pfam
119:1r3j
87:1bl8
58:Pfam
1978:doi
1943:doi
1906:PMC
1896:doi
1884:108
1847:PMC
1837:doi
1825:104
1788:PMC
1780:doi
1731:PMC
1723:doi
1664:PMC
1656:doi
1609:doi
1597:431
1564:PMC
1556:doi
1507:doi
1503:275
1466:PMC
1458:doi
1446:101
1403:doi
1361:PMC
1351:doi
1339:105
1304:doi
1300:124
1264:PMC
1256:doi
1209:doi
1166:doi
1123:doi
1111:413
1078:PMC
1068:doi
1056:108
1019:PMC
1011:doi
957:doi
945:414
914:doi
902:280
836:doi
793:doi
748:doi
736:462
695:PMC
687:doi
455:of
281:EPR
227:by
153:PDB
2005::
1984:.
1974:15
1972:.
1949:.
1939:26
1937:.
1914:.
1904:.
1894:.
1882:.
1878:.
1855:.
1845:.
1835:.
1823:.
1819:.
1796:.
1786:.
1778:.
1768:93
1766:.
1762:.
1739:.
1729:.
1721:.
1713:.
1701:93
1699:.
1695:.
1672:.
1662:.
1652:60
1650:.
1646:.
1623:.
1615:.
1607:.
1595:.
1572:.
1562:.
1554:.
1542:.
1538:.
1515:.
1501:.
1497:.
1474:.
1464:.
1456:.
1444:.
1440:.
1417:.
1409:.
1399:13
1397:.
1383:^
1369:.
1359:.
1349:.
1337:.
1333:.
1310:.
1298:.
1286:^
1272:.
1262:.
1252:61
1250:.
1246:.
1223:.
1215:.
1203:.
1180:.
1172:.
1160:.
1137:.
1129:.
1121:.
1109:.
1086:.
1076:.
1066:.
1054:.
1050:.
1027:.
1017:.
1007:14
1005:.
1001:.
985:^
971:.
963:.
955:.
943:.
920:.
912:.
900:.
882:^
872:.
856:^
842:.
832:34
830:.
807:.
799:.
789:57
787:.
764:.
756:.
746:.
734:.
730:.
703:.
693:.
683:14
681:.
677:.
663:^
503:Mg
495:Cs
464:.
304:.
208:pH
164:;
160:;
143:/
93:/
89:/
1992:.
1980::
1957:.
1945::
1922:.
1898::
1890::
1863:.
1839::
1831::
1804:.
1782::
1774::
1747:.
1725::
1717::
1707::
1680:.
1658::
1631:.
1611::
1603::
1580:.
1558::
1550::
1544:2
1523:.
1509::
1482:.
1460::
1452::
1425:.
1405::
1377:.
1353::
1345::
1318:.
1306::
1280:.
1258::
1231:.
1211::
1205:5
1188:.
1168::
1162:3
1145:.
1125::
1117::
1094:.
1070::
1062::
1035:.
1013::
979:.
959::
951::
928:.
916::
908::
850:.
838::
815:.
795::
772:.
750::
742::
728:"
711:.
689::
107:8
52:?
38:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.