716:
1365:
hydrolytic enzymes and perform the classic lysosomal activity, which is autophagy. These vacuoles are therefore seen as fulfilling the role of the animal lysosome. Based on de Duve's description that "only when considered as part of a system involved directly or indirectly in intracellular digestion does the term lysosome describe a physiological unit", some botanists strongly argued that these vacuoles are lysosomes. However, this is not universally accepted as the vacuoles are strictly not similar to lysosomes, such as in their specific enzymes and lack of phagocytic functions. Vacuoles do not have catabolic activity and do not undergo
497:
817:, a relatively acidic organelle with an approximate pH of 5.5. This acidic environment causes dissociation of the lysosomal enzymes from the mannose 6-phosphate receptors. The enzymes are packed into vesicles for further transport to established lysosomes. The late endosome itself can eventually grow into a mature lysosome, as evidenced by the transport of endosomal membrane components from the lysosomes back to the endosomes.
257:
1309:. However, high tissue concentrations and long elimination half-lives are explained also by lipophilicity and absorption of drugs to fatty tissue structures. Important lysosomal enzymes, such as acid sphingomyelinase, may be inhibited by lysosomally accumulated drugs. Such compounds are termed FIASMAs (functional inhibitor of acid sphingomyelinase) and include for example
544:
many times they repeated the estimation, and led to the conclusion that a membrane-like barrier limited the accessibility of the enzyme to its substrate, and that the enzymes were able to diffuse after a few days (and react with their substrate). They described this membrane-like barrier as a "saclike structure surrounded by a membrane and containing acid phosphatase."
66:
731:), a portion of the cell's plasma membrane pinches off to form vesicles that will eventually fuse with an organelle within the cell. Without active replenishment, the plasma membrane would continuously decrease in size. It is thought that lysosomes participate in this dynamic membrane exchange system and are formed by a gradual maturation process from
4248:
863:. Other conditions are due to defects in lysosomal membrane proteins that fail to transport the enzyme, non-enzymatic soluble lysosomal proteins. The initial effect of such disorders is accumulation of specific macromolecules or monomeric compounds inside the endosomal–autophagic–lysosomal system. This results in abnormal signaling pathways,
543:
and found that the activity was only 10% of the expected value. One day, the enzyme activity of purified cell fractions which had been refrigerated for five days was measured. Surprisingly, the enzyme activity was increased to normal of that of the fresh sample. The result was the same no matter how
1364:
is applied to those in plants, fungi and algae (some animal cells also have vacuoles). Discoveries in plant cells since the 1970s started to challenge this definition. Plant vacuoles are found to be much more diverse in structure and function than previously thought. Some vacuoles contain their own
260:
Lysosomes digest material. Step one shows material entering a food vacuole through the plasma membrane, a process known as endocytosis. In step two a lysosome with an active hydrolytic enzyme comes into the pictures as the food vacuole moves away from the plasma membrane. Step three consists of the
1296:
A significant part of the clinically approved drugs are lipophilic weak bases with lysosomotropic properties. This explains a number of pharmacological properties of these drugs, such as high tissue-to-blood concentration gradients or long tissue elimination half-lives; these properties have been
1288:
properties accumulate in acidic intracellular compartments like lysosomes. While the plasma and lysosomal membranes are permeable for neutral and uncharged species of weak bases, the charged protonated species of weak bases do not permeate biomembranes and accumulate within lysosomes. The
1351:
Impaired lysosome function is prominent in systemic lupus erythematosus preventing macrophages and monocytes from degrading neutrophil extracellular traps and immune complexes. The failure to degrade internalized immune complexes stems from chronic mTORC2 activity, which impairs lysosome
661:
that drain into the cytosol, as these enzymes are pH-sensitive and do not function well or at all in the alkaline environment of the cytosol. This ensures that cytosolic molecules and organelles are not destroyed in case there is leakage of the hydrolytic enzymes from the lysosome.
830:
in order to gain entry into the cell. The lysosome prevents easy entry into the cell by hydrolyzing the biomolecules of pathogens necessary for their replication strategies; reduced lysosomal activity results in an increase in viral infectivity, including HIV. In addition,
858:
caused by a dysfunction of one of the enzymes. The rate of incidence is estimated to be 1 in 5,000 births, and the true figure expected to be higher as many cases are likely to be undiagnosed or misdiagnosed. The primary cause is deficiency of an
500:
TEM views of various vesicular compartments. Lysosomes are denoted by "Ly". They are dyed dark due to their acidity; in the center of the top image, a Golgi
Apparatus can be seen, distal from the cell membrane relative to the lysosome
547:
It became clear that this enzyme from the cell fraction came from membranous fractions, which were definitely cell organelles, and in 1955 De Duve named them "lysosomes" to reflect their digestive properties. The same year,
4252:
3418:
Kornhuber J, Tripal P, Reichel M, Mühle C, Rhein C, Muehlbacher M, et al. (2010). "Functional
Inhibitors of Acid Sphingomyelinase (FIASMAs): a novel pharmacological group of drugs with broad clinical applications".
3313:
Kornhuber J, Tripal P, Reichel M, Terfloth L, Bleich S, Wiltfang J, Gulbins E (January 2008). "Identification of new functional inhibitors of acid sphingomyelinase using a structure-property-activity relation model".
3454:
Fois G, Hobi N, Felder E, Ziegler A, Miklavc P, Walther P, et al. (December 2015). "A new role for an old drug: Ambroxol triggers lysosomal exocytosis via pH-dependent Ca²⁺ release from acidic Ca²⁺ stores".
825:
As the endpoint of endocytosis, the lysosome also acts as a safeguard in preventing pathogens from being able to reach the cytoplasm before being degraded. Pathogens often hijack endocytotic pathways such as
715:
719:
The lysosome is shown in purple, as an endpoint in endocytotic sorting. AP2 is necessary for vesicle formation, whereas the mannose-6-receptor is necessary for sorting hydrolase into the lysosome's lumen.
1352:
acidification. As a result, immune complexes in the lysosome recycle to the surface of macrophages causing an accumulation of nuclear antigens upstream of multiple lupus-associated pathologies.
1327:
is a lysosomotropic drug of clinical use to treat conditions of productive cough for its mucolytic action. Ambroxol triggers the exocytosis of lysosomes via neutralization of lysosomal pH and
2568:
Falcone S, Cocucci E, Podini P, Kirchhausen T, Clementi E, Meldolesi J (November 2006). "Macropinocytosis: regulated coordination of endocytic and exocytic membrane traffic events".
614:
In addition to being able to break down polymers, lysosomes are capable of fusing with other organelles & digesting large structures or cellular debris; through cooperation with
3270:
Kornhuber J, Quack G, Danysz W, Jellinger K, Danielczyk W, Gsell W, Riederer P (July 1995). "Therapeutic brain concentration of the NMDA receptor antagonist amantadine".
524:. However, even after a series of experiments, they failed to purify and isolate the enzyme from the cellular extracts. Therefore, they tried a more arduous procedure of
3094:
Traganos F, Darzynkiewicz Z (1994). "Chapter 12 Lysosomal Proton Pump
Activity: Supravital Cell Staining with Acridine Orange Differentiates Leukocyte Subpopulations".
3736:
Kávai M, Csipö I, Sonkoly I, Csongor J, Szegedi GY (November 1986). "Defective immune complex degradation by monocytes in patients with systemic lupus erythematosus".
277:. The sizes of the organelles vary greatly—the larger ones can be more than 10 times the size of the smaller ones. They were discovered and named by Belgian biologist
3227:
Kornhuber J, Weigmann H, Röhrich J, Wiltfang J, Bleich S, Meineke I, et al. (March 2006). "Region specific distribution of levomepromazine in the human brain".
261:
lysosome fusing with the food vacuole and hydrolytic enzymes entering the food vacuole. In the final step, step four, hydrolytic enzymes digest the food particles.
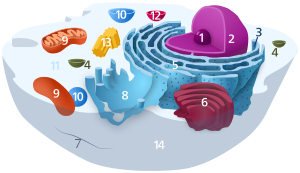
4326:
315:
In 2009, Marco
Sardiello and co-workers discovered that the synthesis of most lysosomal enzymes and membrane proteins is controlled by transcription factor EB (
539:
fraction. This was the crucial step in the serendipitous discovery of lysosomes. To estimate this enzyme activity, they used that of the standardized enzyme
520:, which is the first crucial enzyme in sugar metabolism and the target of insulin. They already suspected that this enzyme played a key role in regulating
1293:, "acid trapping" or "proton pump" effect. The amount of accumulation of lysosomotropic compounds may be estimated using a cell-based mathematical model.
3184:
Kornhuber J, Schultz A, Wiltfang J, Meineke I, Gleiter CH, Zöchling R, et al. (June 1999). "Persistence of haloperidol in human brain tissue".
4228:
4201:
3111:
3035:
2619:
2552:
2387:
1660:
569:
282:
288:
Lysosomes contain more than 60 different enzymes, and have more than 50 membrane proteins. Enzymes of the lysosomes are synthesized in the
4763:
509:
2961:
1611:
1289:
concentration within lysosomes may reach levels 100 to 1000 fold higher than extracellular concentrations. This phenomenon is called
308:
to lysosomes in small vesicles, which fuse with larger acidic vesicles. Enzymes destined for a lysosome are tagged with the molecule
4319:
4257:
4753:
738:
The production of lysosomal proteins suggests one method of lysosome sustainment. Lysosomal protein genes are transcribed in the
902:, which in turn affects spleen, liver, kidneys, lungs, brain and bone marrow. The disease is characterized by bruises, fatigue,
575:
Originally, De Duve had termed the organelles the "suicide bags" or "suicide sacs" of the cells, for their hypothesized role in
4388:
723:
Many components of animal cells are recycled by transferring them inside or embedded in sections of membrane. For instance, in
211:
89:
2165:
4285:
Lysosomal
Disease Network, a research consortium funded by the NIH through its NCATS/Rare Diseases Clinical Research Network
1857:
Saftig P, Klumperman J (September 2009). "Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function".
3830:"Apoptotic Debris Accumulates on Hematopoietic Cells and Promotes Disease in Murine and Human Systemic Lupus Erythematosus"
3781:"mTORC2 Activity Disrupts Lysosome Acidification in Systemic Lupus Erythematosus by Impairing Caspase-1 Cleavage of Rab39a"
3701:
Kavai M, Szegedi G (August 2007). "Immune complex clearance by monocytes and macrophages in systemic lupus erythematosus".
810:
in the Golgi apparatus, a phenomenon that is crucial for proper packaging into vesicles destined for the lysosomal system.
4490:
871:
and degradation and intracellular trafficking, ultimately leading to pathogenetic disorders. The organs most affected are
265:
Lysosomes are degradative organelles that act as the waste disposal system of the cell by digesting used materials in the
4312:
4137:"Barley aleurone cells contain two types of vacuoles. Characterization Of lytic organelles by use of fluorescent probes"
924:
807:
233:
proteins. The lumen's pH (~4.5–5.0) is optimal for the enzymes involved in hydrolysis, analogous to the activity of the
110:
3059:
de Duve C, de Barsy T, Poole B, Trouet A, Tulkens P, Van Hoof F (September 1974). "Commentary. Lysosomotropic agents".
560:
of the new organelle. Using a staining method for acid phosphatase, de Duve and
Novikoff confirmed the location of the
1486:"Fluorescence probe measurement of the intralysosomal pH in living cells and the perturbation of pH by various agents"
855:
340:
289:
94:
3096:
Lysosomal proton pump activity: supravital cell staining with acridine orange differentiates leukocyte subpopulations
1360:
By scientific convention, the term lysosome is applied to these vesicular organelles only in animals, and the term
911:
851:
591:
Lysosomes contain a variety of enzymes, enabling the cell to break down various biomolecules it engulfs, including
692:
It sources its versatile capacity for degradation by import of enzymes with specificity for different substrates;
1340:
332:
4096:"A novel protein RLS1 with NB-ARM domains is involved in chloroplast degradation during leaf senescence in rice"
4485:
203:
4274:
3879:"BAFF Induces Tertiary Lymphoid Structures and Positions T Cells within the Glomeruli during Lupus Nephritis"
3644:"Defects in lysosomal maturation facilitate the activation of innate sensors in systemic lupus erythematosus"
2905:"Lysosomal enzyme replacement therapies: Historical development, clinical outcomes, and future perspectives"
928:
344:
4450:
3359:
Kornhuber J, Muehlbacher M, Trapp S, Pechmann S, Friedl A, Reichel M, et al. (2011). Riezman H (ed.).
496:
4425:
3323:
1388:
1336:
1328:
918:
872:
697:
622:, clearing out damaged structures. Similarly, they are able to break down virus particles or bacteria in
4748:
4689:
4475:
4368:
3533:"Ambroxol improves lysosomal biochemistry in glucocerebrosidase mutation-linked Parkinson disease cells"
2016:"The cell biology of disease: lysosomal storage disorders: the cellular impact of lysosomal dysfunction"
755:
553:
517:
336:
2640:"Inhibition of lysosome and proteasome function enhances human immunodeficiency virus type 1 infection"
685:
are responsible for transport of protons, while the counter transport of chloride ions is performed by
4722:
3655:
3593:
3372:
1967:
1497:
1393:
887:
3983:
Samaj J, Read ND, Volkmann D, Menzel D, Baluska F (August 2005). "The endocytic network in plants".
3328:
763:
4758:
4348:
864:
803:
611:). The enzymes responsible for this hydrolysis require an acidic environment for optimal activity.
557:
485:
484:
form of the prefix but are often treated by readers and editors as mere unthinking replications of
309:
1954:
Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, et al. (July 2009).
4156:
3947:
3761:
3295:
3252:
3209:
2212:
1993:
1882:
1629:
891:
654:
525:
521:
438:
1754:
di Ronza A, Bajaj L, Sharma J, Sanagasetti D, Lotfi P, Adamski CJ, et al. (December 2018).
2491:
Medina DL, Di Paola S, Peluso I, Armani A, De
Stefani D, Venditti R, et al. (March 2015).
4743:
4672:
4510:
4393:
4224:
4197:
4174:
4117:
4076:
4000:
3965:
3908:
3859:
3810:
3753:
3718:
3683:
3621:
3562:
3513:
3472:
3436:
3400:
3341:
3287:
3244:
3201:
3166:
3117:
3107:
3076:
3041:
3031:
3008:
2957:
2934:
2885:
2844:
2809:
2767:
2718:
2669:
2615:
2585:
2548:
2522:
2473:
2424:
2383:
2360:
2296:
2261:
2204:
2146:
2094:
2045:
1985:
1933:
1874:
1839:
1785:
1707:
1656:
1574:
1525:
1450:
899:
751:
645:
ranging from ~4.5–5.0, the interior of the lysosomes is acidic compared to the slightly basic
505:
278:
250:
1725:
850:
Lysosomes are involved in a group of genetically inherited deficiencies, or mutations called
4164:
4148:
4107:
4066:
4058:
4027:
3992:
3955:
3939:
3898:
3890:
3849:
3841:
3800:
3792:
3745:
3710:
3673:
3663:
3611:
3601:
3582:"Impairment of neutrophil extracellular trap degradation is associated with lupus nephritis"
3552:
3544:
3503:
3464:
3428:
3390:
3380:
3333:
3279:
3236:
3193:
3156:
3148:
3099:
3068:
2998:
2990:
2924:
2916:
2875:
2836:
2799:
2757:
2749:
2708:
2700:
2659:
2651:
2577:
2512:
2504:
2463:
2455:
2416:
2350:
2340:
2288:
2251:
2243:
2196:
2136:
2128:
2084:
2076:
2035:
2027:
1975:
1923:
1913:
1866:
1829:
1819:
1775:
1767:
1697:
1689:
1619:
1564:
1556:
1515:
1505:
1440:
1430:
1281:
895:
728:
707:
Recent research also indicates that lysosomes can act as a source of intracellular calcium.
678:
608:
565:
549:
540:
401:
366:
328:
269:, from both inside and outside the cell. Material from outside the cell is taken up through
237:. Besides degradation of polymers, the lysosome is involved in cell processes of secretion,
226:
167:
1655:(Online-Ausg. 1 ed.). Georgetown, Tex.: Landes Bioscience/Eurekah.com. pp. 1–16.
4373:
4299:
2420:
1693:
1545:"Signals from the lysosome: a control centre for cellular clearance and energy metabolism"
1302:
1290:
868:
799:
305:
293:
238:
230:
99:
4294:
4289:
3642:
Monteith AJ, Kang S, Scott E, Hillman K, Rajfur Z, Jacobson K, et al. (April 2016).
2736:
Lieberman AP, Puertollano R, Raben N, Slaugenhaupt S, Walkley SU, Ballabio A (May 2012).
2279:
Klionsky DJ (August 2008). "Autophagy revisited: a conversation with
Christian de Duve".
938:, and age-related diseases such as Alzheimer's, Parkinson's, and cardiovascular disease.
758:, where they are modified. Lysosomal soluble proteins exit the endoplasmic reticulum via
4031:
3779:
Monteith AJ, Vincent HA, Kang S, Li P, Claiborne TM, Rajfur Z, et al. (July 2018).
3659:
3597:
3376:
1971:
1501:
1335:
was also found to improve cellular function in some disease of lysosomal origin such as
339:, due to the inability to break them down. These genetic defects are related to several
4336:
4267:
3903:
3878:
3854:
3829:
3828:
Kang S, Rogers JL, Monteith AJ, Jiang C, Schmitz J, Clarke SH, et al. (May 2016).
3805:
3780:
3749:
3678:
3643:
3616:
3581:
3580:
Hakkim A, Fürnrohr BG, Amann K, Laube B, Abed UA, Brinkmann V, et al. (May 2010).
3557:
3532:
3395:
3360:
3161:
3136:
3003:
2978:
2929:
2904:
2880:
2863:
2762:
2737:
2713:
2688:
2664:
2639:
2517:
2492:
2468:
2443:
2355:
2328:
2256:
2231:
2141:
2116:
2089:
2064:
2040:
2015:
1928:
1901:
1834:
1807:
1780:
1755:
1702:
1677:
1569:
1544:
1445:
1418:
860:
650:
529:
462:
246:
207:
4304:
4169:
4136:
4071:
4046:
3960:
3927:
3103:
1520:
1485:
4737:
4619:
4579:
4554:
4470:
4383:
4358:
3283:
3072:
2608:
1806:
Bajaj L, Sharma J, di Ronza A, Zhang P, Eblimit A, Pal R, et al. (August 2020).
1633:
1318:
1285:
839:
670:
649:(pH 7.2). The lysosomal membrane protects the cytosol, and therefore the rest of the
149:
115:
3765:
3531:
McNeill A, Magalhaes J, Shen C, Chau KY, Hughes D, Mehta A, et al. (May 2014).
3299:
3256:
2840:
2655:
2216:
1997:
1886:
1756:"CLN8 is an endoplasmic reticulum cargo receptor that regulates lysosome biogenesis"
746:). mRNA transcripts exit the nucleus into the cytosol, where they are translated by
4599:
4571:
4549:
4542:
4522:
4462:
4378:
4363:
3213:
1383:
907:
813:
Upon leaving the Golgi apparatus, the lysosomal enzyme-filled vesicle fuses with a
739:
623:
600:
596:
320:
256:
105:
78:
46:
4047:"Autophagy in Tobacco Suspension-Cultured Cells in Response to Sucrose Starvation"
456:
4218:
3714:
3385:
1648:
4701:
4667:
4594:
4589:
4480:
2994:
1298:
827:
724:
674:
657:
within the lysosome. The cell is additionally protected from any lysosomal acid
327:
in the genes for these enzymes are responsible for more than 50 different human
270:
222:
55:
3648:
Proceedings of the
National Academy of Sciences of the United States of America
3586:
Proceedings of the
National Academy of Sciences of the United States of America
3468:
2920:
2704:
2493:"Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB"
1490:
Proceedings of the National Academy of Sciences of the United States of America
4696:
4636:
4629:
4614:
4609:
4604:
4559:
4505:
4495:
4445:
4440:
4430:
3996:
3240:
3152:
2132:
1771:
1624:
1378:
1366:
1314:
1310:
1306:
658:
638:
634:
627:
580:
215:
144:
4295:
Mutations in the Lysosomal Enzyme–Targeting Pathway and Persistent Stuttering
3894:
3845:
3796:
3045:
2827:
Rosenbloom BE, Weinreb NJ (2013). "Gaucher disease: a comprehensive review".
1808:"A CLN6-CLN8 complex recruits lysosomal enzymes at the ER for Golgi transfer"
934:
Dysfunctional lysosome activity is also heavily implicated in the biology of
466:, "body", yielding "body that lyses" or "lytic body". The adjectival form is
4717:
4679:
4652:
4624:
4532:
4517:
4500:
4435:
4408:
4340:
3877:
Kang S, Fedoriw Y, Brenneman EK, Truong YK, Kikly K, Vilen BJ (April 2017).
3668:
3606:
2345:
1980:
1955:
1435:
880:
832:
806:, is added to the peptides. The presence of these tags allow for binding to
747:
732:
693:
619:
615:
576:
561:
536:
450:
434:
274:
266:
242:
133:
129:
73:
4121:
4080:
4004:
3969:
3912:
3863:
3814:
3722:
3687:
3625:
3566:
3548:
3517:
3508:
3491:
3476:
3440:
3404:
3345:
3248:
3205:
3170:
3012:
2938:
2889:
2848:
2813:
2804:
2787:
2771:
2722:
2673:
2589:
2526:
2477:
2428:
2364:
2300:
2265:
2208:
2150:
2098:
2049:
1989:
1937:
1878:
1843:
1789:
1711:
1578:
1510:
1454:
579:. However, it has since been concluded that they only play a minor role in
480:
445:
4284:
4178:
4062:
3757:
3291:
3197:
3121:
3080:
2459:
2031:
886:
There is no direct medical treatment to cure LSDs. The most common LSD is
516:
in liver cells. By 1949, he and his team had focused on the enzyme called
4662:
4418:
4403:
2788:"New strategies for the treatment of lysosomal storage diseases (review)"
2080:
1468:
1398:
1332:
1324:
814:
682:
592:
324:
83:
31:
17:
4112:
4095:
3361:"Identification of novel functional inhibitors of acid sphingomyelinase"
3137:"Quantitative modeling of selective lysosomal targeting for drug design"
2247:
2200:
2115:
Carmona-Gutierrez D, Hughes AL, Madeo F, Ruckenstuhl C (December 2016).
1918:
1529:
689:
Cl/H antiporter. In this way a steady acidic environment is maintained.
528:, by which cellular components are separated based on their sizes using
65:
4684:
4584:
4537:
4413:
4160:
3951:
2753:
1361:
903:
876:
646:
513:
234:
125:
120:
35:
3432:
3337:
2638:
Wei BL, Denton PW, O'Neill E, Luo T, Foster JL, Garcia JV (May 2005).
2581:
2292:
1824:
4527:
2110:
2108:
935:
666:
348:
218:
4152:
3943:
2508:
1870:
1560:
2404:
842:
hijack the endosomal pathway while evading lysosomal degradation.
759:
714:
686:
604:
556:
visited de Duve's laboratory, and successfully obtained the first
495:
255:
273:, while material from the inside of the cell is digested through
4290:
Self-Destructive Behavior in Cells May Hold Key to a Longer Life
4263:
795:
791:
743:
701:
316:
301:
297:
4308:
4094:
Jiao BB, Wang JJ, Zhu XD, Zeng LJ, Li Q, He ZH (January 2012).
917:
The most severe and rarely found, lysosomal storage disease is
798:
proteins. COPII vesicles then deliver lysosomal enzymes to the
4657:
4279:
2232:"Electron microscopy of lysosomerich fractions from rat liver"
3135:
Trapp S, Rosania GR, Horobin RW, Kornhuber J (October 2008).
2380:
Color Atlas of Cytology, Histology, & Microscopic Anatomy
2329:"Lysosomes as "suicide bags" in cell death: myth or reality?"
1956:"A gene network regulating lysosomal biogenesis and function"
4300:
Animation showing how lysosomes are made, and their function
742:
in a process that is controlled by transcription factor EB (
312:, so that they are properly sorted into acidified vesicles.
4275:
3D structures of proteins associated with lysosome membrane
3098:. Methods in Cell Biology. Vol. 41. pp. 185–194.
2687:
Schultz ML, Tecedor L, Chang M, Davidson BL (August 2011).
704:
is necessary to release phosphate groups of phospholipids.
642:
422:
416:
410:
387:
381:
375:
191:
182:
176:
4018:
Matile P (1978). "Biochemistry and Function of Vacuoles".
2864:"Gaucher disease: insights from a rare Mendelian disorder"
2786:
Parenti G, Pignata C, Vajro P, Salerno M (January 2013).
1543:
Settembre C, Fraldi A, Medina DL, Ballabio A (May 2013).
665:
The lysosome maintains its pH differential by pumping in
535:
They succeeded in detecting the enzyme activity from the
512:
in Belgium, had been studying the mechanism of action of
2633:
2631:
2187:
de Duve C (September 2005). "The lysosome turns fifty".
2117:"The crucial impact of lysosomes in aging and longevity"
1331:
from acidic calcium stores. Presumably for this reason,
1048:
Polysaccharides/ mucopolysaccharides hydrolyzing enzymes
2979:"The Autophagy Lysosomal Pathway and Neurodegeneration"
927:
is another lysosomal storage disease that also affects
910:, and enlargement of the liver and spleen. As of 2017,
335:. These diseases result in an accumulation of specific
2014:
Platt FM, Boland B, van der Spoel AC (November 2012).
508:, at the Laboratory of Physiological Chemistry at the
454:, meaning "to loosen", via Ancient Greek λύσις ), and
2614:(4th ed.). New York: Scientific American Books.
894:. Consequently, the enzyme substrate, the fatty acid
419:
413:
384:
378:
225:. A lysosome has a specific composition, of both its
188:
185:
179:
2442:
Ishida Y, Nayak S, Mindell JA, Grabe M (June 2013).
2313:
Hayashi, Teru, and others. "Subcellular Particles."
2065:"Autophagy in ageing and ageing-associated diseases"
1749:
1747:
1419:"Lysosomal Calcium Channels in Autophagy and Cancer"
914:
is available for treating 8 of the 50-60 known LDs.
407:
372:
173:
4710:
4645:
4570:
4461:
4347:
2236:
The Journal of Biophysical and Biochemical Cytology
425:
404:
390:
369:
194:
170:
45:
2607:
1902:"Lysosomal exocytosis and lipid storage disorders"
568:and electron microscopic studies. de Duve won the
1467:By convention similar cells in plants are called
696:are the major class of hydrolytic enzymes, while
1605:
1603:
488:, which has no doubt been true as often as not.
4280:Hide and Seek Foundation For Lysosomal Research
3030:(3rd ed.). New Delhi: Pathfinder Academy.
2230:Novikoff AB, Beaufay H, De Duve C (July 1956).
1949:
1947:
1801:
1799:
304:proteins. The enzymes are transported from the
2956:(4th ed.). Garland Science. p. 744.
1612:"When the brain's waste disposal system fails"
4320:
1596:(8th AP ed.). Pearson Benjamin Cummings.
1109:Bacterial cell walls and mucopolysaccharides
8:
4135:Swanson SJ, Bethke PC, Jones RL (May 1998).
2792:International Journal of Molecular Medicine
2547:(4th ed.). New York: Garland Science.
890:, which is due to deficiency of the enzyme
4327:
4313:
4305:
4196:. New York: Plenum Press. pp. 7, 15.
2983:Cold Spring Harbor Perspectives in Biology
2738:"Autophagy in lysosomal storage disorders"
2538:
2536:
2166:"The Discovery of Lysosomes and Autophagy"
1472:
1256:B- Glucosamine (N-acetyl)-6-Sulfatase/GNS
762:-coated vesicles after recruitment by the
296:upon recruitment by a complex composed of
4168:
4111:
4070:
3959:
3902:
3853:
3804:
3677:
3667:
3615:
3605:
3556:
3507:
3394:
3384:
3327:
3160:
3002:
2928:
2879:
2803:
2761:
2712:
2663:
2601:
2599:
2516:
2467:
2354:
2344:
2255:
2140:
2088:
2039:
1979:
1927:
1917:
1833:
1823:
1779:
1701:
1623:
1568:
1519:
1509:
1444:
1434:
3492:"Magic shotgun for Parkinson's disease?"
3028:Life Sciences: Fundamentals and practice
2009:
2007:
1099:Polysaccharides and mucopolysaccharides
947:
2689:"Clarifying lysosomal storage diseases"
1409:
1119:Hyaluronic acids, chondroitin sulfates
991:Oligonucleotides and phosphodiesterase
319:), which promotes the transcription of
1859:Nature Reviews. Molecular Cell Biology
1549:Nature Reviews. Molecular Cell Biology
942:Different enzymes present in Lysosomes
700:is responsible for carbohydrates, and
633:The size of lysosomes varies from 0.1
42:
3637:
3635:
2421:10.1146/annurev-physiol-012110-142317
1812:The Journal of Clinical Investigation
1694:10.1146/annurev-physiol-021014-071649
1417:Wu Y, Huang P, Dong XP (March 2021).
669:(H ions) from the cytosol across the
570:Nobel Prize in Physiology or Medicine
283:Nobel Prize in Physiology or Medicine
70:Components of a typical animal cell:
7:
4220:Plant Cell Vacuoles: An Introduction
4045:Moriyasu Y, Ohsumi Y (August 1996).
3421:Cellular Physiology and Biochemistry
2903:Solomon M, Muro S (September 2017).
2444:"A model of lysosomal pH regulation"
2405:"Lysosomal acidification mechanisms"
2382:(4th ed.). Thieme. p. 34.
1649:"History and Morphology of Lysosome"
4032:10.1146/annurev.pp.29.060178.001205
2333:The Journal of Biological Chemistry
3750:10.1111/j.1365-3083.1986.tb02167.x
3738:Scandinavian Journal of Immunology
3186:The American Journal of Psychiatry
1592:Holtzclaw FW, et al. (2008).
802:, where a specific lysosomal tag,
25:
4020:Annual Review of Plant Physiology
2448:The Journal of General Physiology
2063:He LQ, Lu JH, Yue ZY (May 2013).
1594:AP* Biology: to Accompany Biology
750:. The nascent peptide chains are
4251: This article incorporates
4246:
400:
365:
166:
64:
4223:. Australia: Csiro Publishing.
3490:Albin RL, Dauer WT (May 2014).
2841:10.1615/CritRevOncog.2013006060
2829:Critical Reviews in Oncogenesis
2656:10.1128/jvi.79.9.5705-5712.2005
2543:Alberts B, et al. (2002).
1484:Ohkuma S, Poole B (July 1978).
1246:A- Arylsulfatase(A, B & G)
3316:Journal of Medicinal Chemistry
3229:Journal of Neural Transmission
2909:Advanced Drug Delivery Reviews
2606:Lodish H, et al. (2000).
2327:Turk B, Turk V (August 2009).
2164:Susana Castro-Obregon (2010).
510:Catholic University of Louvain
281:, who eventually received the
1:
4491:Microtubule organizing center
3104:10.1016/S0091-679X(08)61717-3
2954:Molecular Biology of the Cell
2545:Molecular biology of the cell
1266:C- Iduronate 2-Sulfatase/IDS
898:accumulates, particularly in
808:mannose 6-phosphate receptors
790:ystem), which is composed of
478:are much rarer; they use the
3715:10.1016/j.autrev.2007.01.017
3386:10.1371/journal.pone.0023852
3284:10.1016/0028-3908(95)00056-c
3073:10.1016/0006-2952(74)90174-9
2862:Sidransky E (October 2012).
1900:Samie MA, Xu H (June 2014).
1473:§ Controversy in botany
1347:Systemic lupus erythematosus
925:Metachromatic leukodystrophy
572:in 1974 for this discovery.
111:Smooth endoplasmic reticulum
3141:European Biophysics Journal
2995:10.1101/cshperspect.a033993
2977:Finkbeiner S (March 2020).
2409:Annual Review of Physiology
2020:The Journal of Cell Biology
1682:Annual Review of Physiology
856:inborn errors of metabolism
698:lysosomal alpha-glucosidase
618:, they are able to conduct
448:and derived from the Latin
355:Etymology and pronunciation
341:neurodegenerative disorders
290:rough endoplasmic reticulum
95:Rough endoplasmic reticulum
4780:
4764:Lysosomal storage diseases
3469:10.1016/j.ceca.2015.10.002
2921:10.1016/j.addr.2017.05.004
2705:10.1016/j.tins.2011.05.006
2069:Acta Pharmacologica Sinica
1089:Mannosides, glycoproteins
1028:B- Acid deoxyribonuclease
988:B- Acid phosphodiesterase
912:enzyme replacement therapy
852:lysosomal storage diseases
702:lysosomal acid phosphatase
333:lysosomal storage diseases
221:that digest many kinds of
29:
3997:10.1016/j.tcb.2005.06.006
3241:10.1007/s00702-005-0331-3
3153:10.1007/s00249-008-0338-4
2133:10.1016/j.arr.2016.04.009
1906:Journal of Lipid Research
1772:10.1038/s41556-018-0228-7
1647:Lüllmznn-Rauch R (2005).
1625:10.1146/knowable-121118-1
1341:lysosomal storage disease
63:
54:
4486:Prokaryotic cytoskeleton
3895:10.4049/jimmunol.1600281
3846:10.4049/jimmunol.1500418
3797:10.4049/jimmunol.1701712
3061:Biochemical Pharmacology
3026:Kumar P, Mina U (2013).
1297:found for drugs such as
1269:O- and N-Sulfate esters
1249:O- and N-Sulfate esters
204:membrane-bound organelle
132:; with which, comprises
30:Not to be confused with
4754:Eukaryotic cell anatomy
3669:10.1073/pnas.1513943113
3607:10.1073/pnas.0909927107
2693:Trends in Neurosciences
2570:Journal of Cell Science
2346:10.1074/jbc.R109.023820
2121:Ageing Research Reviews
1981:10.1126/science.1174447
1436:10.3390/cancers13061299
1196:Lipid degrading enzymes
981:Most phosphomonoesters
929:sphingolipid metabolism
906:, low blood platelets,
345:cardiovascular diseases
27:Cell membrane organelle
4253:public domain material
3985:Trends in Cell Biology
3926:Marty F (April 1999).
2805:10.3892/ijmm.2012.1187
2610:Molecular cell biology
2315:Subcellular Particles.
1678:"Lysosomal physiology"
1511:10.1073/pnas.75.7.3327
1389:Antimicrobial peptides
919:inclusion cell disease
720:
587:Function and structure
502:
331:collectively known as
262:
4476:Intermediate filament
4369:Endoplasmic reticulum
4063:10.1104/pp.111.4.1233
3883:Journal of Immunology
3834:Journal of Immunology
3785:Journal of Immunology
3198:10.1176/ajp.156.6.885
2460:10.1085/jgp.201210930
2032:10.1083/jcb.201208152
1651:. In Zaftig P (ed.).
1356:Controversy in botany
1018:A- Acid ribonuclease
846:Clinical significance
756:endoplasmic reticulum
718:
679:chloride ion channels
554:University of Vermont
518:glucose 6-phosphatase
499:
259:
210:. They are spherical
206:found in many animal
128:(fluid that contains
4723:Extracellular matrix
3703:Autoimmunity Reviews
3549:10.1093/brain/awu020
3509:10.1093/brain/awu076
2576:(Pt 22): 4758–4769.
2242:(4 Suppl): 179–184.
2081:10.1038/aps.2012.188
1676:Xu H, Ren D (2015).
1610:Underwood E (2018).
1394:Innate immune system
1096:D- β- Glucoronidase
978:A- Acid phosphatase
727:(more specifically,
558:electron micrographs
292:and exported to the
4426:Cytoplasmic granule
4270:on 8 December 2009.
4192:Holtzman E (1989).
3660:2016PNAS..113E2142M
3654:(15): E2142–E2151.
3598:2010PNAS..107.9813H
3543:(Pt 5): 1481–1495.
3502:(Pt 5): 1274–1275.
3377:2011PLoSO...623852K
2644:Journal of Virology
2497:Nature Cell Biology
2403:Mindell JA (2012).
2339:(33): 21783–21787.
2248:10.1083/jcb.2.4.179
2201:10.1038/ncb0905-847
2189:Nature Cell Biology
1972:2009Sci...325..473S
1919:10.1194/jlr.R046896
1760:Nature Cell Biology
1726:"Lysosomal Enzymes"
1502:1978PNAS...75.3327O
1259:Glycosaminoglycans
1058:A- β-Galactosidase
949:
865:calcium homeostasis
804:mannose 6-phosphate
655:degradative enzymes
564:of lysosomes using
310:mannose 6-phosphate
86:(dots as part of 5)
57:Animal cell diagram
4451:Weibel–Palade body
4335:Structures of the
2952:Alberts B (2002).
2868:Discovery Medicine
2754:10.4161/auto.19469
2378:Kuehnel W (2003).
1730:www.rndsystems.com
1209:Fatty acyl esters
1126:H- Arylsulphatase
948:
892:glucocerebrosidase
869:lipid biosynthesis
721:
562:hydrolytic enzymes
526:cell fractionation
522:blood sugar levels
503:
263:
4731:
4730:
4511:Spindle pole body
4230:978-0-643-09944-9
4203:978-0306-4-3126-5
4113:10.1093/mp/ssr081
3840:(10): 4030–4039.
3592:(21): 9813–9818.
3433:10.1159/000315101
3338:10.1021/jm070524a
3272:Neuropharmacology
3113:978-0-12-564142-5
3067:(18): 2495–2531.
3037:978-81-906427-0-5
2621:978-0-7167-3136-8
2582:10.1242/jcs.03238
2554:978-0-8153-3218-3
2389:978-1-58890-175-0
2293:10.4161/auto.6398
1966:(5939): 473–477.
1825:10.1172/JCI130955
1766:(12): 1370–1377.
1732:. R&D Systems
1662:978-0-387-28957-1
1616:Knowable Magazine
1369:as lysosomes do.
1273:
1272:
1216:B- Phospholipase
1129:Organic sulfates
1116:F- Hyaluronidase
900:white blood cells
888:Gaucher's disease
506:Christian de Duve
329:genetic disorders
279:Christian de Duve
251:energy metabolism
227:membrane proteins
158:
157:
16:(Redirected from
4771:
4329:
4322:
4315:
4306:
4271:
4266:. Archived from
4250:
4249:
4235:
4234:
4214:
4208:
4207:
4189:
4183:
4182:
4172:
4132:
4126:
4125:
4115:
4091:
4085:
4084:
4074:
4057:(4): 1233–1241.
4051:Plant Physiology
4042:
4036:
4035:
4015:
4009:
4008:
3980:
3974:
3973:
3963:
3928:"Plant vacuoles"
3923:
3917:
3916:
3906:
3889:(7): 2602–2611.
3874:
3868:
3867:
3857:
3825:
3819:
3818:
3808:
3776:
3770:
3769:
3733:
3727:
3726:
3698:
3692:
3691:
3681:
3671:
3639:
3630:
3629:
3619:
3609:
3577:
3571:
3570:
3560:
3528:
3522:
3521:
3511:
3487:
3481:
3480:
3451:
3445:
3444:
3415:
3409:
3408:
3398:
3388:
3356:
3350:
3349:
3331:
3310:
3304:
3303:
3267:
3261:
3260:
3224:
3218:
3217:
3181:
3175:
3174:
3164:
3147:(8): 1317–1328.
3132:
3126:
3125:
3091:
3085:
3084:
3056:
3050:
3049:
3023:
3017:
3016:
3006:
2974:
2968:
2967:
2949:
2943:
2942:
2932:
2900:
2894:
2893:
2883:
2859:
2853:
2852:
2824:
2818:
2817:
2807:
2782:
2776:
2775:
2765:
2733:
2727:
2726:
2716:
2684:
2678:
2677:
2667:
2650:(9): 5705–5712.
2635:
2626:
2625:
2613:
2603:
2594:
2593:
2565:
2559:
2558:
2540:
2531:
2530:
2520:
2488:
2482:
2481:
2471:
2439:
2433:
2432:
2400:
2394:
2393:
2375:
2369:
2368:
2358:
2348:
2324:
2318:
2311:
2305:
2304:
2276:
2270:
2269:
2259:
2227:
2221:
2220:
2184:
2178:
2177:
2170:Nature Education
2161:
2155:
2154:
2144:
2112:
2103:
2102:
2092:
2060:
2054:
2053:
2043:
2011:
2002:
2001:
1983:
1951:
1942:
1941:
1931:
1921:
1897:
1891:
1890:
1854:
1848:
1847:
1837:
1827:
1818:(8): 4118–4132.
1803:
1794:
1793:
1783:
1751:
1742:
1741:
1739:
1737:
1722:
1716:
1715:
1705:
1673:
1667:
1666:
1644:
1638:
1637:
1627:
1607:
1598:
1597:
1589:
1583:
1582:
1572:
1540:
1534:
1533:
1523:
1513:
1496:(7): 3327–3331.
1481:
1475:
1465:
1459:
1458:
1448:
1438:
1414:
1156:A- Cathepsin(s)
950:
896:glucosylceramide
782:nzymes of the ly
729:macropinocytosis
683:Vacuolar-ATPases
609:lysosomal lipase
550:Alex B. Novikoff
541:acid phosphatase
432:
431:
428:
427:
424:
421:
418:
415:
412:
409:
406:
397:
396:
393:
392:
389:
386:
383:
380:
377:
374:
371:
201:
200:
197:
196:
193:
190:
187:
184:
181:
178:
175:
172:
102:(or, Golgi body)
68:
58:
49:
43:
21:
4779:
4778:
4774:
4773:
4772:
4770:
4769:
4768:
4734:
4733:
4732:
4727:
4706:
4641:
4566:
4457:
4374:Golgi apparatus
4350:
4343:
4333:
4256:
4247:
4243:
4238:
4231:
4216:
4215:
4211:
4204:
4191:
4190:
4186:
4153:10.2307/3870657
4134:
4133:
4129:
4100:Molecular Plant
4093:
4092:
4088:
4044:
4043:
4039:
4017:
4016:
4012:
3982:
3981:
3977:
3944:10.2307/3870886
3925:
3924:
3920:
3876:
3875:
3871:
3827:
3826:
3822:
3778:
3777:
3773:
3735:
3734:
3730:
3700:
3699:
3695:
3641:
3640:
3633:
3579:
3578:
3574:
3530:
3529:
3525:
3489:
3488:
3484:
3453:
3452:
3448:
3417:
3416:
3412:
3358:
3357:
3353:
3329:10.1.1.324.8854
3312:
3311:
3307:
3269:
3268:
3264:
3226:
3225:
3221:
3183:
3182:
3178:
3134:
3133:
3129:
3114:
3093:
3092:
3088:
3058:
3057:
3053:
3038:
3025:
3024:
3020:
2976:
2975:
2971:
2964:
2951:
2950:
2946:
2902:
2901:
2897:
2874:(77): 273–281.
2861:
2860:
2856:
2826:
2825:
2821:
2785:
2783:
2779:
2735:
2734:
2730:
2686:
2685:
2681:
2637:
2636:
2629:
2622:
2605:
2604:
2597:
2567:
2566:
2562:
2555:
2542:
2541:
2534:
2509:10.1038/ncb3114
2490:
2489:
2485:
2441:
2440:
2436:
2402:
2401:
2397:
2390:
2377:
2376:
2372:
2326:
2325:
2321:
2312:
2308:
2278:
2277:
2273:
2229:
2228:
2224:
2186:
2185:
2181:
2163:
2162:
2158:
2114:
2113:
2106:
2062:
2061:
2057:
2013:
2012:
2005:
1953:
1952:
1945:
1912:(6): 995–1009.
1899:
1898:
1894:
1871:10.1038/nrm2745
1856:
1855:
1851:
1805:
1804:
1797:
1753:
1752:
1745:
1735:
1733:
1724:
1723:
1719:
1675:
1674:
1670:
1663:
1646:
1645:
1641:
1609:
1608:
1601:
1591:
1590:
1586:
1561:10.1038/nrm3565
1542:
1541:
1537:
1483:
1482:
1478:
1466:
1462:
1416:
1415:
1411:
1407:
1375:
1358:
1349:
1329:calcium release
1303:levomepromazine
1291:lysosomotropism
1278:
1276:Lysosomotropism
1166:B- Collagenase
944:
848:
838:toxins such as
836:
823:
800:Golgi apparatus
754:into the rough
713:
589:
494:
439:combining forms
403:
399:
368:
364:
357:
306:Golgi apparatus
294:Golgi apparatus
239:plasma membrane
169:
165:
154:
100:Golgi apparatus
56:
47:
39:
28:
23:
22:
15:
12:
11:
5:
4777:
4775:
4767:
4766:
4761:
4756:
4751:
4746:
4736:
4735:
4729:
4728:
4726:
4725:
4720:
4714:
4712:
4708:
4707:
4705:
4704:
4699:
4694:
4693:
4692:
4687:
4677:
4676:
4675:
4670:
4665:
4655:
4649:
4647:
4646:Other internal
4643:
4642:
4640:
4639:
4634:
4633:
4632:
4627:
4622:
4617:
4612:
4607:
4602:
4597:
4592:
4582:
4576:
4574:
4568:
4567:
4565:
4564:
4563:
4562:
4557:
4547:
4546:
4545:
4540:
4535:
4530:
4520:
4515:
4514:
4513:
4508:
4503:
4498:
4488:
4483:
4478:
4473:
4467:
4465:
4459:
4458:
4456:
4455:
4454:
4453:
4448:
4443:
4438:
4433:
4423:
4422:
4421:
4416:
4411:
4406:
4401:
4396:
4386:
4381:
4376:
4371:
4366:
4361:
4355:
4353:
4345:
4344:
4334:
4332:
4331:
4324:
4317:
4309:
4303:
4302:
4297:
4292:
4287:
4282:
4277:
4272:
4259:Science Primer
4242:
4241:External links
4239:
4237:
4236:
4229:
4217:De DN (2000).
4209:
4202:
4184:
4147:(5): 685–698.
4141:The Plant Cell
4127:
4106:(1): 205–217.
4086:
4037:
4026:(1): 193–213.
4010:
3991:(8): 425–433.
3975:
3938:(4): 587–600.
3932:The Plant Cell
3918:
3869:
3820:
3791:(2): 371–382.
3771:
3744:(5): 527–532.
3728:
3709:(7): 497–502.
3693:
3631:
3572:
3523:
3482:
3463:(6): 628–637.
3446:
3410:
3351:
3322:(2): 219–237.
3305:
3278:(7): 713–721.
3262:
3235:(3): 387–397.
3219:
3192:(6): 885–890.
3176:
3127:
3112:
3086:
3051:
3036:
3018:
2989:(3): a033993.
2969:
2963:978-0815340720
2962:
2944:
2895:
2854:
2835:(3): 163–175.
2819:
2777:
2748:(5): 719–730.
2728:
2699:(8): 401–410.
2679:
2627:
2620:
2595:
2560:
2553:
2532:
2503:(3): 288–299.
2483:
2454:(6): 705–720.
2434:
2395:
2388:
2370:
2319:
2306:
2287:(6): 740–743.
2271:
2222:
2195:(9): 847–849.
2179:
2156:
2104:
2075:(5): 605–611.
2055:
2026:(5): 723–734.
2003:
1943:
1892:
1865:(9): 623–635.
1849:
1795:
1743:
1717:
1668:
1661:
1639:
1599:
1584:
1555:(5): 283–296.
1535:
1476:
1460:
1408:
1406:
1403:
1402:
1401:
1396:
1391:
1386:
1381:
1374:
1371:
1357:
1354:
1348:
1345:
1277:
1274:
1271:
1270:
1267:
1264:
1261:
1260:
1257:
1254:
1251:
1250:
1247:
1244:
1241:
1240:
1238:
1233:
1229:
1228:
1226:
1224:
1221:
1220:
1219:Phospholipids
1217:
1214:
1211:
1210:
1207:
1204:
1201:
1200:
1198:
1193:
1189:
1188:
1186:
1184:
1181:
1180:
1177:
1174:
1171:
1170:
1167:
1164:
1161:
1160:
1157:
1154:
1151:
1150:
1148:
1143:
1139:
1138:
1136:
1134:
1131:
1130:
1127:
1124:
1121:
1120:
1117:
1114:
1111:
1110:
1107:
1104:
1101:
1100:
1097:
1094:
1091:
1090:
1087:
1080:
1077:
1076:
1073:
1066:
1063:
1062:
1059:
1056:
1053:
1052:
1050:
1045:
1041:
1040:
1038:
1036:
1033:
1032:
1029:
1026:
1023:
1022:
1019:
1016:
1013:
1012:
1010:
1005:
1001:
1000:
998:
996:
993:
992:
989:
986:
983:
982:
979:
976:
973:
972:
970:
965:
961:
960:
957:
954:
943:
940:
861:acid hydrolase
847:
844:
834:
822:
821:Pathogen entry
819:
764:EGRESS complex
712:
709:
588:
585:
530:centrifugation
493:
490:
444:(referring to
437:that uses the
356:
353:
247:cell signaling
202:) is a single
156:
155:
153:
152:
147:
142:
137:
123:
118:
113:
108:
103:
97:
92:
87:
81:
76:
69:
61:
60:
52:
51:
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
4776:
4765:
4762:
4760:
4757:
4755:
4752:
4750:
4747:
4745:
4742:
4741:
4739:
4724:
4721:
4719:
4716:
4715:
4713:
4709:
4703:
4700:
4698:
4695:
4691:
4688:
4686:
4683:
4682:
4681:
4678:
4674:
4671:
4669:
4666:
4664:
4661:
4660:
4659:
4656:
4654:
4651:
4650:
4648:
4644:
4638:
4635:
4631:
4628:
4626:
4623:
4621:
4620:Proteinoplast
4618:
4616:
4613:
4611:
4608:
4606:
4603:
4601:
4598:
4596:
4593:
4591:
4588:
4587:
4586:
4583:
4581:
4580:Mitochondrion
4578:
4577:
4575:
4573:
4572:Endosymbionts
4569:
4561:
4558:
4556:
4555:Lamellipodium
4553:
4552:
4551:
4548:
4544:
4541:
4539:
4536:
4534:
4531:
4529:
4526:
4525:
4524:
4521:
4519:
4516:
4512:
4509:
4507:
4504:
4502:
4499:
4497:
4494:
4493:
4492:
4489:
4487:
4484:
4482:
4479:
4477:
4474:
4472:
4471:Microfilament
4469:
4468:
4466:
4464:
4460:
4452:
4449:
4447:
4444:
4442:
4439:
4437:
4434:
4432:
4429:
4428:
4427:
4424:
4420:
4417:
4415:
4412:
4410:
4407:
4405:
4402:
4400:
4397:
4395:
4392:
4391:
4390:
4387:
4385:
4384:Autophagosome
4382:
4380:
4377:
4375:
4372:
4370:
4367:
4365:
4362:
4360:
4359:Cell membrane
4357:
4356:
4354:
4352:
4349:Endomembrane
4346:
4342:
4338:
4330:
4325:
4323:
4318:
4316:
4311:
4310:
4307:
4301:
4298:
4296:
4293:
4291:
4288:
4286:
4283:
4281:
4278:
4276:
4273:
4269:
4265:
4261:
4260:
4254:
4245:
4244:
4240:
4232:
4226:
4222:
4221:
4213:
4210:
4205:
4199:
4195:
4188:
4185:
4180:
4176:
4171:
4166:
4162:
4158:
4154:
4150:
4146:
4142:
4138:
4131:
4128:
4123:
4119:
4114:
4109:
4105:
4101:
4097:
4090:
4087:
4082:
4078:
4073:
4068:
4064:
4060:
4056:
4052:
4048:
4041:
4038:
4033:
4029:
4025:
4021:
4014:
4011:
4006:
4002:
3998:
3994:
3990:
3986:
3979:
3976:
3971:
3967:
3962:
3957:
3953:
3949:
3945:
3941:
3937:
3933:
3929:
3922:
3919:
3914:
3910:
3905:
3900:
3896:
3892:
3888:
3884:
3880:
3873:
3870:
3865:
3861:
3856:
3851:
3847:
3843:
3839:
3835:
3831:
3824:
3821:
3816:
3812:
3807:
3802:
3798:
3794:
3790:
3786:
3782:
3775:
3772:
3767:
3763:
3759:
3755:
3751:
3747:
3743:
3739:
3732:
3729:
3724:
3720:
3716:
3712:
3708:
3704:
3697:
3694:
3689:
3685:
3680:
3675:
3670:
3665:
3661:
3657:
3653:
3649:
3645:
3638:
3636:
3632:
3627:
3623:
3618:
3613:
3608:
3603:
3599:
3595:
3591:
3587:
3583:
3576:
3573:
3568:
3564:
3559:
3554:
3550:
3546:
3542:
3538:
3534:
3527:
3524:
3519:
3515:
3510:
3505:
3501:
3497:
3493:
3486:
3483:
3478:
3474:
3470:
3466:
3462:
3458:
3450:
3447:
3442:
3438:
3434:
3430:
3426:
3422:
3414:
3411:
3406:
3402:
3397:
3392:
3387:
3382:
3378:
3374:
3371:(8): e23852.
3370:
3366:
3362:
3355:
3352:
3347:
3343:
3339:
3335:
3330:
3325:
3321:
3317:
3309:
3306:
3301:
3297:
3293:
3289:
3285:
3281:
3277:
3273:
3266:
3263:
3258:
3254:
3250:
3246:
3242:
3238:
3234:
3230:
3223:
3220:
3215:
3211:
3207:
3203:
3199:
3195:
3191:
3187:
3180:
3177:
3172:
3168:
3163:
3158:
3154:
3150:
3146:
3142:
3138:
3131:
3128:
3123:
3119:
3115:
3109:
3105:
3101:
3097:
3090:
3087:
3082:
3078:
3074:
3070:
3066:
3062:
3055:
3052:
3047:
3043:
3039:
3033:
3029:
3022:
3019:
3014:
3010:
3005:
3000:
2996:
2992:
2988:
2984:
2980:
2973:
2970:
2965:
2959:
2955:
2948:
2945:
2940:
2936:
2931:
2926:
2922:
2918:
2914:
2910:
2906:
2899:
2896:
2891:
2887:
2882:
2877:
2873:
2869:
2865:
2858:
2855:
2850:
2846:
2842:
2838:
2834:
2830:
2823:
2820:
2815:
2811:
2806:
2801:
2797:
2793:
2789:
2781:
2778:
2773:
2769:
2764:
2759:
2755:
2751:
2747:
2743:
2739:
2732:
2729:
2724:
2720:
2715:
2710:
2706:
2702:
2698:
2694:
2690:
2683:
2680:
2675:
2671:
2666:
2661:
2657:
2653:
2649:
2645:
2641:
2634:
2632:
2628:
2623:
2617:
2612:
2611:
2602:
2600:
2596:
2591:
2587:
2583:
2579:
2575:
2571:
2564:
2561:
2556:
2550:
2546:
2539:
2537:
2533:
2528:
2524:
2519:
2514:
2510:
2506:
2502:
2498:
2494:
2487:
2484:
2479:
2475:
2470:
2465:
2461:
2457:
2453:
2449:
2445:
2438:
2435:
2430:
2426:
2422:
2418:
2414:
2410:
2406:
2399:
2396:
2391:
2385:
2381:
2374:
2371:
2366:
2362:
2357:
2352:
2347:
2342:
2338:
2334:
2330:
2323:
2320:
2316:
2310:
2307:
2302:
2298:
2294:
2290:
2286:
2282:
2275:
2272:
2267:
2263:
2258:
2253:
2249:
2245:
2241:
2237:
2233:
2226:
2223:
2218:
2214:
2210:
2206:
2202:
2198:
2194:
2190:
2183:
2180:
2175:
2171:
2167:
2160:
2157:
2152:
2148:
2143:
2138:
2134:
2130:
2126:
2122:
2118:
2111:
2109:
2105:
2100:
2096:
2091:
2086:
2082:
2078:
2074:
2070:
2066:
2059:
2056:
2051:
2047:
2042:
2037:
2033:
2029:
2025:
2021:
2017:
2010:
2008:
2004:
1999:
1995:
1991:
1987:
1982:
1977:
1973:
1969:
1965:
1961:
1957:
1950:
1948:
1944:
1939:
1935:
1930:
1925:
1920:
1915:
1911:
1907:
1903:
1896:
1893:
1888:
1884:
1880:
1876:
1872:
1868:
1864:
1860:
1853:
1850:
1845:
1841:
1836:
1831:
1826:
1821:
1817:
1813:
1809:
1802:
1800:
1796:
1791:
1787:
1782:
1777:
1773:
1769:
1765:
1761:
1757:
1750:
1748:
1744:
1731:
1727:
1721:
1718:
1713:
1709:
1704:
1699:
1695:
1691:
1687:
1683:
1679:
1672:
1669:
1664:
1658:
1654:
1650:
1643:
1640:
1635:
1631:
1626:
1621:
1617:
1613:
1606:
1604:
1600:
1595:
1588:
1585:
1580:
1576:
1571:
1566:
1562:
1558:
1554:
1550:
1546:
1539:
1536:
1531:
1527:
1522:
1517:
1512:
1507:
1503:
1499:
1495:
1491:
1487:
1480:
1477:
1474:
1470:
1464:
1461:
1456:
1452:
1447:
1442:
1437:
1432:
1428:
1424:
1420:
1413:
1410:
1404:
1400:
1397:
1395:
1392:
1390:
1387:
1385:
1382:
1380:
1377:
1376:
1372:
1370:
1368:
1363:
1355:
1353:
1346:
1344:
1342:
1338:
1334:
1330:
1326:
1322:
1320:
1319:amitriptyline
1316:
1312:
1308:
1304:
1300:
1294:
1292:
1287:
1283:
1275:
1268:
1265:
1263:
1262:
1258:
1255:
1253:
1252:
1248:
1245:
1243:
1242:
1239:
1237:
1234:
1231:
1230:
1227:
1225:
1223:
1222:
1218:
1215:
1213:
1212:
1208:
1205:
1203:
1202:
1199:
1197:
1194:
1191:
1190:
1187:
1185:
1183:
1182:
1178:
1176:C- Peptidase
1175:
1173:
1172:
1168:
1165:
1163:
1162:
1158:
1155:
1153:
1152:
1149:
1147:
1144:
1141:
1140:
1137:
1135:
1133:
1132:
1128:
1125:
1123:
1122:
1118:
1115:
1113:
1112:
1108:
1106:E- Lysozymes
1105:
1103:
1102:
1098:
1095:
1093:
1092:
1088:
1085:
1081:
1079:
1078:
1074:
1071:
1067:
1065:
1064:
1061:Galactosides
1060:
1057:
1055:
1054:
1051:
1049:
1046:
1043:
1042:
1039:
1037:
1035:
1034:
1030:
1027:
1025:
1024:
1020:
1017:
1015:
1014:
1011:
1009:
1006:
1003:
1002:
999:
997:
995:
994:
990:
987:
985:
984:
980:
977:
975:
974:
971:
969:
966:
963:
962:
958:
955:
952:
951:
946:
941:
939:
937:
932:
930:
926:
922:
920:
915:
913:
909:
905:
901:
897:
893:
889:
884:
882:
878:
874:
870:
866:
862:
857:
853:
845:
843:
841:
837:
829:
820:
818:
816:
815:late endosome
811:
809:
805:
801:
797:
793:
789:
785:
781:
777:
773:
769:
765:
761:
757:
753:
749:
745:
741:
736:
734:
730:
726:
717:
710:
708:
705:
703:
699:
695:
690:
688:
684:
680:
676:
672:
668:
663:
660:
656:
652:
648:
644:
640:
636:
631:
629:
625:
621:
617:
612:
610:
606:
602:
601:carbohydrates
598:
597:nucleic acids
594:
586:
584:
582:
578:
573:
571:
567:
563:
559:
555:
551:
545:
542:
538:
533:
531:
527:
523:
519:
515:
511:
507:
498:
491:
489:
487:
483:
482:
477:
473:
469:
465:
464:
459:
458:
453:
452:
447:
443:
440:
436:
430:
395:
362:
354:
352:
350:
349:aging-related
346:
342:
338:
334:
330:
326:
322:
321:nuclear genes
318:
313:
311:
307:
303:
299:
295:
291:
286:
284:
280:
276:
272:
268:
258:
254:
252:
248:
244:
240:
236:
232:
228:
224:
220:
217:
214:that contain
213:
209:
205:
199:
163:
151:
150:Cell membrane
148:
146:
143:
141:
138:
135:
131:
127:
124:
122:
119:
117:
116:Mitochondrion
114:
112:
109:
107:
104:
101:
98:
96:
93:
91:
88:
85:
82:
80:
77:
75:
72:
71:
67:
62:
59:
53:
50:
44:
41:
37:
33:
19:
4749:Cell anatomy
4600:Gerontoplast
4550:Pseudopodium
4543:Radial spoke
4523:Undulipodium
4463:Cytoskeleton
4398:
4379:Parenthesome
4268:the original
4258:
4219:
4212:
4193:
4187:
4144:
4140:
4130:
4103:
4099:
4089:
4054:
4050:
4040:
4023:
4019:
4013:
3988:
3984:
3978:
3935:
3931:
3921:
3886:
3882:
3872:
3837:
3833:
3823:
3788:
3784:
3774:
3741:
3737:
3731:
3706:
3702:
3696:
3651:
3647:
3589:
3585:
3575:
3540:
3536:
3526:
3499:
3495:
3485:
3460:
3457:Cell Calcium
3456:
3449:
3424:
3420:
3413:
3368:
3364:
3354:
3319:
3315:
3308:
3275:
3271:
3265:
3232:
3228:
3222:
3189:
3185:
3179:
3144:
3140:
3130:
3095:
3089:
3064:
3060:
3054:
3027:
3021:
2986:
2982:
2972:
2953:
2947:
2912:
2908:
2898:
2871:
2867:
2857:
2832:
2828:
2822:
2798:(1): 11–20.
2795:
2791:
2780:
2745:
2741:
2731:
2696:
2692:
2682:
2647:
2643:
2609:
2573:
2569:
2563:
2544:
2500:
2496:
2486:
2451:
2447:
2437:
2415:(1): 69–86.
2412:
2408:
2398:
2379:
2373:
2336:
2332:
2322:
2314:
2309:
2284:
2280:
2274:
2239:
2235:
2225:
2192:
2188:
2182:
2173:
2169:
2159:
2124:
2120:
2072:
2068:
2058:
2023:
2019:
1963:
1959:
1909:
1905:
1895:
1862:
1858:
1852:
1815:
1811:
1763:
1759:
1734:. Retrieved
1729:
1720:
1688:(1): 57–80.
1685:
1681:
1671:
1652:
1642:
1615:
1593:
1587:
1552:
1548:
1538:
1493:
1489:
1479:
1463:
1426:
1422:
1412:
1384:Cathelicidin
1359:
1350:
1323:
1295:
1279:
1235:
1206:A- Esterase
1195:
1145:
1086:Mannosidase
1083:
1072:Glucosidase
1069:
1047:
1007:
967:
945:
933:
923:
916:
908:osteoporosis
885:
849:
824:
812:
787:
783:
779:
775:
771:
767:
752:translocated
737:
722:
706:
691:
675:proton pumps
664:
632:
624:phagocytosis
613:
590:
574:
546:
534:
504:
479:
475:
471:
470:. The forms
467:
461:
455:
449:
441:
360:
358:
314:
287:
264:
223:biomolecules
161:
159:
139:
106:Cytoskeleton
48:Cell biology
40:
4702:Magnetosome
4668:Spliceosome
4595:Chromoplast
4590:Chloroplast
4481:Microtubule
3427:(1): 9–20.
2915:: 109–134.
1429:(6): 1299.
1299:haloperidol
879:, bone and
828:pinocytosis
778:elaying of
725:endocytosis
653:, from the
628:macrophages
343:, cancers,
271:endocytosis
4759:Organelles
4738:Categories
4697:Proteasome
4690:Inclusions
4637:Nitroplast
4630:Apicoplast
4615:Elaioplast
4610:Amyloplast
4605:Leucoplast
4560:Filopodium
4506:Basal body
4496:Centrosome
4446:Peroxisome
4441:Glyoxysome
4431:Melanosome
4341:organelles
1405:References
1379:Peroxisome
1367:exocytosis
1315:sertraline
1311:fluoxetine
1307:amantadine
1286:lipophilic
1236:Sulfatases
968:Phosphates
959:Substrate
694:cathepsins
659:hydrolases
616:phagosomes
581:cell death
537:microsomal
351:diseases.
337:substrates
216:hydrolytic
145:Centrosome
130:organelles
4718:Cell wall
4680:Cytoplasm
4653:Nucleolus
4625:Tannosome
4533:Flagellum
4518:Myofibril
4501:Centriole
4436:Microbody
4409:Phagosome
4194:Lysosomes
3324:CiteSeerX
3046:857764171
2742:Autophagy
2281:Autophagy
1736:4 October
1653:Lysosomes
1634:187669426
1337:Parkinson
1179:Peptides
1169:Collagen
1159:Proteins
1146:Proteases
1075:Glycogen
1008:Nucleases
881:cartilage
748:ribosomes
733:endosomes
711:Formation
641:. With a
620:autophagy
577:apoptosis
552:from the
492:Discovery
476:*lyosomal
468:lysosomal
435:Neo-Latin
359:The word
325:Mutations
285:in 1974.
275:autophagy
267:cytoplasm
243:apoptosis
134:cytoplasm
74:Nucleolus
18:Lysosomal
4744:Vesicles
4711:External
4663:Ribosome
4419:Acrosome
4404:Endosome
4399:Lysosome
4122:21980143
4081:12226358
4005:16006126
3970:10213780
3913:28235864
3864:27059595
3815:29866702
3766:23685272
3723:17643939
3688:27035940
3626:20439745
3567:24574503
3518:24771397
3477:26560688
3441:20502000
3405:21909365
3365:PLOS ONE
3346:18027916
3300:25784783
3257:24735371
3249:15997416
3206:10360127
3171:18504571
3013:30936119
2939:28502768
2890:23114583
2849:23510062
2814:23165354
2772:22647656
2723:21723623
2674:15827185
2590:17077125
2527:25720963
2478:23712550
2429:22335796
2365:19473965
2301:18567941
2266:13357540
2217:30307451
2209:16136179
2176:(9): 49.
2151:27125853
2127:: 2–12.
2099:23416930
2050:23185029
1998:20353685
1990:19556463
1938:24668941
1887:24493663
1879:19672277
1844:32597833
1790:30397314
1712:25668017
1579:23609508
1469:vacuoles
1455:33803964
1399:TMEM106B
1373:See also
1333:Ambroxol
1325:Ambroxol
956:Enzymes
671:membrane
593:peptides
472:*lyosome
361:lysosome
241:repair,
229:and its
212:vesicles
162:lysosome
140:Lysosome
84:Ribosome
32:Lysozyme
4685:Cytosol
4585:Plastid
4538:Axoneme
4414:Vacuole
4394:Exosome
4389:Vesicle
4364:Nucleus
4179:9596630
4161:3870657
3952:3870886
3904:5360485
3855:4868781
3806:6039264
3758:3787186
3679:4839468
3656:Bibcode
3617:2906830
3594:Bibcode
3558:3999713
3396:3166082
3373:Bibcode
3292:8532138
3214:7258546
3162:2711917
3122:7532261
3081:4606365
3004:6773515
2930:5828774
2881:4141347
2763:3378416
2714:3153126
2665:1082736
2518:4801004
2469:3664703
2356:2755904
2317:, 1959.
2257:2229688
2142:5081277
2090:3647216
2041:3514785
1968:Bibcode
1960:Science
1929:4031951
1835:7410054
1781:6277210
1703:4524569
1570:4387238
1498:Bibcode
1446:8001254
1423:Cancers
1362:vacuole
953:Sr. No
904:anaemia
877:viscera
854:(LSD),
840:cholera
786:osomal
740:nucleus
667:protons
647:cytosol
637:to 1.2
514:insulin
460:, from
235:stomach
231:lumenal
219:enzymes
126:Cytosol
121:Vacuole
90:Vesicle
79:Nucleus
36:lectins
4528:Cilium
4351:system
4227:
4200:
4177:
4170:144374
4167:
4159:
4120:
4079:
4072:161001
4069:
4003:
3968:
3961:144210
3958:
3950:
3911:
3901:
3862:
3852:
3813:
3803:
3764:
3756:
3721:
3686:
3676:
3624:
3614:
3565:
3555:
3516:
3475:
3439:
3403:
3393:
3344:
3326:
3298:
3290:
3255:
3247:
3212:
3204:
3169:
3159:
3120:
3110:
3079:
3044:
3034:
3011:
3001:
2960:
2937:
2927:
2888:
2878:
2847:
2812:
2770:
2760:
2721:
2711:
2672:
2662:
2618:
2588:
2551:
2525:
2515:
2476:
2466:
2427:
2386:
2363:
2353:
2299:
2264:
2254:
2215:
2207:
2149:
2139:
2097:
2087:
2048:
2038:
1996:
1988:
1936:
1926:
1885:
1877:
1842:
1832:
1788:
1778:
1710:
1700:
1659:
1632:
1577:
1567:
1528:
1521:392768
1518:
1471:, see
1453:
1443:
1339:'s or
1305:, and
605:lipids
603:, and
347:, and
249:, and
4673:Vault
4255:from
4157:JSTOR
3948:JSTOR
3762:S2CID
3537:Brain
3496:Brain
3296:S2CID
3253:S2CID
3210:S2CID
2213:S2CID
1994:S2CID
1883:S2CID
1630:S2CID
1530:28524
1317:, or
1284:with
1282:bases
1280:Weak
936:aging
873:brain
774:olgi
770:R-to-
760:COPII
687:ClC-7
566:light
486:typos
457:-some
451:lysis
446:lysis
442:lyso-
433:) is
208:cells
4337:cell
4264:NCBI
4225:ISBN
4198:ISBN
4175:PMID
4118:PMID
4077:PMID
4001:PMID
3966:PMID
3909:PMID
3860:PMID
3811:PMID
3754:PMID
3719:PMID
3684:PMID
3622:PMID
3563:PMID
3514:PMID
3473:PMID
3437:PMID
3401:PMID
3342:PMID
3288:PMID
3245:PMID
3202:PMID
3167:PMID
3118:PMID
3108:ISBN
3077:PMID
3042:OCLC
3032:ISBN
3009:PMID
2958:ISBN
2935:PMID
2886:PMID
2845:PMID
2810:PMID
2768:PMID
2719:PMID
2670:PMID
2616:ISBN
2586:PMID
2549:ISBN
2523:PMID
2474:PMID
2425:PMID
2384:ISBN
2361:PMID
2297:PMID
2262:PMID
2205:PMID
2147:PMID
2095:PMID
2046:PMID
1986:PMID
1934:PMID
1875:PMID
1840:PMID
1786:PMID
1738:2016
1708:PMID
1657:ISBN
1575:PMID
1526:PMID
1451:PMID
1082:C- α
1068:B- α
1031:DNA
1021:RNA
796:CLN8
794:and
792:CLN6
744:TFEB
677:and
673:via
651:cell
481:lyo-
474:and
463:soma
317:TFEB
302:CLN8
300:and
298:CLN6
4658:RNA
4165:PMC
4149:doi
4108:doi
4067:PMC
4059:doi
4055:111
4028:doi
3993:doi
3956:PMC
3940:doi
3899:PMC
3891:doi
3887:198
3850:PMC
3842:doi
3838:196
3801:PMC
3793:doi
3789:201
3746:doi
3711:doi
3674:PMC
3664:doi
3652:113
3612:PMC
3602:doi
3590:107
3553:PMC
3545:doi
3541:137
3504:doi
3500:137
3465:doi
3429:doi
3391:PMC
3381:doi
3334:doi
3280:doi
3237:doi
3233:113
3194:doi
3190:156
3157:PMC
3149:doi
3100:doi
3069:doi
2999:PMC
2991:doi
2925:PMC
2917:doi
2913:118
2876:PMC
2837:doi
2800:doi
2758:PMC
2750:doi
2709:PMC
2701:doi
2660:PMC
2652:doi
2578:doi
2574:119
2513:PMC
2505:doi
2464:PMC
2456:doi
2452:141
2417:doi
2351:PMC
2341:doi
2337:284
2289:doi
2252:PMC
2244:doi
2197:doi
2137:PMC
2129:doi
2085:PMC
2077:doi
2036:PMC
2028:doi
2024:199
1976:doi
1964:325
1924:PMC
1914:doi
1867:doi
1830:PMC
1820:doi
1816:130
1776:PMC
1768:doi
1698:PMC
1690:doi
1620:doi
1565:PMC
1557:doi
1516:PMC
1506:doi
1441:PMC
1431:doi
626:of
34:or
4740::
4339:/
4262:.
4173:.
4163:.
4155:.
4145:10
4143:.
4139:.
4116:.
4102:.
4098:.
4075:.
4065:.
4053:.
4049:.
4024:29
4022:.
3999:.
3989:15
3987:.
3964:.
3954:.
3946:.
3936:11
3934:.
3930:.
3907:.
3897:.
3885:.
3881:.
3858:.
3848:.
3836:.
3832:.
3809:.
3799:.
3787:.
3783:.
3760:.
3752:.
3742:24
3740:.
3717:.
3705:.
3682:.
3672:.
3662:.
3650:.
3646:.
3634:^
3620:.
3610:.
3600:.
3588:.
3584:.
3561:.
3551:.
3539:.
3535:.
3512:.
3498:.
3494:.
3471:.
3461:58
3459:.
3435:.
3425:26
3423:.
3399:.
3389:.
3379:.
3367:.
3363:.
3340:.
3332:.
3320:51
3318:.
3294:.
3286:.
3276:34
3274:.
3251:.
3243:.
3231:.
3208:.
3200:.
3188:.
3165:.
3155:.
3145:37
3143:.
3139:.
3116:.
3106:.
3075:.
3065:23
3063:.
3040:.
3007:.
2997:.
2987:12
2985:.
2981:.
2933:.
2923:.
2911:.
2907:.
2884:.
2872:14
2870:.
2866:.
2843:.
2833:18
2831:.
2808:.
2796:31
2794:.
2790:.
2766:.
2756:.
2744:.
2740:.
2717:.
2707:.
2697:34
2695:.
2691:.
2668:.
2658:.
2648:79
2646:.
2642:.
2630:^
2598:^
2584:.
2572:.
2535:^
2521:.
2511:.
2501:17
2499:.
2495:.
2472:.
2462:.
2450:.
2446:.
2423:.
2413:74
2411:.
2407:.
2359:.
2349:.
2335:.
2331:.
2295:.
2283:.
2260:.
2250:.
2238:.
2234:.
2211:.
2203:.
2191:.
2172:.
2168:.
2145:.
2135:.
2125:32
2123:.
2119:.
2107:^
2093:.
2083:.
2073:34
2071:.
2067:.
2044:.
2034:.
2022:.
2018:.
2006:^
1992:.
1984:.
1974:.
1962:.
1958:.
1946:^
1932:.
1922:.
1910:55
1908:.
1904:.
1881:.
1873:.
1863:10
1861:.
1838:.
1828:.
1814:.
1810:.
1798:^
1784:.
1774:.
1764:20
1762:.
1758:.
1746:^
1728:.
1706:.
1696:.
1686:77
1684:.
1680:.
1628:.
1618:.
1614:.
1602:^
1573:.
1563:.
1553:14
1551:.
1547:.
1524:.
1514:.
1504:.
1494:75
1492:.
1488:.
1449:.
1439:.
1427:13
1425:.
1421:.
1343:.
1321:.
1313:,
1301:,
1232:6
1192:5
1142:4
1044:3
1004:2
964:1
931:.
921:.
883:.
875:,
867:,
833:AB
735:.
681:.
643:pH
639:μm
635:μm
630:.
599:,
595:,
583:.
532:.
423:oʊ
411:aɪ
398:,
388:oʊ
382:oʊ
376:aɪ
323:.
253:.
245:,
192:oʊ
177:aɪ
160:A
4328:e
4321:t
4314:v
4233:.
4206:.
4181:.
4151::
4124:.
4110::
4104:5
4083:.
4061::
4034:.
4030::
4007:.
3995::
3972:.
3942::
3915:.
3893::
3866:.
3844::
3817:.
3795::
3768:.
3748::
3725:.
3713::
3707:6
3690:.
3666::
3658::
3628:.
3604::
3596::
3569:.
3547::
3520:.
3506::
3479:.
3467::
3443:.
3431::
3407:.
3383::
3375::
3369:6
3348:.
3336::
3302:.
3282::
3259:.
3239::
3216:.
3196::
3173:.
3151::
3124:.
3102::
3083:.
3071::
3048:.
3015:.
2993::
2966:.
2941:.
2919::
2892:.
2851:.
2839::
2816:.
2802::
2784:.
2774:.
2752::
2746:8
2725:.
2703::
2676:.
2654::
2624:.
2592:.
2580::
2557:.
2529:.
2507::
2480:.
2458::
2431:.
2419::
2392:.
2367:.
2343::
2303:.
2291::
2285:4
2268:.
2246::
2240:2
2219:.
2199::
2193:7
2174:3
2153:.
2131::
2101:.
2079::
2052:.
2030::
2000:.
1978::
1970::
1940:.
1916::
1889:.
1869::
1846:.
1822::
1792:.
1770::
1740:.
1714:.
1692::
1665:.
1636:.
1622::
1581:.
1559::
1532:.
1508::
1500::
1457:.
1433::
1084:-
1070:-
835:5
788:s
784:s
780:e
776:r
772:G
768:E
766:(
607:(
501:.
429:/
426:m
420:z
417:ə
414:z
408:l
405:ˈ
402:/
394:/
391:m
385:s
379:s
373:l
370:ˈ
367:/
363:(
198:/
195:m
189:s
186:ˌ
183:ə
180:s
174:l
171:ˈ
168:/
164:(
136:)
38:.
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.