987:
834:
677:
512:
379:
213:
32:
1651:
Schubert WD, Gobel G, Diepholz M, Darji A, Kloer D, Hain T, Chakraborty T, Wehland J, Domann E, Heinz DW (September 2001). "Internalins from the human pathogen
Listeria monocytogenes combine three distinct folds into a contiguous internalin domain".
1323:. Their function appears to be mainly structural: They are fused to the C-terminal end of leucine-rich repeats, significantly stabilising the LRR, and forming a common rigid entity with the LRR. They are themselves not involved in
1193:, composed of many such repeats, has a horseshoe shape with an interior parallel beta sheet and an exterior array of helices. One face of the beta sheet and one side of the helix array are exposed to
1736:
Freiberg A, Machner MP, Pfeil W, Schubert WD, Heinz DW, Seckler R (March 2004). "Folding and stability of the leucine-rich repeat domain of internalin B from
Listeri monocytogenes".
2438:
1890:
1099:
945:
792:
635:
470:
337:
171:
2101:
881:
728:
559:
2431:
2298:
1952:
1047:
893:
740:
571:
273:
107:
1883:
682:
1566:"slit: an extracellular protein necessary for development of midline glia and commissural axon pathways contains both EGF and LRR domains"
1437:
Enkhbayar P, Kamiya M, Osaki M, Matsumoto T, Matsushima N (February 2004). "Structural principles of leucine-rich repeat (LRR) proteins".
2424:
2348:
1858:
1853:
2293:
1783:
1224:
Leucine-rich repeat motifs have been identified in a large number of functionally unrelated proteins. The best-known example is the
1608:
Peters JW, Stowell MH, Rees DC (December 1996). "A leucine-rich repeat variant with a novel repetitive protein structural motif".
1358:
have a two-domain structure, composed of a small N-terminal domain containing a cluster of four
Cysteine residues that houses the
1844:
1324:
1213:
2731:
2308:
1876:
1517:"A leucine-rich repeat peptide derived from the Drosophila Toll receptor forms extended filaments with a beta-sheet structure"
2741:
2216:
1689:"Structure of internalin, a major invasion protein of Listeria monocytogenes, in complex with its human receptor E-cadherin"
1119:
965:
812:
655:
490:
357:
191:
2171:
2094:
1687:
Schubert WD, Urbanke C, Ziehm T, Beier V, Machner MP, Domann E, Wehland J, Chakraborty T, Heinz DW (December 2002).
2447:
2360:
2087:
1987:
1247:
Although the canonical LRR protein contains approximately one helix for every beta strand, variants that form
1107:
953:
800:
643:
478:
345:
179:
2068:
1928:
1170:
1248:
1244:
possesses 10 successive LRR motifs which serve to bind pathogen- and danger-associated molecular patterns.
2002:
1899:
1319:
1225:
1166:
37:
2736:
2523:
2251:
1103:
949:
796:
639:
474:
341:
175:
886:
733:
564:
1060:
906:
753:
584:
286:
120:
1982:
1863:
1359:
1347:
1343:
is, however, much smaller than in most standard Ig-like domains, making it somewhat of an outlier.
1182:
591:
426:
293:
127:
2400:
2318:
1967:
1718:
1633:
1546:
1462:
1241:
1237:
1150:
2579:
2471:
2395:
2016:
1992:
1826:
1779:
1753:
1710:
1669:
1625:
1587:
1538:
1497:
1454:
1419:
1190:
1094:
940:
787:
630:
465:
332:
166:
1480:
Kobe B, Kajava AV (December 2001). "The leucine-rich repeat as a protein recognition motif".
2574:
2497:
2385:
1947:
1816:
1806:
1745:
1700:
1661:
1617:
1577:
1528:
1489:
1446:
1411:
1402:
Kobe B, Deisenhofer J (October 1994). "The leucine-rich repeat: a versatile binding motif".
1255:
1202:
1143:
1848:
1086:
932:
779:
622:
457:
324:
158:
2600:
2564:
2466:
88:
1795:"LRRML: a conformational database and an XML description of leucine-rich repeats (LRRs)"
2605:
2533:
2502:
2461:
2323:
2186:
2110:
2021:
1942:
1937:
1918:
1821:
1794:
1383:
1332:
1328:
1290:
1705:
1688:
1493:
2725:
2569:
2256:
1957:
1533:
1516:
1415:
1339:
that follow the classical connectivity of Ig-domains. The beta strands in one of the
1251:
folds sometimes have long loops rather than helices linking successive beta strands.
1052:
898:
745:
576:
278:
112:
1722:
1637:
1550:
1466:
1015:
862:
709:
540:
407:
241:
63:
2487:
2338:
2261:
2246:
2206:
2141:
2052:
1363:
1304:
1233:
1082:
928:
775:
618:
453:
320:
154:
17:
1028:
596:
431:
254:
76:
2717:
2703:
2689:
2675:
2661:
2647:
1040:
874:
721:
552:
419:
266:
100:
2528:
2492:
2405:
2390:
2313:
2303:
2283:
2278:
2273:
2236:
2191:
2166:
2151:
2146:
2131:
2036:
2026:
2011:
1972:
1913:
1336:
1300:
1229:
1198:
1186:
1158:
2610:
2554:
2538:
2507:
2380:
2375:
2370:
2355:
2343:
2328:
2268:
2241:
2231:
2226:
2221:
2211:
2181:
2176:
2161:
2156:
2136:
2126:
2031:
1962:
1749:
1340:
1312:
1308:
1283:
1279:
1267:
1259:
1178:
1154:
991:
a leucine-rich repeat variant with a novel repetitive protein structural motif
217:
a leucine-rich repeat variant with a novel repetitive protein structural motif
2416:
298:
132:
2626:
2595:
2365:
2288:
2201:
2196:
2121:
1315:
1830:
1811:
1793:
Wei T, Gong J, Jamitzky F, Heckl WM, Stark RW, Roessle SC (November 2008).
1757:
1714:
1673:
1665:
1582:
1565:
1501:
1458:
1629:
1591:
1564:
Rothberg JM, Jacobs JR, Goodman CS, Artavanis-Tsakonas S (December 1990).
1542:
1423:
1056:
986:
902:
833:
749:
676:
580:
511:
378:
282:
212:
116:
31:
2713:
2699:
2685:
2671:
2657:
2643:
1868:
1286:
1035:
869:
716:
547:
414:
261:
95:
1621:
1450:
1355:
1351:
1294:
1194:
1162:
1140:
1367:
1206:
1201:
residues. The region between the helices and sheets is the protein's
1114:
960:
807:
650:
485:
352:
186:
1327:
but help to present the adjacent LRR-domain for this purpose. These
1254:
One leucine-rich repeat variant domain (LRV) has a novel repetitive
1299:
They also co-occur with LRR adjacent domains. These are small, all
2079:
1371:
1212:
Leucine-rich repeats are frequently involved in the formation of
2709:
2695:
2681:
2667:
2653:
2639:
1076:
1022:
1010:
922:
857:
769:
704:
612:
535:
447:
402:
314:
248:
236:
148:
83:
70:
58:
2420:
2083:
1872:
1354:
containing the leucine-rich repeat variant domain (LRV). These
1266:
arranged in a right-handed superhelix, with the absence of the
1362:, and a larger C-terminal domain containing the LRV repeats.
383:
internalin h: crystal structure of fused n-terminal domains.
685:), a leucine rich repeat protein involved in plant defense
1366:
studies revealed that the 4Fe:4S cluster is sensitive to
1864:
LRRML: a conformational database of leucine-rich repeats
1515:
Gay NJ, Packman LC, Weldon MA, Barna JC (October 1991).
516:
dimeric bovine tissue-extracted decorin, crystal form 2
36:
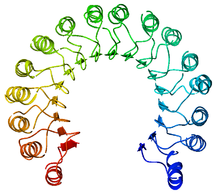
An example of a leucine-rich repeat protein, a porcine
2708:
This article incorporates text from the public domain
2694:
This article incorporates text from the public domain
2680:
This article incorporates text from the public domain
2666:
This article incorporates text from the public domain
2652:
This article incorporates text from the public domain
2638:
This article incorporates text from the public domain
1311:(InlA) and related proteins InlB, InlE, InlH from the
2619:
2588:
2547:
2516:
2480:
2454:
2061:
2045:
2001:
1927:
1906:
1113:
1093:
1075:
1070:
1046:
1034:
1021:
1009:
1001:
996:
979:
959:
939:
921:
916:
892:
880:
868:
856:
848:
843:
826:
806:
786:
768:
763:
739:
727:
715:
703:
695:
690:
669:
649:
629:
611:
606:
590:
570:
558:
546:
534:
526:
521:
504:
484:
464:
446:
441:
425:
413:
401:
393:
388:
371:
351:
331:
313:
308:
292:
272:
260:
247:
235:
227:
222:
205:
185:
165:
147:
142:
126:
106:
94:
82:
69:
57:
49:
44:
1335:domains in that they consist of two sandwiched
1778:(2nd ed.). New York: Garland Publishing.
2432:
2095:
1884:
1603:
1601:
8:
2439:
2425:
2417:
2102:
2088:
2080:
1891:
1877:
1869:
1278:Leucine-rich repeats are often flanked by
1067:
985:
913:
832:
760:
675:
603:
510:
438:
377:
305:
211:
139:
30:
1820:
1810:
1704:
1581:
1532:
1370:, but does not appear to have reversible
1157:stretches that are unusually rich in the
1774:Tooze, John; Brändén, Carl-Ivar (1999).
1394:
1270:present in other leucine-rich repeats.
1859:CATH Alpha-beta horseshoe architecture
976:
823:
666:
501:
368:
202:
26:
1953:Transcription activator-like effector
1293:, but not always as is the case with
1258:consisting of alternating alpha- and
827:Leucine rich repeat C-terminal domain
670:Leucine rich repeat N-terminal domain
505:Leucine rich repeat N-terminal domain
7:
1153:. It is composed of repeating 20–30
683:polygalacturonase inhibiting protein
1350:is found at the N-terminus of some
1240:also share the motif. In fact, the
838:third lrr domain of drosophila slit
1177:. Typically, each repeat unit has
25:
1776:Introduction to Protein Structure
1228:, but other proteins such as the
1169:commonly fold together to form a
1845:Eukaryotic Linear Motif resource
2472:Structure determination methods
1197:and are therefore dominated by
681:the crystal structure of pgip (
1209:packed with leucine residues.
1:
1706:10.1016/S0092-8674(02)01136-4
1494:10.1016/S0959-440X(01)00266-4
1189:structure, and the assembled
1071:Available protein structures:
917:Available protein structures:
764:Available protein structures:
607:Available protein structures:
442:Available protein structures:
309:Available protein structures:
143:Available protein structures:
1534:10.1016/0014-5793(91)81110-T
1416:10.1016/0968-0004(94)90090-6
1325:protein-protein-interactions
1214:protein–protein interactions
206:Leucine rich repeat variant
2758:
2707:
2693:
2679:
2665:
2651:
2637:
2448:Protein tertiary structure
1307:described for the protein
1175:leucine-rich repeat domain
2117:
1750:10.1016/j.jmb.2004.01.044
1303:domains, which have been
1066:
984:
912:
831:
759:
674:
602:
509:
437:
376:
304:
210:
138:
29:
1988:Tetratricopeptide repeat
1482:Curr. Opin. Struct. Biol
1331:belong to the family of
980:LRV protein FeS4 cluster
2069:Repeated sequence (DNA)
1171:solenoid protein domain
2732:Protein tandem repeats
1900:Protein tandem repeats
1812:10.1186/1472-6807-8-47
1666:10.1006/jmbi.2001.4989
1583:10.1101/gad.4.12a.2169
1320:Listeria monocytogenes
1226:ribonuclease inhibitor
38:ribonuclease inhibitor
2742:Protein superfamilies
2524:Immunoglobulin domain
1249:beta-alpha superhelix
1348:iron sulphur cluster
2560:Leucine-rich repeat
1983:Pentapeptide repeat
1978:Leucine-rich repeat
1849:LIG_SCF_Skp2-Cks1_1
1622:10.1038/nsb1296-991
1404:Trends Biochem. Sci
1133:leucine-rich repeat
18:Leucine rich repeat
2046:Beads-on-a-string:
1968:Antifreeze protein
1451:10.1002/prot.10605
1274:Associated domains
1242:toll-like receptor
1238:toll-like receptor
2635:
2634:
2580:Trefoil knot fold
2462:Structural domain
2414:
2413:
2077:
2076:
2017:Beta trefoil fold
1993:Trefoil knot fold
1610:Nat. Struct. Biol
1129:
1128:
1125:
1124:
1120:structure summary
975:
974:
971:
970:
966:structure summary
822:
821:
818:
817:
813:structure summary
665:
664:
661:
660:
656:structure summary
500:
499:
496:
495:
491:structure summary
367:
366:
363:
362:
358:structure summary
201:
200:
197:
196:
192:structure summary
16:(Redirected from
2749:
2620:Irregular folds:
2575:Thioredoxin fold
2498:Homeodomain fold
2441:
2434:
2427:
2418:
2104:
2097:
2090:
2081:
1948:Armadillo repeat
1893:
1886:
1879:
1870:
1834:
1824:
1814:
1799:BMC Struct. Biol
1789:
1762:
1761:
1733:
1727:
1726:
1708:
1684:
1678:
1677:
1648:
1642:
1641:
1605:
1596:
1595:
1585:
1576:(12A): 2169–87.
1561:
1555:
1554:
1536:
1512:
1506:
1505:
1477:
1471:
1470:
1434:
1428:
1427:
1399:
1256:structural motif
1203:hydrophobic core
1144:structural motif
1068:
989:
977:
914:
836:
824:
761:
679:
667:
604:
514:
502:
439:
381:
369:
306:
215:
203:
140:
34:
27:
21:
2757:
2756:
2752:
2751:
2750:
2748:
2747:
2746:
2722:
2721:
2720:
2706:
2692:
2678:
2664:
2650:
2636:
2631:
2615:
2601:Ferredoxin fold
2584:
2565:Flavodoxin fold
2543:
2512:
2476:
2467:Protein folding
2450:
2445:
2415:
2410:
2113:
2111:Protein domains
2108:
2078:
2073:
2057:
2041:
1997:
1923:
1902:
1897:
1841:
1792:
1786:
1773:
1770:
1768:Further reading
1765:
1735:
1734:
1730:
1686:
1685:
1681:
1650:
1649:
1645:
1607:
1606:
1599:
1563:
1562:
1558:
1514:
1513:
1509:
1479:
1478:
1474:
1436:
1435:
1431:
1401:
1400:
1396:
1392:
1380:
1276:
1263:
1222:
1205:and is tightly
992:
839:
686:
517:
384:
218:
40:
23:
22:
15:
12:
11:
5:
2755:
2753:
2745:
2744:
2739:
2734:
2724:
2723:
2633:
2632:
2630:
2629:
2623:
2621:
2617:
2616:
2614:
2613:
2608:
2606:Ribonuclease A
2603:
2598:
2592:
2590:
2586:
2585:
2583:
2582:
2577:
2572:
2567:
2562:
2557:
2551:
2549:
2545:
2544:
2542:
2541:
2536:
2534:Beta-propeller
2531:
2526:
2520:
2518:
2514:
2513:
2511:
2510:
2505:
2503:Alpha solenoid
2500:
2495:
2490:
2484:
2482:
2478:
2477:
2475:
2474:
2469:
2464:
2458:
2456:
2452:
2451:
2446:
2444:
2443:
2436:
2429:
2421:
2412:
2411:
2409:
2408:
2403:
2398:
2393:
2388:
2383:
2378:
2373:
2368:
2363:
2358:
2353:
2352:
2351:
2341:
2336:
2331:
2326:
2321:
2316:
2311:
2306:
2301:
2296:
2291:
2286:
2281:
2276:
2271:
2266:
2265:
2264:
2259:
2254:
2249:
2239:
2234:
2229:
2224:
2219:
2214:
2209:
2204:
2199:
2194:
2189:
2184:
2179:
2174:
2169:
2164:
2159:
2154:
2149:
2144:
2139:
2134:
2129:
2124:
2118:
2115:
2114:
2109:
2107:
2106:
2099:
2092:
2084:
2075:
2074:
2072:
2071:
2065:
2063:
2059:
2058:
2056:
2055:
2049:
2047:
2043:
2042:
2040:
2039:
2034:
2029:
2024:
2022:Beta-propeller
2019:
2014:
2008:
2006:
1999:
1998:
1996:
1995:
1990:
1985:
1980:
1975:
1970:
1965:
1960:
1955:
1950:
1945:
1943:Ankyrin repeat
1940:
1938:Alpha solenoid
1934:
1932:
1925:
1924:
1922:
1921:
1919:Collagen helix
1916:
1910:
1908:
1904:
1903:
1898:
1896:
1895:
1888:
1881:
1873:
1867:
1866:
1861:
1856:
1851:
1840:
1839:External links
1837:
1836:
1835:
1790:
1784:
1769:
1766:
1764:
1763:
1728:
1679:
1643:
1597:
1556:
1507:
1472:
1445:(3): 394–403.
1429:
1410:(10): 415–21.
1393:
1391:
1388:
1387:
1386:
1384:Leucine zipper
1379:
1376:
1360:4Fe:4S cluster
1275:
1272:
1261:
1221:
1218:
1167:tandem repeats
1146:that forms an
1127:
1126:
1123:
1122:
1117:
1111:
1110:
1097:
1091:
1090:
1080:
1073:
1072:
1064:
1063:
1050:
1044:
1043:
1038:
1032:
1031:
1026:
1019:
1018:
1013:
1007:
1006:
1003:
999:
998:
994:
993:
990:
982:
981:
973:
972:
969:
968:
963:
957:
956:
943:
937:
936:
926:
919:
918:
910:
909:
896:
890:
889:
884:
878:
877:
872:
866:
865:
860:
854:
853:
850:
846:
845:
841:
840:
837:
829:
828:
820:
819:
816:
815:
810:
804:
803:
790:
784:
783:
773:
766:
765:
757:
756:
743:
737:
736:
731:
725:
724:
719:
713:
712:
707:
701:
700:
697:
693:
692:
688:
687:
680:
672:
671:
663:
662:
659:
658:
653:
647:
646:
633:
627:
626:
616:
609:
608:
600:
599:
594:
588:
587:
574:
568:
567:
562:
556:
555:
550:
544:
543:
538:
532:
531:
528:
524:
523:
519:
518:
515:
507:
506:
498:
497:
494:
493:
488:
482:
481:
468:
462:
461:
451:
444:
443:
435:
434:
429:
423:
422:
417:
411:
410:
405:
399:
398:
395:
391:
390:
386:
385:
382:
374:
373:
365:
364:
361:
360:
355:
349:
348:
335:
329:
328:
318:
311:
310:
302:
301:
296:
290:
289:
276:
270:
269:
264:
258:
257:
252:
245:
244:
239:
233:
232:
229:
225:
224:
220:
219:
216:
208:
207:
199:
198:
195:
194:
189:
183:
182:
169:
163:
162:
152:
145:
144:
136:
135:
130:
124:
123:
110:
104:
103:
98:
92:
91:
86:
80:
79:
74:
67:
66:
61:
55:
54:
51:
47:
46:
42:
41:
35:
24:
14:
13:
10:
9:
6:
4:
3:
2:
2754:
2743:
2740:
2738:
2735:
2733:
2730:
2729:
2727:
2719:
2715:
2711:
2705:
2701:
2697:
2691:
2687:
2683:
2677:
2673:
2669:
2663:
2659:
2655:
2649:
2645:
2641:
2628:
2625:
2624:
2622:
2618:
2612:
2611:SH2-like fold
2609:
2607:
2604:
2602:
2599:
2597:
2594:
2593:
2591:
2587:
2581:
2578:
2576:
2573:
2571:
2570:Rossmann fold
2568:
2566:
2563:
2561:
2558:
2556:
2553:
2552:
2550:
2546:
2540:
2537:
2535:
2532:
2530:
2527:
2525:
2522:
2521:
2519:
2515:
2509:
2506:
2504:
2501:
2499:
2496:
2494:
2491:
2489:
2486:
2485:
2483:
2479:
2473:
2470:
2468:
2465:
2463:
2460:
2459:
2457:
2453:
2449:
2442:
2437:
2435:
2430:
2428:
2423:
2422:
2419:
2407:
2404:
2402:
2399:
2397:
2394:
2392:
2389:
2387:
2384:
2382:
2379:
2377:
2374:
2372:
2369:
2367:
2364:
2362:
2359:
2357:
2354:
2350:
2347:
2346:
2345:
2342:
2340:
2337:
2335:
2332:
2330:
2327:
2325:
2322:
2320:
2317:
2315:
2312:
2310:
2307:
2305:
2302:
2300:
2297:
2295:
2292:
2290:
2287:
2285:
2282:
2280:
2277:
2275:
2272:
2270:
2267:
2263:
2260:
2258:
2255:
2253:
2250:
2248:
2245:
2244:
2243:
2240:
2238:
2235:
2233:
2230:
2228:
2225:
2223:
2220:
2218:
2215:
2213:
2210:
2208:
2205:
2203:
2200:
2198:
2195:
2193:
2190:
2188:
2185:
2183:
2180:
2178:
2175:
2173:
2170:
2168:
2165:
2163:
2160:
2158:
2155:
2153:
2150:
2148:
2145:
2143:
2140:
2138:
2135:
2133:
2130:
2128:
2125:
2123:
2120:
2119:
2116:
2112:
2105:
2100:
2098:
2093:
2091:
2086:
2085:
2082:
2070:
2067:
2066:
2064:
2060:
2054:
2051:
2050:
2048:
2044:
2038:
2035:
2033:
2030:
2028:
2025:
2023:
2020:
2018:
2015:
2013:
2010:
2009:
2007:
2004:
2000:
1994:
1991:
1989:
1986:
1984:
1981:
1979:
1976:
1974:
1971:
1969:
1966:
1964:
1961:
1959:
1958:Beta solenoid
1956:
1954:
1951:
1949:
1946:
1944:
1941:
1939:
1936:
1935:
1933:
1930:
1926:
1920:
1917:
1915:
1912:
1911:
1909:
1905:
1901:
1894:
1889:
1887:
1882:
1880:
1875:
1874:
1871:
1865:
1862:
1860:
1857:
1855:
1854:SCOP LRR fold
1852:
1850:
1846:
1843:
1842:
1838:
1832:
1828:
1823:
1818:
1813:
1808:
1804:
1800:
1796:
1791:
1787:
1785:0-8153-2305-0
1781:
1777:
1772:
1771:
1767:
1759:
1755:
1751:
1747:
1744:(2): 453–61.
1743:
1739:
1732:
1729:
1724:
1720:
1716:
1712:
1707:
1702:
1699:(6): 825–36.
1698:
1694:
1690:
1683:
1680:
1675:
1671:
1667:
1663:
1660:(4): 783–94.
1659:
1655:
1647:
1644:
1639:
1635:
1631:
1627:
1623:
1619:
1616:(12): 991–4.
1615:
1611:
1604:
1602:
1598:
1593:
1589:
1584:
1579:
1575:
1571:
1567:
1560:
1557:
1552:
1548:
1544:
1540:
1535:
1530:
1526:
1522:
1518:
1511:
1508:
1503:
1499:
1495:
1491:
1488:(6): 725–32.
1487:
1483:
1476:
1473:
1468:
1464:
1460:
1456:
1452:
1448:
1444:
1440:
1433:
1430:
1425:
1421:
1417:
1413:
1409:
1405:
1398:
1395:
1389:
1385:
1382:
1381:
1377:
1375:
1373:
1369:
1365:
1361:
1357:
1353:
1349:
1344:
1342:
1338:
1334:
1330:
1326:
1322:
1321:
1317:
1314:
1310:
1306:
1302:
1297:
1296:
1292:
1288:
1285:
1281:
1273:
1271:
1269:
1265:
1257:
1252:
1250:
1245:
1243:
1239:
1235:
1231:
1227:
1219:
1217:
1215:
1210:
1208:
1204:
1200:
1196:
1192:
1188:
1184:
1180:
1176:
1172:
1168:
1164:
1160:
1156:
1152:
1149:
1148:α/β horseshoe
1145:
1142:
1138:
1134:
1121:
1118:
1116:
1112:
1109:
1105:
1101:
1098:
1096:
1092:
1088:
1084:
1081:
1078:
1074:
1069:
1065:
1062:
1058:
1054:
1051:
1049:
1045:
1042:
1039:
1037:
1033:
1030:
1027:
1024:
1020:
1017:
1014:
1012:
1008:
1004:
1000:
995:
988:
983:
978:
967:
964:
962:
958:
955:
951:
947:
944:
942:
938:
934:
930:
927:
924:
920:
915:
911:
908:
904:
900:
897:
895:
891:
888:
885:
883:
879:
876:
873:
871:
867:
864:
861:
859:
855:
851:
847:
842:
835:
830:
825:
814:
811:
809:
805:
802:
798:
794:
791:
789:
785:
781:
777:
774:
771:
767:
762:
758:
755:
751:
747:
744:
742:
738:
735:
732:
730:
726:
723:
720:
718:
714:
711:
708:
706:
702:
698:
694:
689:
684:
678:
673:
668:
657:
654:
652:
648:
645:
641:
637:
634:
632:
628:
624:
620:
617:
614:
610:
605:
601:
598:
595:
593:
589:
586:
582:
578:
575:
573:
569:
566:
563:
561:
557:
554:
551:
549:
545:
542:
539:
537:
533:
529:
525:
520:
513:
508:
503:
492:
489:
487:
483:
480:
476:
472:
469:
467:
463:
459:
455:
452:
449:
445:
440:
436:
433:
430:
428:
424:
421:
418:
416:
412:
409:
406:
404:
400:
396:
392:
387:
380:
375:
370:
359:
356:
354:
350:
347:
343:
339:
336:
334:
330:
326:
322:
319:
316:
312:
307:
303:
300:
297:
295:
291:
288:
284:
280:
277:
275:
271:
268:
265:
263:
259:
256:
253:
250:
246:
243:
240:
238:
234:
230:
226:
221:
214:
209:
204:
193:
190:
188:
184:
181:
177:
173:
170:
168:
164:
160:
156:
153:
150:
146:
141:
137:
134:
131:
129:
125:
122:
118:
114:
111:
109:
105:
102:
99:
97:
93:
90:
87:
85:
81:
78:
75:
72:
68:
65:
62:
60:
56:
52:
48:
43:
39:
33:
28:
19:
2737:LRR proteins
2559:
2517:All-β folds:
2488:Helix bundle
2481:All-α folds:
2333:
2053:Sushi domain
1977:
1847:motif class
1802:
1798:
1775:
1741:
1738:J. Mol. Biol
1737:
1731:
1696:
1692:
1682:
1657:
1654:J. Mol. Biol
1653:
1646:
1613:
1609:
1573:
1569:
1559:
1527:(1): 87–91.
1524:
1520:
1510:
1485:
1481:
1475:
1442:
1438:
1432:
1407:
1403:
1397:
1345:
1318:
1305:structurally
1298:
1277:
1253:
1246:
1234:tropomodulin
1223:
1211:
1174:
1147:
1136:
1132:
1130:
397:LRR_adjacent
372:LRR adjacent
2529:Beta barrel
2493:Globin fold
2406:zinc finger
2037:WD40 repeat
2027:Kelch motif
2012:Beta barrel
1973:HEAT repeat
1914:Coiled coil
1364:Biochemical
1337:beta sheets
1301:beta strand
1268:beta-sheets
1230:tropomyosin
1199:hydrophilic
1187:alpha helix
1179:beta strand
1161:amino acid
1159:hydrophobic
997:Identifiers
844:Identifiers
691:Identifiers
522:Identifiers
389:Identifiers
223:Identifiers
45:Identifiers
2726:Categories
2589:α+β folds:
2555:TIM barrel
2548:α/β folds:
2539:Beta helix
2508:Death fold
2032:TIM barrel
1963:Beta helix
1390:References
1374:activity.
1313:pathogenic
1309:Internalin
1284:C-terminal
1280:N-terminal
1232:regulator
1207:sterically
1155:amino acid
1083:structures
929:structures
776:structures
619:structures
592:Membranome
454:structures
427:Membranome
321:structures
294:Membranome
155:structures
128:Membranome
2718:IPR004830
2704:IPR004830
2690:IPR000483
2676:IPR000372
2662:IPR013210
2648:IPR012569
2627:Conotoxin
2596:DNA clamp
2294:EcoEI_R_C
2062:See also:
1929:Elongated
1805:(1): 47.
1570:Genes Dev
1521:FEBS Lett
1316:bacterium
1173:, termed
1041:IPR008665
875:IPR000483
722:IPR013210
553:IPR000372
420:IPR012569
267:IPR004830
101:IPR001611
2714:InterPro
2700:InterPro
2686:InterPro
2672:InterPro
2658:InterPro
2644:InterPro
1907:Fibrous:
1831:18986514
1758:15003459
1723:17232767
1715:12526809
1674:11575932
1638:36535731
1551:84294221
1502:11751054
1467:19951452
1459:14747988
1439:Proteins
1378:See also
1356:proteins
1352:proteins
1287:cysteine
1264:-helices
1236:and the
1220:Examples
1165:. These
1100:RCSB PDB
1036:InterPro
946:RCSB PDB
870:InterPro
793:RCSB PDB
717:InterPro
636:RCSB PDB
548:InterPro
471:RCSB PDB
415:InterPro
338:RCSB PDB
262:InterPro
172:RCSB PDB
96:InterPro
2455:General
2324:Kringle
2217:CGI-121
2187:BTB/POZ
1822:2645405
1630:8946850
1592:2176636
1543:1657640
1424:7817399
1333:Ig-like
1329:domains
1295:C5orf36
1291:domains
1195:solvent
1163:leucine
1141:protein
1139:) is a
1016:PF05484
1005:LRV_FeS
863:PF01463
710:PF08263
699:LRRNT_2
541:PF01462
408:PF08191
242:PF01816
89:207.1.1
64:PF00560
2003:Closed
1829:
1819:
1782:
1756:
1721:
1713:
1672:
1636:
1628:
1590:
1549:
1541:
1500:
1465:
1457:
1422:
1368:oxygen
1341:sheets
1289:-rich
1191:domain
1115:PDBsum
1089:
1079:
1061:SUPFAM
1029:CL0020
1002:Symbol
961:PDBsum
935:
925:
907:SUPFAM
849:Symbol
808:PDBsum
782:
772:
754:SUPFAM
696:Symbol
651:PDBsum
625:
615:
585:SUPFAM
527:Symbol
486:PDBsum
460:
450:
394:Symbol
353:PDBsum
327:
317:
287:SUPFAM
255:CL0020
228:Symbol
187:PDBsum
161:
151:
121:SUPFAM
77:CL0022
50:Symbol
2339:NACHT
2262:Pyrin
2242:Death
2207:Cache
2142:ADF-H
1719:S2CID
1634:S2CID
1547:S2CID
1463:S2CID
1372:redox
1057:SCOPe
1048:SCOP2
903:SCOPe
894:SCOP2
887:LRRCT
882:SMART
852:LRRCT
750:SCOPe
741:SCOP2
734:LRRNT
729:SMART
581:SCOPe
572:SCOP2
565:LRRNT
560:SMART
530:LRRNT
283:SCOPe
274:SCOP2
117:SCOPe
108:SCOP2
53:LRR_1
2712:and
2710:Pfam
2698:and
2696:Pfam
2684:and
2682:Pfam
2670:and
2668:Pfam
2656:and
2654:Pfam
2642:and
2640:Pfam
2391:WD40
2386:TRIO
2319:HEAT
2314:FYVE
2309:FGGY
2304:ENTH
2284:DHR2
2279:DHR1
2274:DHHC
2257:CARD
2237:CVHN
2192:BZIP
2167:BESS
2152:ARID
2147:ANTH
2132:ACDC
1827:PMID
1780:ISBN
1754:PMID
1711:PMID
1693:Cell
1670:PMID
1626:PMID
1588:PMID
1539:PMID
1498:PMID
1455:PMID
1420:PMID
1282:and
1183:turn
1151:fold
1108:PDBj
1104:PDBe
1087:ECOD
1077:Pfam
1053:1lrv
1025:clan
1023:Pfam
1011:Pfam
954:PDBj
950:PDBe
933:ECOD
923:Pfam
899:1m10
858:Pfam
801:PDBj
797:PDBe
780:ECOD
770:Pfam
746:1m10
705:Pfam
644:PDBj
640:PDBe
623:ECOD
613:Pfam
577:1m10
536:Pfam
479:PDBj
475:PDBe
458:ECOD
448:Pfam
403:Pfam
346:PDBj
342:PDBe
325:ECOD
315:Pfam
279:1lrv
251:clan
249:Pfam
237:Pfam
180:PDBj
176:PDBe
159:ECOD
149:Pfam
113:2bnh
84:ECOD
73:clan
71:Pfam
59:Pfam
2401:YTH
2381:SUN
2376:SH3
2371:SH2
2356:PDZ
2349:LOV
2344:PAS
2334:LRR
2329:LIM
2299:EF1
2269:DEP
2252:DED
2232:CUT
2227:CUB
2222:CRM
2212:CBS
2182:BPS
2177:BMC
2172:BIR
2162:BEN
2157:BAR
2137:ACT
2127:ABM
1817:PMC
1807:doi
1746:doi
1742:337
1701:doi
1697:111
1662:doi
1658:312
1618:doi
1578:doi
1529:doi
1525:291
1490:doi
1447:doi
1412:doi
1346:An
1137:LRR
1095:PDB
941:PDB
788:PDB
631:PDB
597:127
466:PDB
432:341
333:PDB
299:737
231:LRV
167:PDB
133:605
2728::
2716::
2702::
2688::
2674::
2660::
2646::
2396:X8
2366:PX
2361:PH
2289:DM
2247:DD
2202:C2
2197:C1
2122:3H
1825:.
1815:.
1801:.
1797:.
1752:.
1740:.
1717:.
1709:.
1695:.
1691:.
1668:.
1656:.
1632:.
1624:.
1612:.
1600:^
1586:.
1572:.
1568:.
1545:.
1537:.
1523:.
1519:.
1496:.
1486:11
1484:.
1461:.
1453:.
1443:54
1441:.
1418:.
1408:19
1406:.
1262:10
1216:.
1131:A
1106:;
1102:;
1085:/
1059:/
1055:/
952:;
948:;
931:/
905:/
901:/
799:;
795:;
778:/
752:/
748:/
642:;
638:;
621:/
583:/
579:/
477:;
473:;
456:/
344:;
340:;
323:/
285:/
281:/
178:;
174:;
157:/
119:/
115:/
2440:e
2433:t
2426:v
2103:e
2096:t
2089:v
2005::
1931::
1892:e
1885:t
1878:v
1833:.
1809::
1803:8
1788:.
1760:.
1748::
1725:.
1703::
1676:.
1664::
1640:.
1620::
1614:3
1594:.
1580::
1574:4
1553:.
1531::
1504:.
1492::
1469:.
1449::
1426:.
1414::
1260:3
1185:-
1181:-
1135:(
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.