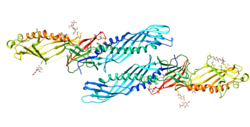
1228:(BPI), the encoded protein binds LPS and interacts with the CD14 receptor, probably playing a role in regulating LPS-dependent monocyte responses. Studies in mice suggest that the encoded protein is necessary for the rapid acute-phase response to LPS but not for the clearance of LPS from circulation. This protein is part of a family of structurally and functionally related proteins, including BPI, plasma cholesteryl ester transfer protein (CETP), and phospholipid transfer protein (PLTP). Finally, this gene is found on chromosome 20, immediately downstream of the BPI gene.
304:
281:
178:
203:
555:
562:
310:
209:
31:
57:
1549:
Thomas CJ, Kapoor Mili, Sharma Shilpi, Bausinger
Huguette, Zyilan Umit, Lipsker Dan, Hanau Daniel, Surolia Avadhesha (November 2002). "Evidence of a trimolecular complex involving LPS, LPS binding protein and soluble CD14 as an effector of LPS response".
1418:
Muta T, Takeshige K (2001). "Essential roles of CD14 and lipopolysaccharide-binding protein for activation of toll-like receptor (TLR)2 as well as TLR4 Reconstitution of TLR2- and TLR4-activation by distinguishable ligands in LPS preparations".
1365:
Gray PW, Corcorran AE, Eddy RL Jr, Byers MG, Shows TB (March 1993). "The genes for the lipopolysaccharide binding protein (LBP) and the bactericidal permeability increasing protein (BPI) are encoded in the same region of human chromosome 20".
1223:
The protein encoded by this gene is involved in the acute-phase immunologic response to gram-negative bacterial infections. Gram-negative bacteria contain a glycolipid, lipopolysaccharide (LPS), on their outer cell wall. Together with
1893:
Kirschning CJ, Au-Young J, Lamping N, et al. (1998). "Similar organization of the lipopolysaccharide-binding protein (LBP) and phospholipid transfer protein (PLTP) genes suggests a common gene family of lipid-binding proteins".
2341:
Reyes O, Vallespi MG, Garay HE, et al. (2002). "Identification of single amino acid residues essential for the binding of lipopolysaccharide (LPS) to LPS binding protein (LBP) residues 86-99 by using an Ala-scanning library".
2118:
Dentener MA, Vreugdenhil AC, Hoet PH, et al. (2000). "Production of the acute-phase protein lipopolysaccharide-binding protein by respiratory type II epithelial cells: implications for local defense to bacterial endotoxins".
1818:
Hubacek JA, Büchler C, Aslanidis C, Schmitz G (1997). "The genomic organization of the genes for human lipopolysaccharide binding protein (LBP) and bactericidal permeability increasing protein (BPI) is highly conserved".
1995:
Vreugdenhil AC, Dentener MA, Snoek AM, et al. (1999). "Lipopolysaccharide binding protein and serum amyloid A secretion by human intestinal epithelial cells during the acute phase response".
1236:
LPS exposure induces LBP production. LBP is synthesized by the liver, adipose tissue, and intestinal cells. Dietary glucose and saturated fats acutely increase plasma LBP.
1512:
Tuomi K, Logomarsino JV (2016). "Bacterial
Lipopolysaccharide, Lipopolysaccharide-Binding Protein, and Other Inflammatory Markers in Obesity and After Bariatric Surgery".
317:
216:
2434:
1966:
Sato M, Saeki Y, Tanaka K, Kaneda Y (1999). "Ribosome-associated protein LBP/p40 binds to S21 protein of 40S ribosome: analysis using a yeast two-hybrid system".
2304:
Kaden J, Zwerenz P, Lambrecht HG, Dostatni R (2002). "Lipopolysaccharide-binding protein as a new and reliable infection marker after kidney transplantation".
1721:"Bactericidal/permeability-increasing protein and lipopolysaccharide (LPS)-binding protein. LPS binding properties and effects on LPS-mediated cell activation"
1225:
1756:"Plasma lipopolysaccharide-binding protein is found associated with a particle containing apolipoprotein A-I, phospholipid, and factor H-related proteins"
2399:
1848:
Jack RS, Fan X, Bernheiden M, et al. (1997). "Lipopolysaccharide-binding protein is required to combat a murine gram-negative bacterial infection".
920:
901:
139:
2150:"Cathelicidin family of antibacterial peptides CAP18 and CAP11 inhibit the expression of TNF-alpha by blocking the binding of LPS to CD14(+) cells"
2185:"Dual Role of Lipopolysaccharide (LPS)-Binding Protein in Neutralization of LPS and Enhancement of LPS-Induced Activation of Mononuclear Cells"
2644:
1347:
1329:
2228:"The carboxyl-terminal domain of closely related endotoxin-binding proteins determines the target of protein-lipopolysaccharide complexes"
2034:"Lipopolysaccharide-Binding Protein and Phospholipid Transfer Protein Release Lipopolysaccharides from Gram-Negative Bacterial Membranes"
2427:
2077:"Innate Recognition of Bacteria in Human Milk Is Mediated by a Milk-Derived Highly Expressed Pattern Recognition Receptor, Soluble Cd14"
303:
2560:
2545:
1789:
Nanbo A, Nishimura H, Nagasawa S (1997). "Lipopolysaccharide binding protein from normal human plasma purified with high efficiency".
1127:
1120:
280:
1467:
1404:
1638:"Lipopolysaccharides of Bacteroides fragilis, Chlamydia trachomatis and Pseudomonas aeruginosa signal via toll-like receptor 2"
1316:
1295:
2420:
202:
177:
1312:
54:
2649:
1291:
1209:
119:
1682:
Schumann RR, Leong SR, Flaggs GW, et al. (1990). "Structure and function of lipopolysaccharide binding protein".
1255:
1463:"Obesity-associated cancer risk: the role of intestinal microbiota in the etiology of the host proinflammatory state"
316:
215:
309:
208:
2390:
965:
127:
2386:
946:
2550:
2514:
2509:
191:
2504:
2499:
2494:
2489:
2484:
2479:
2474:
2469:
2464:
2274:
1857:
1691:
1601:
Yu B, Wright S D (1995). "LPS-dependent interaction of Mac-2-binding protein with immobilized CD14".
106:
2443:
1201:
2524:
2519:
2456:
2447:
2367:
2329:
2020:
1881:
1583:
1217:
1205:
151:
1103:
1082:
1056:
1035:
2359:
2321:
2292:
2249:
2214:
2171:
2136:
2106:
2063:
2012:
1983:
1954:
1911:
1873:
1836:
1806:
1777:
1742:
1707:
1659:
1618:
1610:
1575:
1567:
1531:
1494:
1436:
1383:
99:
47:
2351:
2313:
2282:
2239:
2204:
2196:
2161:
2128:
2096:
2088:
2053:
2045:
2004:
1975:
1944:
1936:
1903:
1865:
1828:
1798:
1767:
1732:
1699:
1649:
1559:
1523:
1514:
1484:
1476:
1428:
1375:
1239:
The proinflammatory activity of plasma LPS is increased by LBP, which is higher in obesity.
396:
327:
271:
226:
2412:
554:
371:
147:
2278:
1861:
1695:
1242:
Plasma LBP is used as a better biomarker of plasma LPS than LPS itself due to the short
561:
2101:
2076:
1949:
1924:
1489:
1462:
2209:
2184:
1737:
1720:
1563:
835:
830:
825:
820:
815:
810:
805:
800:
795:
790:
785:
780:
775:
770:
765:
760:
755:
750:
745:
740:
735:
730:
725:
720:
715:
710:
705:
700:
695:
690:
685:
680:
664:
659:
654:
649:
644:
628:
623:
618:
613:
608:
603:
2638:
2403:
2200:
2058:
2033:
1885:
1432:
590:
2371:
2333:
2049:
2024:
1587:
1208:(or LPS) to elicit immune responses by presenting the LPS to important cell surface
131:
389:
168:
155:
2166:
2149:
2008:
1352:
National Center for
Biotechnology Information, U.S. National Library of Medicine
1334:
National Center for
Biotechnology Information, U.S. National Library of Medicine
1480:
472:
2317:
288:
185:
135:
2132:
1772:
1755:
1614:
1571:
2092:
1925:"The BPI/LBP family of proteins: a structural analysis of conserved regions"
1703:
1243:
865:
532:
410:
355:
342:
254:
241:
143:
2363:
2325:
2296:
2253:
2244:
2227:
2218:
2175:
2140:
2110:
2067:
2016:
1987:
1979:
1940:
1907:
1832:
1802:
1663:
1579:
1535:
1498:
1440:
1379:
1958:
1915:
1877:
1840:
1810:
1781:
1746:
1711:
1654:
1637:
1622:
1527:
1387:
1167:
1162:
786:
positive regulation of respiratory burst involved in inflammatory response
1151:
1010:
991:
30:
1187:
977:
932:
76:
2618:
2608:
2593:
1135:
887:
2355:
2287:
2262:
2623:
2613:
2598:
2583:
2578:
2573:
2568:
2263:"The DNA sequence and comparative analysis of human chromosome 20"
1869:
850:
846:
2603:
2588:
2540:
1267:
1263:
1259:
1213:
1194:
123:
2416:
570:
741:
positive regulation of toll-like receptor 4 signaling pathway
1636:
Erridge C, Pridmore A, Eley A, Stewart J, Poxton IR (2004).
2261:
Deloukas P, Matthews LH, Ashurst J, et al. (2002).
1719:
Wilde CG, Seilhamer JJ, McGrogan M, et al. (1994).
379:
2032:
Vesy CJ, Kitchens RL, Wolfbauer G, et al. (2000).
771:
positive regulation of tumor necrosis factor production
751:
negative regulation of tumor necrosis factor production
2183:
Gutsmann T, Müller M, Carroll SF, et al. (2001).
2148:
Nagaoka I, Hirota S, Niyonsaba F, et al. (2001).
701:
leukocyte chemotaxis involved in inflammatory response
2226:
Iovine N, Eastvold J, Elsbach P, et al. (2002).
1405:"Entrez Gene: LBP lipopolysaccharide binding protein"
1254:
Lipopolysaccharide-binding protein has been shown to
544:
2559:
2533:
2455:
1096:
1075:
1049:
1028:
2075:Labéta MO, Vidal K, Nores JE, et al. (2000).
1308:
1306:
1304:
1287:
1285:
1283:
781:macrophage activation involved in immune response
326:
225:
731:positive regulation of interleukin-6 production
726:positive regulation of interleukin-8 production
1456:
1454:
1452:
1450:
1313:GRCm38: Ensembl release 89: ENSMUSG00000016024
2428:
756:lipopolysaccharide-mediated signaling pathway
8:
1226:bactericidal permeability-increasing protein
811:positive regulation of macrophage activation
736:positive regulation of neutrophil chemotaxis
110:, BPIFD2, lipopolysaccharide binding protein
1923:Beamer LJ, Carroll SF, Eisenberg D (1998).
1292:GRCh38: Ensembl release 89: ENSG00000129988
801:defense response to Gram-positive bacterium
766:defense response to Gram-negative bacterium
746:positive regulation of chemokine production
2435:
2421:
2413:
2400:United States National Library of Medicine
861:
586:
367:
266:
163:
65:
2389:at the U.S. National Library of Medicine
2286:
2243:
2208:
2165:
2100:
2057:
1948:
1771:
1736:
1653:
1488:
681:detection of molecule of bacterial origin
2398:This article incorporates text from the
1515:Metabolic Syndrome and Related Disorders
1399:
1397:
1279:
806:cellular response to lipopolysaccharide
711:cellular response to lipoteichoic acid
691:toll-like receptor 4 signaling pathway
20:
331:
292:
287:
230:
189:
184:
7:
816:toll-like receptor signaling pathway
836:cytokine-mediated signaling pathway
2387:lipopolysaccharide-binding+protein
1180:Lipopolysaccharide binding protein
1093:
1072:
1046:
1025:
1001:
982:
956:
937:
911:
892:
549:
487:choroid plexus of fourth ventricle
467:
405:
384:
14:
1190:that in humans is encoded by the
2201:10.1128/IAI.69.11.6942-6950.2001
1433:10.1046/j.1432-1327.2001.02385.x
826:positive regulation of cytolysis
560:
553:
315:
308:
302:
279:
214:
207:
201:
176:
29:
2050:10.1128/IAI.68.5.2410-2417.2000
1642:Journal of Medical Microbiology
776:response to lipopolysaccharide
571:More reference expression data
533:More reference expression data
1:
2121:Am. J. Respir. Cell Mol. Biol
1968:Biochem. Biophys. Res. Commun
1821:Biochem. Biophys. Res. Commun
1738:10.1016/S0021-9258(17)32454-7
1564:10.1016/S0014-5793(02)03499-3
1210:pattern recognition receptors
716:defense response to bacterium
300:
199:
2645:Genes on human chromosome 20
721:lipopolysaccharide transport
495:Epithelium of choroid plexus
2167:10.4049/jimmunol.167.6.3329
2009:10.4049/jimmunol.163.5.2792
1754:Park CT, Wright SD (1996).
457:right lobe of thyroid gland
449:subcutaneous adipose tissue
2666:
1481:10.1016/j.trsl.2016.07.017
1270:and the co-receptor MD-2.
821:macromolecule localization
624:lipopolysaccharide binding
619:signaling receptor binding
2318:10.1007/s00147-002-0392-2
1348:"Mouse PubMed Reference:"
1330:"Human PubMed Reference:"
1166:
1161:
1157:
1150:
1134:
1128:Chr 2: 158.15 – 158.17 Mb
1115:
1100:
1079:
1068:
1053:
1032:
1021:
1008:
1004:
989:
985:
976:
963:
959:
944:
940:
931:
918:
914:
899:
895:
886:
871:
864:
860:
844:
706:cellular defense response
604:lipoteichoic acid binding
589:
585:
568:
552:
543:
530:
479:
470:
417:
408:
378:
370:
366:
349:
336:
299:
278:
269:
265:
248:
235:
198:
175:
166:
162:
117:
114:
104:
97:
92:
73:
68:
51:
46:
41:
37:
28:
23:
2391:Medical Subject Headings
2133:10.1165/ajrcmb.23.2.3855
1773:10.1074/jbc.271.30.18054
1204:that binds to bacterial
1121:Chr 20: 38.35 – 38.38 Mb
429:tibialis anterior muscle
2093:10.1084/jem.191.10.1807
1704:10.1126/science.2402637
2245:10.1074/jbc.M109622200
1980:10.1006/bbrc.1999.0343
1941:10.1002/pro.5560070408
1908:10.1006/geno.1997.5030
1833:10.1006/bbrc.1997.6970
1803:10.1006/prep.1996.0712
1468:Translational Research
1380:10.1006/geno.1993.1030
796:innate immune response
499:lumbar spinal ganglion
2561:Downstream signalling
2154:Journal of Immunology
1997:Journal of Immunology
1655:10.1099/jmm.0.45598-0
1528:10.1089/met.2015.0170
1232:Clinical significance
696:immune system process
650:extracellular exosome
511:stroma of bone marrow
192:Chromosome 20 (human)
2650:Acute-phase proteins
791:acute-phase response
655:extracellular region
433:gastrocnemius muscle
333:2 H1|2 78.72 cM
294:Chromosome 2 (mouse)
69:List of PDB id codes
42:Available structures
2279:2001Natur.414..865D
1862:1997Natur.389..742J
1791:Protein Expr. Purif
1696:1990Sci...249.1429S
1202:acute-phase protein
665:extracellular space
629:lipopeptide binding
421:right lobe of liver
2402:, which is in the
1206:lipopolysaccharide
966:ENSMUSG00000016024
674:Biological process
638:Cellular component
597:Molecular function
515:left lobe of liver
2632:
2631:
2450:signaling pathway
2444:Signaling pathway
1690:(4975): 1429–31.
1461:Djuric Z (2017).
1200:LBP is a soluble
1177:
1176:
1173:
1172:
1146:
1145:
1111:
1110:
1090:
1089:
1064:
1063:
1043:
1042:
1017:
1016:
998:
997:
972:
971:
953:
952:
927:
926:
908:
907:
856:
855:
581:
580:
577:
576:
539:
538:
526:
525:
464:
463:
362:
361:
261:
260:
88:
87:
84:
83:
52:Ortholog search:
18:Protein in humans
2657:
2437:
2430:
2423:
2414:
2375:
2337:
2300:
2290:
2273:(6866): 865–71.
2257:
2247:
2222:
2212:
2179:
2169:
2144:
2114:
2104:
2071:
2061:
2028:
1991:
1962:
1952:
1919:
1889:
1844:
1814:
1785:
1775:
1766:(30): 18054–60.
1750:
1740:
1715:
1668:
1667:
1657:
1648:(Pt 8): 735–40.
1633:
1627:
1626:
1598:
1592:
1591:
1546:
1540:
1539:
1509:
1503:
1502:
1492:
1458:
1445:
1444:
1415:
1409:
1408:
1401:
1392:
1391:
1362:
1356:
1355:
1344:
1338:
1337:
1326:
1320:
1310:
1299:
1289:
1159:
1158:
1130:
1123:
1106:
1094:
1085:
1073:
1069:RefSeq (protein)
1059:
1047:
1038:
1026:
1002:
983:
957:
938:
912:
893:
862:
587:
573:
564:
557:
550:
535:
475:
473:Top expressed in
468:
413:
411:Top expressed in
406:
385:
368:
358:
345:
334:
319:
312:
306:
295:
283:
267:
257:
244:
233:
218:
211:
205:
194:
180:
164:
158:
109:
102:
79:
66:
60:
39:
38:
33:
21:
2665:
2664:
2660:
2659:
2658:
2656:
2655:
2654:
2635:
2634:
2633:
2628:
2555:
2534:Other receptors
2529:
2451:
2441:
2410:
2383:
2378:
2356:10.1002/psc.375
2340:
2303:
2288:10.1038/414865a
2260:
2225:
2195:(11): 6942–50.
2182:
2147:
2117:
2087:(10): 1807–12.
2074:
2031:
1994:
1965:
1922:
1892:
1856:(6652): 742–5.
1847:
1817:
1788:
1753:
1731:(26): 17411–6.
1718:
1681:
1677:
1675:Further reading
1672:
1671:
1635:
1634:
1630:
1600:
1599:
1595:
1548:
1547:
1543:
1511:
1510:
1506:
1460:
1459:
1448:
1421:Eur. J. Biochem
1417:
1416:
1412:
1403:
1402:
1395:
1364:
1363:
1359:
1346:
1345:
1341:
1328:
1327:
1323:
1311:
1302:
1290:
1281:
1276:
1252:
1234:
1168:View/Edit Mouse
1163:View/Edit Human
1126:
1119:
1116:Location (UCSC)
1102:
1081:
1055:
1034:
947:ENSG00000129988
840:
686:lipid transport
669:
633:
609:protein binding
569:
559:
558:
531:
522:
517:
513:
509:
505:
501:
497:
493:
489:
485:
471:
460:
455:
451:
447:
443:
439:
437:muscle of thigh
435:
431:
427:
423:
409:
353:
340:
332:
322:
321:
320:
313:
293:
270:Gene location (
252:
239:
231:
221:
220:
219:
212:
190:
167:Gene location (
156:LBP - orthologs
118:
105:
98:
75:
53:
19:
12:
11:
5:
2663:
2661:
2653:
2652:
2647:
2637:
2636:
2630:
2629:
2627:
2626:
2621:
2616:
2611:
2606:
2601:
2596:
2591:
2586:
2581:
2576:
2571:
2565:
2563:
2557:
2556:
2554:
2553:
2548:
2543:
2537:
2535:
2531:
2530:
2528:
2527:
2522:
2517:
2512:
2507:
2502:
2497:
2492:
2487:
2482:
2477:
2472:
2467:
2461:
2459:
2453:
2452:
2442:
2440:
2439:
2432:
2425:
2417:
2395:
2394:
2382:
2381:External links
2379:
2377:
2376:
2338:
2301:
2258:
2238:(10): 7970–8.
2223:
2180:
2160:(6): 3329–38.
2145:
2115:
2072:
2029:
1992:
1963:
1920:
1890:
1845:
1815:
1786:
1751:
1716:
1678:
1676:
1673:
1670:
1669:
1628:
1593:
1541:
1522:(6): 279–288.
1504:
1446:
1427:(16): 4580–9.
1410:
1393:
1357:
1339:
1321:
1300:
1278:
1277:
1275:
1272:
1251:
1248:
1233:
1230:
1175:
1174:
1171:
1170:
1165:
1155:
1154:
1148:
1147:
1144:
1143:
1141:
1139:
1132:
1131:
1124:
1117:
1113:
1112:
1109:
1108:
1098:
1097:
1091:
1088:
1087:
1077:
1076:
1070:
1066:
1065:
1062:
1061:
1051:
1050:
1044:
1041:
1040:
1030:
1029:
1023:
1019:
1018:
1015:
1014:
1006:
1005:
999:
996:
995:
987:
986:
980:
974:
973:
970:
969:
961:
960:
954:
951:
950:
942:
941:
935:
929:
928:
925:
924:
916:
915:
909:
906:
905:
897:
896:
890:
884:
883:
878:
873:
869:
868:
858:
857:
854:
853:
842:
841:
839:
838:
833:
828:
823:
818:
813:
808:
803:
798:
793:
788:
783:
778:
773:
768:
763:
758:
753:
748:
743:
738:
733:
728:
723:
718:
713:
708:
703:
698:
693:
688:
683:
677:
675:
671:
670:
668:
667:
662:
657:
652:
647:
641:
639:
635:
634:
632:
631:
626:
621:
616:
611:
606:
600:
598:
594:
593:
583:
582:
579:
578:
575:
574:
566:
565:
547:
541:
540:
537:
536:
528:
527:
524:
523:
521:
520:
516:
512:
508:
504:
500:
496:
492:
488:
484:
480:
477:
476:
465:
462:
461:
459:
458:
454:
450:
446:
445:deltoid muscle
442:
438:
434:
430:
426:
422:
418:
415:
414:
402:
401:
393:
382:
376:
375:
372:RNA expression
364:
363:
360:
359:
351:
347:
346:
338:
335:
330:
324:
323:
314:
307:
301:
297:
296:
291:
285:
284:
276:
275:
263:
262:
259:
258:
250:
246:
245:
237:
234:
229:
223:
222:
213:
206:
200:
196:
195:
188:
182:
181:
173:
172:
160:
159:
116:
112:
111:
103:
95:
94:
90:
89:
86:
85:
82:
81:
71:
70:
62:
61:
50:
44:
43:
35:
34:
26:
25:
17:
13:
10:
9:
6:
4:
3:
2:
2662:
2651:
2648:
2646:
2643:
2642:
2640:
2625:
2622:
2620:
2617:
2615:
2612:
2610:
2607:
2605:
2602:
2600:
2597:
2595:
2592:
2590:
2587:
2585:
2582:
2580:
2577:
2575:
2572:
2570:
2567:
2566:
2564:
2562:
2558:
2552:
2549:
2547:
2544:
2542:
2539:
2538:
2536:
2532:
2526:
2523:
2521:
2518:
2516:
2513:
2511:
2508:
2506:
2503:
2501:
2498:
2496:
2493:
2491:
2488:
2486:
2483:
2481:
2478:
2476:
2473:
2471:
2468:
2466:
2463:
2462:
2460:
2458:
2454:
2449:
2445:
2438:
2433:
2431:
2426:
2424:
2419:
2418:
2415:
2411:
2408:
2407:
2405:
2404:public domain
2401:
2392:
2388:
2385:
2384:
2380:
2373:
2369:
2365:
2361:
2357:
2353:
2350:(4): 144–50.
2349:
2345:
2339:
2335:
2331:
2327:
2323:
2319:
2315:
2312:(4): 163–72.
2311:
2307:
2302:
2298:
2294:
2289:
2284:
2280:
2276:
2272:
2268:
2264:
2259:
2255:
2251:
2246:
2241:
2237:
2233:
2232:J. Biol. Chem
2229:
2224:
2220:
2216:
2211:
2206:
2202:
2198:
2194:
2190:
2189:Infect. Immun
2186:
2181:
2177:
2173:
2168:
2163:
2159:
2155:
2151:
2146:
2142:
2138:
2134:
2130:
2127:(2): 146–53.
2126:
2122:
2116:
2112:
2108:
2103:
2098:
2094:
2090:
2086:
2082:
2078:
2073:
2069:
2065:
2060:
2055:
2051:
2047:
2044:(5): 2410–7.
2043:
2039:
2038:Infect. Immun
2035:
2030:
2026:
2022:
2018:
2014:
2010:
2006:
2003:(5): 2792–8.
2002:
1998:
1993:
1989:
1985:
1981:
1977:
1974:(2): 385–90.
1973:
1969:
1964:
1960:
1956:
1951:
1946:
1942:
1938:
1935:(4): 906–14.
1934:
1930:
1926:
1921:
1917:
1913:
1909:
1905:
1902:(3): 416–25.
1901:
1897:
1891:
1887:
1883:
1879:
1875:
1871:
1870:10.1038/39622
1867:
1863:
1859:
1855:
1851:
1846:
1842:
1838:
1834:
1830:
1827:(2): 427–30.
1826:
1822:
1816:
1812:
1808:
1804:
1800:
1796:
1792:
1787:
1783:
1779:
1774:
1769:
1765:
1761:
1760:J. Biol. Chem
1757:
1752:
1748:
1744:
1739:
1734:
1730:
1726:
1725:J. Biol. Chem
1722:
1717:
1713:
1709:
1705:
1701:
1697:
1693:
1689:
1685:
1680:
1679:
1674:
1665:
1661:
1656:
1651:
1647:
1643:
1639:
1632:
1629:
1624:
1620:
1616:
1612:
1609:(2): 115–25.
1608:
1604:
1597:
1594:
1589:
1585:
1581:
1577:
1573:
1569:
1565:
1561:
1557:
1553:
1545:
1542:
1537:
1533:
1529:
1525:
1521:
1517:
1516:
1508:
1505:
1500:
1496:
1491:
1486:
1482:
1478:
1474:
1470:
1469:
1464:
1457:
1455:
1453:
1451:
1447:
1442:
1438:
1434:
1430:
1426:
1422:
1414:
1411:
1406:
1400:
1398:
1394:
1389:
1385:
1381:
1377:
1374:(1): 188–90.
1373:
1369:
1361:
1358:
1353:
1349:
1343:
1340:
1335:
1331:
1325:
1322:
1318:
1314:
1309:
1307:
1305:
1301:
1297:
1293:
1288:
1286:
1284:
1280:
1273:
1271:
1269:
1265:
1261:
1257:
1249:
1247:
1245:
1240:
1237:
1231:
1229:
1227:
1221:
1219:
1215:
1211:
1207:
1203:
1198:
1196:
1193:
1189:
1185:
1181:
1169:
1164:
1160:
1156:
1153:
1149:
1142:
1140:
1137:
1133:
1129:
1125:
1122:
1118:
1114:
1107:
1105:
1099:
1095:
1092:
1086:
1084:
1078:
1074:
1071:
1067:
1060:
1058:
1052:
1048:
1045:
1039:
1037:
1031:
1027:
1024:
1022:RefSeq (mRNA)
1020:
1013:
1012:
1007:
1003:
1000:
994:
993:
988:
984:
981:
979:
975:
968:
967:
962:
958:
955:
949:
948:
943:
939:
936:
934:
930:
923:
922:
917:
913:
910:
904:
903:
898:
894:
891:
889:
885:
882:
879:
877:
874:
870:
867:
863:
859:
852:
848:
843:
837:
834:
832:
829:
827:
824:
822:
819:
817:
814:
812:
809:
807:
804:
802:
799:
797:
794:
792:
789:
787:
784:
782:
779:
777:
774:
772:
769:
767:
764:
762:
759:
757:
754:
752:
749:
747:
744:
742:
739:
737:
734:
732:
729:
727:
724:
722:
719:
717:
714:
712:
709:
707:
704:
702:
699:
697:
694:
692:
689:
687:
684:
682:
679:
678:
676:
673:
672:
666:
663:
661:
658:
656:
653:
651:
648:
646:
643:
642:
640:
637:
636:
630:
627:
625:
622:
620:
617:
615:
614:lipid binding
612:
610:
607:
605:
602:
601:
599:
596:
595:
592:
591:Gene ontology
588:
584:
572:
567:
563:
556:
551:
548:
546:
542:
534:
529:
518:
514:
510:
506:
502:
498:
494:
490:
486:
482:
481:
478:
474:
469:
466:
456:
452:
448:
444:
440:
436:
432:
428:
424:
420:
419:
416:
412:
407:
404:
403:
400:
398:
394:
392:
391:
387:
386:
383:
381:
377:
373:
369:
365:
357:
352:
348:
344:
339:
329:
325:
318:
311:
305:
298:
290:
286:
282:
277:
273:
268:
264:
256:
251:
247:
243:
238:
228:
224:
217:
210:
204:
197:
193:
187:
183:
179:
174:
170:
165:
161:
157:
153:
149:
145:
141:
137:
133:
129:
125:
121:
113:
108:
101:
96:
91:
80:
78:
72:
67:
64:
63:
59:
56:
49:
45:
40:
36:
32:
27:
22:
16:
2409:
2397:
2396:
2347:
2344:J. Pept. Sci
2343:
2309:
2306:Transpl. Int
2305:
2270:
2266:
2235:
2231:
2192:
2188:
2157:
2153:
2124:
2120:
2084:
2080:
2041:
2037:
2000:
1996:
1971:
1967:
1932:
1928:
1899:
1895:
1853:
1849:
1824:
1820:
1797:(1): 55–60.
1794:
1790:
1763:
1759:
1728:
1724:
1687:
1683:
1645:
1641:
1631:
1606:
1602:
1596:
1558:(2): 184–8.
1555:
1551:
1544:
1519:
1513:
1507:
1472:
1466:
1424:
1420:
1413:
1371:
1367:
1360:
1351:
1342:
1333:
1324:
1253:
1250:Interactions
1241:
1238:
1235:
1222:
1199:
1191:
1183:
1179:
1178:
1101:
1080:
1054:
1033:
1009:
990:
964:
945:
919:
900:
880:
875:
761:opsonization
660:cell surface
395:
388:
354:158,174,772
341:158,148,413
115:External IDs
74:
15:
2081:J. Exp. Med
1929:Protein Sci
1475:: 155–167.
507:ankle joint
253:38,377,013
240:38,346,482
93:Identifiers
2639:Categories
1603:J. Inflamm
1319:, May 2017
1298:, May 2017
1274:References
399:(ortholog)
136:HomoloGene
1886:205027151
1615:1078-7852
1572:0014-5793
1552:FEBS Lett
1244:half-life
1104:NP_032515
1083:NP_004130
1057:NM_008489
1036:NM_004139
866:Orthologs
831:transport
144:GeneCards
2372:28803013
2364:11991204
2334:23593715
2326:11976738
2297:11780052
2254:11773072
2219:11598069
2176:11544322
2141:10919979
2111:10811873
2068:10768924
2025:25321198
2017:10453023
1988:10079194
1896:Genomics
1664:15272059
1588:25135963
1580:12417309
1536:27228236
1499:27522986
1441:11502220
1368:Genomics
1315:–
1294:–
1256:interact
1246:of LPS.
1152:Wikidata
845:Sources:
645:membrane
483:gastrula
441:appendix
425:testicle
232:20q11.23
2275:Bibcode
2102:2193148
1959:9568897
1950:2143972
1916:9441745
1878:9338787
1858:Bibcode
1841:9240454
1811:9179291
1782:8663389
1747:7517398
1712:2402637
1692:Bibcode
1684:Science
1623:7583357
1490:5164980
1388:8432532
1317:Ensembl
1296:Ensembl
1212:called
1188:protein
1186:) is a
978:UniProt
933:Ensembl
872:Species
851:QuickGO
491:decidua
374:pattern
132:1098776
100:Aliases
2619:TOLLIP
2609:TICAM1
2594:MAP3K7
2393:(MeSH)
2370:
2362:
2332:
2324:
2295:
2267:Nature
2252:
2217:
2210:100074
2207:
2174:
2139:
2109:
2099:
2066:
2056:
2023:
2015:
1986:
1957:
1947:
1914:
1884:
1876:
1850:Nature
1839:
1809:
1780:
1745:
1710:
1662:
1621:
1613:
1586:
1578:
1570:
1534:
1497:
1487:
1439:
1386:
1138:search
1136:PubMed
1011:Q61805
992:P18428
888:Entrez
545:BioGPS
519:uterus
453:glutes
124:151990
2624:TRAF6
2614:TIRAP
2599:MYD88
2584:IRAK4
2579:IRAK3
2574:IRAK2
2569:IRAK1
2525:TLR13
2520:TLR12
2515:TLR11
2510:TLR10
2368:S2CID
2330:S2CID
2059:97439
2021:S2CID
1882:S2CID
1584:S2CID
1258:with
921:16803
881:Mouse
876:Human
847:Amigo
503:ankle
397:Mouse
390:Human
337:Start
272:Mouse
236:Start
169:Human
2604:TAB1
2589:IRF3
2541:CD14
2505:TLR9
2500:TLR8
2495:TLR7
2490:TLR6
2485:TLR5
2480:TLR4
2475:TLR3
2470:TLR2
2465:TLR1
2457:TLRs
2360:PMID
2322:PMID
2293:PMID
2250:PMID
2215:PMID
2172:PMID
2137:PMID
2107:PMID
2064:PMID
2013:PMID
1984:PMID
1955:PMID
1912:PMID
1874:PMID
1837:PMID
1807:PMID
1778:PMID
1743:PMID
1708:PMID
1660:PMID
1619:PMID
1611:ISSN
1576:PMID
1568:ISSN
1532:PMID
1495:PMID
1437:PMID
1384:PMID
1268:TLR4
1264:TLR2
1260:CD14
1218:TLR4
1216:and
1214:CD14
1195:gene
902:3929
380:Bgee
328:Band
289:Chr.
227:Band
186:Chr.
140:3055
120:OMIM
77:4M4D
58:RCSB
55:PDBe
2551:MD2
2546:LBP
2448:TLR
2352:doi
2314:doi
2283:doi
2271:414
2240:doi
2236:277
2205:PMC
2197:doi
2162:doi
2158:167
2129:doi
2097:PMC
2089:doi
2085:191
2054:PMC
2046:doi
2005:doi
2001:163
1976:doi
1972:256
1945:PMC
1937:doi
1904:doi
1866:doi
1854:389
1829:doi
1825:236
1799:doi
1768:doi
1764:271
1733:doi
1729:269
1700:doi
1688:249
1650:doi
1560:doi
1556:531
1524:doi
1485:PMC
1477:doi
1473:179
1429:doi
1425:268
1376:doi
1192:LBP
1184:LBP
350:End
249:End
152:OMA
148:LBP
128:MGI
107:LBP
48:PDB
24:LBP
2641::
2446::
2366:.
2358:.
2346:.
2328:.
2320:.
2310:15
2308:.
2291:.
2281:.
2269:.
2265:.
2248:.
2234:.
2230:.
2213:.
2203:.
2193:69
2191:.
2187:.
2170:.
2156:.
2152:.
2135:.
2125:23
2123:.
2105:.
2095:.
2083:.
2079:.
2062:.
2052:.
2042:68
2040:.
2036:.
2019:.
2011:.
1999:.
1982:.
1970:.
1953:.
1943:.
1931:.
1927:.
1910:.
1900:46
1898:.
1880:.
1872:.
1864:.
1852:.
1835:.
1823:.
1805:.
1795:10
1793:.
1776:.
1762:.
1758:.
1741:.
1727:.
1723:.
1706:.
1698:.
1686:.
1658:.
1646:53
1644:.
1640:.
1617:.
1607:45
1605:.
1582:.
1574:.
1566:.
1554:.
1530:.
1520:14
1518:.
1493:.
1483:.
1471:.
1465:.
1449:^
1435:.
1423:.
1396:^
1382:.
1372:15
1370:.
1350:.
1332:.
1303:^
1282:^
1266:,
1262:,
1220:.
1197:.
849:/
356:bp
343:bp
255:bp
242:bp
150:;
146::
142:;
138::
134:;
130::
126:;
122::
2436:e
2429:t
2422:v
2406:.
2374:.
2354::
2348:8
2336:.
2316::
2299:.
2285::
2277::
2256:.
2242::
2221:.
2199::
2178:.
2164::
2143:.
2131::
2113:.
2091::
2070:.
2048::
2027:.
2007::
1990:.
1978::
1961:.
1939::
1933:7
1918:.
1906::
1888:.
1868::
1860::
1843:.
1831::
1813:.
1801::
1784:.
1770::
1749:.
1735::
1714:.
1702::
1694::
1666:.
1652::
1625:.
1590:.
1562::
1538:.
1526::
1501:.
1479::
1443:.
1431::
1407:.
1390:.
1378::
1354:.
1336:.
1182:(
274:)
171:)
154::
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.