473:
450:
347:
372:
3704:
3674:
724:
3629:
3614:
3644:
479:
378:
40:
3719:
3599:
3584:
3689:
3659:
3403:
Bonifati V, Dekker MC, Vanacore N, Fabbrini G, Squitieri F, Marconi R, Antonini A, Brustenghi P, Dalla Libera A, De Mari M, Stocchi F, Montagna P, Gallai V, Rizzu P, van
Swieten JC, Oostra B, van Duijn CM, Meco G, Heutink P (Sep 2002). "Autosomal recessive early onset parkinsonism is linked to three
2830:
Bonifati V, Rizzu P, van Baren MJ, Schaap O, Breedveld GJ, Krieger E, Dekker MC, Squitieri F, Ibanez P, Joosse M, van Dongen JW, Vanacore N, van
Swieten JC, Brice A, Meco G, van Duijn CM, Oostra BA, Heutink P (Jan 2003). "Mutations in the DJ-1 gene associated with autosomal recessive early-onset
66:
2933:
Niki T, Takahashi-Niki K, Taira T, Iguchi-Ariga SM, Ariga H (Feb 2003). "DJBP: a novel DJ-1-binding protein, negatively regulates the androgen receptor by recruiting histone deacetylase complex, and DJ-1 antagonizes this inhibition by abrogation of this complex".
2104:
has been reported for decades. Recently evidence has been presented that defective DNA repair is linked specifically to DJ-1 mutation, and thus DJ-1 mutation likely contributes to
Parkinson's disease pathogenesis.
3441:
Dekker M, Bonifati V, van
Swieten J, Leenders N, Galjaard RJ, Snijders P, Horstink M, Heutink P, Oostra B, van Duijn C (Jul 2003). "Clinical features and neuroimaging of PARK7-linked parkinsonism".
2624:
Nunome K, Miyazaki S, Nakano M, Iguchi-Ariga S, Ariga H (Jul 2008). "Pyrroloquinoline quinone prevents oxidative stress-induced neuronal death probably through changes in oxidative status of DJ-1".
3296:
Taira T, Takahashi K, Kitagawa R, Iguchi-Ariga SM, Ariga H (Jan 2001). "Molecular cloning of human and mouse DJ-1 genes and identification of Sp1-dependent activation of the human DJ-1 promoter".
3325:
van Duijn CM, Dekker MC, Bonifati V, Galjaard RJ, Houwing-Duistermaat JJ, Snijders PJ, Testers L, Breedveld GJ, Horstink M, Sandkuijl LA, van
Swieten JC, Oostra BA, Heutink P (Sep 2001).
3703:
3267:
Nagakubo D, Taira T, Kitaura H, Ikeda M, Tamai K, Iguchi-Ariga SM, Ariga H (Feb 1997). "DJ-1, a novel oncogene which transforms mouse NIH3T3 cells in cooperation with ras".
3673:
486:
385:
3478:
Miller DW, Ahmad R, Hague S, Baptista MJ, Canet-Aviles R, McLendon C, Carter DM, Zhu PP, Stadler J, Chandran J, Klinefelter GR, Blackstone C, Cookson MR (Sep 2003).
3049:
Bonifati V, Oostra BA, Heutink P (Mar 2004). "Linking DJ-1 to neurodegeneration offers novel insights for understanding the pathogenesis of
Parkinson's disease".
3740:
1617:
1598:
308:
2512:
Martín-Nieto J, Uribe ML, Esteve-Rudd J, Herrero MT, Campello L (August 2019). "A role for DJ-1 against oxidative stress in the mammalian retina".
2002:. Accordingly, DJ-1 apparently protects neurons against oxidative stress and cell death. In parallel, protein DJ-1 acts as a positive regulator of
2161:
2477:
Zhou W, Zhu M, Wilson MA, Petsko GA, Fink AL (Mar 2006). "The oxidation state of DJ-1 regulates its chaperone activity toward alpha-synuclein".
3628:
3613:
3563:
3212:
2250:
2232:
3643:
3760:
2148:
2725:
Richarme G, Liu C, Mihoub M, Abdallah J, Leger T, Joly N, Liebart JC, Jurkunas UV, Nadal M, Bouloc P, Dairou J, Lamouri A (July 2017).
472:
1832:
2166:
1825:
449:
2275:
2010:
of mammals, where it exerts a neuroprotective role against oxidative stress under both physiological and pathological conditions.
2085:
2219:
2198:
3718:
2215:
371:
346:
63:
2194:
3598:
3583:
288:
3688:
2129:
3480:"L166P mutant DJ-1, causative for recessive Parkinson's disease, is degraded through the ubiquitin-proteasome system"
485:
384:
2093:
2016:(PQQ) has been shown to reduce the self-oxidation of the DJ-1 protein, an early step in the onset of some forms of
2007:
3658:
478:
377:
3556:
2097:
2013:
1662:
296:
3230:
Lev N, Roncevic D, Roncevich D, Ickowicz D, Melamed E, Offen D (2007). "Role of DJ-1 in
Parkinson's disease".
2387:"Crystal structures of human DJ-1 and Escherichia coli Hsp31, which share an evolutionarily conserved domain"
2669:
Björkblom B, Adilbayeva A, Maple-Grødem J, Piston D, Ökvist M, Xu XM, Brede C, Larsen JP, Møller SG (2013).
1643:
2114:
2101:
2072:. Deglycase-deficient bacterial mutants with reduced ability to repair glycated bases in DNA show strong
2023:
Functional DJ-1 protein has been shown to bind metals and protect against metal-induced cytotoxicity from
2017:
3370:"DJ-1 positively regulates the androgen receptor by impairing the binding of PIASx alpha to the receptor"
2963:"DJ-1 positively regulates the androgen receptor by impairing the binding of PIASx alpha to the receptor"
3549:
2840:
2738:
2578:
360:
275:
2385:
Lee SJ, Kim SJ, Kim IK, Ko J, Jeong CS, Kim GH, Park C, Kang SO, Suh PG, Lee HS, Cha SS (Nov 2003).
1979:
1783:
1732:
1063:
negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway
3115:
Abou-Sleiman PM, Healy DG, Wood NW (Oct 2004). "Causes of
Parkinson's disease: genetics of DJ-1".
2779:
Wang ZX, Liu Y, Li YL, Wei Q, Lin RR, Kang R, Ruan Y, Lin ZH, Xue NJ, Zhang BR, Pu JL (May 2023).
3466:
3429:
3255:
3140:
3074:
3037:
2864:
2547:
2428:"DJ-1 is a redox-dependent molecular chaperone that inhibits alpha-synuclein aggregate formation"
1208:
negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway
320:
1998:
via its chaperone activity, thus functioning as a redox-sensitive chaperone and as a sensor for
1808:
1787:
1757:
1736:
3521:
3501:
3458:
3421:
3391:
3356:
3313:
3284:
3247:
3218:
3208:
3183:
3132:
3103:
3066:
3029:
2984:
2943:
2915:
2856:
2812:
2756:
2702:
2651:
2606:
2539:
2494:
2459:
2408:
2367:
2326:
2073:
2028:
2003:
1931:
268:
56:
3195:
Heutink P (2006). "PINK-1 and DJ-1 — new genes for autosomal recessive
Parkinson's disease".
2884:"Neuron-specific protein interactions of Drosophila CASK-β are revealed by mass spectrometry"
2671:"Parkinson disease protein DJ-1 binds metals and protects against metal-induced cytotoxicity"
1173:
negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway
3712:: The Crystal Structure of DJ-1, a Protein Related to Male Fertility and Parkinson's Disease
3491:
3450:
3413:
3381:
3346:
3338:
3305:
3276:
3239:
3200:
3173:
3165:
3124:
3095:
3058:
3019:
2974:
2905:
2895:
2848:
2802:
2792:
2746:
2692:
2682:
2641:
2633:
2596:
2586:
2529:
2521:
2486:
2449:
2439:
2398:
2357:
2346:"Crystal structure of human DJ-1, a protein associated with early onset Parkinson's disease"
2316:
2305:"The crystal structure of DJ-1, a protein related to male fertility and Parkinson's disease"
1999:
1975:
1188:
negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway
565:
496:
440:
395:
3327:"Park7, a novel locus for autosomal recessive early-onset parkinsonism, on chromosome 1p36"
723:
316:
540:
2844:
2807:
2780:
2742:
2582:
3351:
3326:
2910:
2883:
2697:
2670:
2601:
2567:"DJ-1-dependent regulation of oxidative stress in the retinal pigment epithelium (RPE)"
2566:
2171:
2088:
response protein that is recruited to sites of DNA damage where it participates in the
1995:
1478:
negative regulation of nitrosative stress-induced intrinsic apoptotic signaling pathway
39:
3309:
3178:
3153:
3024:
3007:
2454:
2427:
1532:
1527:
1522:
1517:
1512:
1507:
1502:
1497:
1492:
1487:
1482:
1477:
1472:
1467:
1462:
1457:
1452:
1447:
1442:
1437:
1432:
1427:
1422:
1417:
1412:
1407:
1402:
1397:
1392:
1387:
1382:
1377:
1372:
1367:
1362:
1357:
1352:
1347:
1342:
1337:
1332:
1327:
1322:
1317:
1312:
1307:
1302:
1297:
1292:
1287:
1282:
1277:
1272:
1267:
1262:
1257:
1252:
1247:
1242:
1237:
1232:
1227:
1222:
1217:
1212:
1207:
1202:
1197:
1192:
1187:
1182:
1177:
1172:
1167:
1162:
1157:
1152:
1147:
1142:
1137:
1132:
1127:
1122:
1117:
1112:
1107:
1102:
1097:
1092:
1087:
1082:
1077:
1072:
1067:
1062:
1057:
1052:
1047:
1031:
1026:
1021:
1016:
1011:
1006:
1001:
996:
991:
986:
981:
976:
971:
966:
961:
956:
951:
946:
941:
936:
920:
915:
910:
905:
900:
895:
890:
885:
880:
875:
870:
865:
860:
855:
850:
845:
840:
835:
830:
825:
820:
815:
810:
805:
800:
795:
790:
785:
780:
775:
770:
765:
3754:
3744:
3530:
2551:
2039:
1963:
1951:
1403:
positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway
752:
3470:
3433:
3259:
3078:
3041:
2868:
300:
3144:
2057:
558:
337:
1959:
3204:
2591:
2525:
2444:
2303:
Honbou K, Suzuki NN, Horiuchi M, Niki T, Taira T, Ariga H, Inagaki F (Aug 2003).
1994:
Under an oxidative condition, protein deglycase DJ-1 inhibits the aggregation of
324:
2255:
National Center for
Biotechnology Information, U.S. National Library of Medicine
2237:
National Center for Biotechnology Information, U.S. National Library of Medicine
1955:
1083:
negative regulation of proteasomal ubiquitin-dependent protein catabolic process
3243:
3099:
3086:
Le W, Appel SH (Feb 2004). "Mutant genes responsible for Parkinson's disease".
2781:"Nuclear DJ-1 Regulates DNA Damage Repair via the Regulation of PARP1 Activity"
2056:, either in the form of a free nucleotide or as a nucleotide incorporated into
641:
3682:: Crystal structure of DJ-1/RS and implication on familial Parkinson's disease
3128:
3062:
2490:
2089:
2049:
457:
354:
304:
3199:. Journal of Neural Transmission. Supplementa. Vol. 70. pp. 215–9.
2900:
3368:
Takahashi K, Taira T, Niki T, Seino C, Iguchi-Ariga SM, Ariga H (Oct 2001).
3169:
2961:
Takahashi K, Taira T, Niki T, Seino C, Iguchi-Ariga SM, Ariga H (Oct 2001).
2852:
2751:
2726:
2687:
2046:
1967:
1562:
701:
579:
524:
511:
423:
410:
312:
3505:
3496:
3479:
3462:
3425:
3395:
3386:
3369:
3360:
3317:
3280:
3251:
3222:
3187:
3136:
3107:
3070:
3033:
2988:
2979:
2962:
2947:
2919:
2860:
2816:
2760:
2706:
2655:
2610:
2543:
2498:
2463:
2412:
2403:
2386:
2371:
2362:
2345:
2330:
2321:
2304:
1168:
positive regulation of mitochondrial electron transport, NADH to ubiquinone
3417:
3288:
2006:-dependent transcription. DJ-1 is expressed in both the neural retina and
1872:
1867:
2797:
2289:
1856:
1707:
1688:
2637:
1483:
positive regulation of acute inflammatory response to antigenic stimulus
3535:
3525:
2534:
2426:
Shendelman S, Jonason A, Martinat C, Leete T, Abeliovich A (Nov 2004).
2053:
2043:
1983:
1892:
1674:
1629:
3454:
2646:
2113:
Defects in this gene are the cause of autosomal recessive early-onset
245:
241:
237:
233:
229:
225:
221:
217:
213:
209:
205:
201:
197:
193:
189:
185:
181:
177:
173:
169:
165:
161:
157:
153:
149:
145:
141:
137:
133:
129:
125:
121:
117:
113:
109:
105:
101:
97:
93:
89:
85:
2142:
2024:
1916:
1840:
1584:
17:
3342:
2727:"Guanine glycation repair by DJ-1/Park7 and its bacterial homologs"
1533:
negative regulation of reactive oxygen species biosynthetic process
1108:
positive regulation of oxidative phosphorylation uncoupler activity
1103:
positive regulation of reactive oxygen species biosynthetic process
1348:
negative regulation of TRAIL-activated apoptotic signaling pathway
1053:
positive regulation of transcription regulatory region DNA binding
1547:
1543:
1418:
positive regulation of pyrroline-5-carboxylate reductase activity
2136:
1939:
1899:
1243:
negative regulation of death-inducing signaling complex assembly
1238:
positive regulation of DNA-binding transcription factor activity
732:
292:
279:, DJ-1, DJ1, HEL-S-67p, Parkinsonism associated deglycase, GATD2
3545:
3541:
2065:
2061:
2038:
and its bacterial homologs: Hsp31, YhbO, and YajL can repair
1398:
negative regulation of ubiquitin-protein transferase activity
1278:
negative regulation of hydrogen peroxide-induced neuron death
3520:
Overview of all the structural information available in the
2882:
Mukherjee K, Slawson JB, Christmann BL, Griffith LC (2014).
1218:
negative regulation of oxidative stress-induced neuron death
1133:
negative regulation of extrinsic apoptotic signaling pathway
2565:
Shadrach KG, Rayborn ME, Hollyfield JG, Bonilha VL (2013).
1358:
negative regulation of ubiquitin-specific protease activity
1293:
negative regulation of hydrogen peroxide-induced cell death
1258:
negative regulation of protein K48-linked deubiquitination
1178:
negative regulation of oxidative stress-induced cell death
1423:
positive regulation of transcription by RNA polymerase II
1268:
positive regulation of tyrosine 3-monooxygenase activity
1148:
positive regulation of protein homodimerization activity
548:
3637:: Crystal structure of human DJ-1, P 1 21 1 space group
3622:: Crystal structure of human DJ-1, P 31 2 1 space group
871:
oxidoreductase activity, acting on peroxide as acceptor
1323:
positive regulation of protein localization to nucleus
1163:
positive regulation of peptidyl-serine phosphorylation
3652:: Crystal structure of the K130R mutant of human DJ-1
713:
1318:
positive regulation of dopamine biosynthetic process
1313:
positive regulation of superoxide dismutase activity
1273:
positive regulation of L-dopa decarboxylase activity
3269:
Biochemical and Biophysical Research Communications
1954:in size and composed of 189 amino acids with seven
1801:
1776:
1750:
1725:
2211:
2209:
2207:
2190:
2188:
2186:
1393:negative regulation of protein export from nucleus
1328:positive regulation of L-dopa biosynthetic process
1413:positive regulation of androgen receptor activity
1298:positive regulation of autophagy of mitochondrion
1288:regulation of androgen receptor signaling pathway
1263:dopamine uptake involved in synaptic transmission
1113:positive regulation of protein kinase B signaling
836:ubiquitin-like protein conjugating enzyme binding
495:
394:
2100:. Evidence for a linkage between DNA damage and
1118:negative regulation of protein catabolic process
2720:
2718:
2716:
1493:positive regulation of NAD(P)H oxidase activity
1193:regulation of supramolecular fiber organization
1078:positive regulation of interleukin-8 production
1048:negative regulation of neuron apoptotic process
2216:GRCm38: Ensembl release 89: ENSMUSG00000028964
1973:The protein structures of human protein DJ-1,
1373:regulation of mitochondrial membrane potential
1213:negative regulation of protein kinase activity
1203:negative regulation of protein phosphorylation
881:superoxide dismutase copper chaperone activity
3557:
3534:(Protein/nucleic acid deglycase DJ-1) at the
1378:negative regulation of protein ubiquitination
8:
1368:cellular response to reactive oxygen species
2774:
2772:
2770:
2195:GRCh38: Ensembl release 89: ENSG00000116288
801:tyrosine 3-monooxygenase activator activity
3741:United States National Library of Medicine
3564:
3550:
3542:
1558:
1523:guanine deglycation, methylglyoxal removal
1463:protein deglycation, methylglyoxal removal
1098:negative regulation of protein acetylation
1058:negative regulation of protein sumoylation
748:
536:
435:
332:
74:
3495:
3385:
3350:
3197:Parkinson's Disease and Related Disorders
3177:
3023:
2978:
2909:
2899:
2806:
2796:
2750:
2696:
2686:
2645:
2600:
2590:
2533:
2453:
2443:
2402:
2361:
2320:
972:mitochondrial respiratory chain complex I
3739:This article incorporates text from the
2626:Biological & Pharmaceutical Bulletin
2270:
2268:
2266:
2264:
2068:), if glycated, can be repaired by DJ-1/
1942:and locates at chromosome band 1p36.23.
1503:cellular response to DNA damage stimulus
1248:negative regulation of apoptotic process
614:Skeletal muscle tissue of biceps brachii
3579:
2182:
1986:protease are evolutionarily conserved.
1950:The human protein deglycase DJ-1 is 20
1333:activation of protein kinase B activity
826:small protein activating enzyme binding
786:L-dopa decarboxylase activator activity
1428:cellular response to hydrogen peroxide
1388:positive regulation of gene expression
1153:negative regulation of gene expression
1128:regulation of neuron apoptotic process
1068:negative regulation of protein binding
29:
3727:: Pre-oxidation Complex of Human DJ-1
1408:cellular response to oxidative stress
500:
461:
456:
399:
358:
353:
7:
2162:Animal models of Parkinson's disease
1528:guanine deglycation, glyoxal removal
1458:protein deglycation, glyoxal removal
3484:The Journal of Biological Chemistry
3374:The Journal of Biological Chemistry
2967:The Journal of Biological Chemistry
2888:Frontiers in Molecular Neuroscience
2391:The Journal of Biological Chemistry
2350:The Journal of Biological Chemistry
2309:The Journal of Biological Chemistry
1383:hydrogen peroxide metabolic process
1283:regulation of inflammatory response
1198:negative regulation of neuron death
1073:synaptic transmission, dopaminergic
896:ubiquitin-specific protease binding
664:medial head of gastrocnemius muscle
606:dorsal motor nucleus of vagus nerve
27:Protein-coding gene found in humans
3331:American Journal of Human Genetics
2090:repair of DNA double-strand breaks
1895:which in humans is encoded by the
1798:
1773:
1747:
1722:
1698:
1679:
1653:
1634:
1608:
1589:
921:protein-containing complex binding
866:transcription coactivator activity
718:
636:
574:
553:
25:
3607:: Crystal Structure of Human DJ-1
3592:: Crystal Structure of Human DJ-1
3232:Journal of Molecular Neuroscience
3152:Pankratz N, Foroud T (Apr 2004).
2167:Mitochondria associated membranes
1138:negative regulation of cell death
952:mitochondrial intermembrane space
816:protein homodimerization activity
3717:
3702:
3687:
3672:
3657:
3642:
3627:
3612:
3597:
3582:
3404:loci: PARK2, PARK6, and PARK7".
722:
688:extensor digitorum longus muscle
484:
477:
471:
448:
383:
376:
370:
345:
38:
3697:: Human DJ-1 with sulfinic acid
3154:"Genetics of Parkinson disease"
3088:Current Opinion in Pharmacology
2675:Journal of Biological Chemistry
2290:"Uniprot: Q99497 - PARK7_HUMAN"
1433:methylglyoxal metabolic process
1353:cellular oxidant detoxification
1088:Ras protein signal transduction
1022:perinuclear region of cytoplasm
1143:glycolate biosynthetic process
733:More reference expression data
702:More reference expression data
1:
3310:10.1016/S0378-1119(00)00590-4
3051:Journal of Molecular Medicine
3025:10.1016/S0896-6273(02)01166-2
1982:Hsp31, YhbO, and YajL and an
1962:in total and is present as a
1448:peptidyl-arginine deglycation
1443:peptidyl-cysteine deglycation
1343:detoxification of mercury ion
1303:response to hydrogen peroxide
469:
368:
3205:10.1007/978-3-211-45295-0_33
2592:10.1371/journal.pone.0067983
2526:10.1016/j.neulet.2019.134361
2479:Journal of Molecular Biology
2445:10.1371/journal.pbio.0020362
1438:lactate biosynthetic process
1308:detoxification of copper ion
1093:cellular response to glyoxal
781:transcription factor binding
602:Epithelium of choroid plexus
3761:Genes on human chromosome 1
3667:: crystal structure of DJ-1
1930:, encodes a protein of the
1889:Parkinson disease protein 7
1453:peptidyl-lysine deglycation
851:double-stranded DNA binding
831:single-stranded DNA binding
3777:
3100:10.1016/j.coph.2003.09.005
3008:"Pathways to Parkinsonism"
2344:Tao X, Tong L (Aug 2003).
2094:non-homologous end joining
2008:retinal pigment epithelium
1914:
1338:membrane hyperpolarization
1233:mitochondrion organization
916:protein deglycase activity
766:signaling receptor binding
676:retinal pigment epithelium
3577:
3129:10.1007/s00441-004-0922-6
3063:10.1007/s00109-003-0512-1
2936:Molecular Cancer Research
2491:10.1016/j.jmb.2005.12.030
2251:"Mouse PubMed Reference:"
2233:"Human PubMed Reference:"
2092:through the processes of
1871:
1866:
1862:
1855:
1839:
1820:
1805:
1780:
1769:
1754:
1729:
1718:
1705:
1701:
1686:
1682:
1673:
1660:
1656:
1641:
1637:
1628:
1615:
1611:
1596:
1592:
1583:
1568:
1561:
1557:
1541:
1473:glyoxal metabolic process
1123:adult locomotory behavior
891:identical protein binding
876:androgen receptor binding
751:
747:
730:
721:
712:
699:
648:
639:
586:
577:
547:
539:
535:
518:
505:
468:
447:
438:
434:
417:
404:
367:
344:
335:
331:
286:
283:
273:
266:
261:
82:
77:
60:
55:
50:
46:
37:
32:
3117:Cell and Tissue Research
2901:10.3389/fnmol.2014.00058
2098:homologous recombination
2014:Pyrroloquinoline quinone
1970:C56 family of proteins.
821:scaffold protein binding
672:tibialis anterior muscle
610:tibialis anterior muscle
3170:10.1602/neurorx.1.2.235
3006:Cookson MR (Jan 2003).
2853:10.1126/science.1077209
2752:10.1126/science.aag1095
2688:10.1074/jbc.M113.482091
1934:family. The human gene
1488:protein deglycosylation
1468:glutathione deglycation
1223:membrane depolarization
656:vastus lateralis muscle
622:vastus lateralis muscle
3497:10.1074/jbc.M304272200
3387:10.1074/jbc.M101730200
3281:10.1006/bbrc.1997.6132
2980:10.1074/jbc.M101730200
2404:10.1074/jbc.M304517200
2363:10.1074/jbc.M304221200
2322:10.1074/jbc.M305878200
1885:Protein deglycase DJ-1
1833:Chr 4: 150.98 – 151 Mb
846:peroxiredoxin activity
652:triceps brachii muscle
3418:10.1007/s100720200069
3406:Neurological Sciences
2109:Clinical significance
1966:. It belongs to the
1826:Chr 1: 7.95 – 7.99 Mb
1228:protein stabilization
1183:inflammatory response
992:extracellular exosome
977:endoplasmic reticulum
3244:10.1385/JMN:29:3:215
2798:10.3390/ijms24108651
2276:"Entrez Gene: PARK7"
1158:single fertilization
1002:mitochondrial matrix
668:facial motor nucleus
660:gastrocnemius muscle
463:Chromosome 4 (mouse)
361:Chromosome 1 (human)
78:List of PDB id codes
51:Available structures
3412:(Suppl 2): S59-60.
2845:2003Sci...299..256B
2743:2017Sci...357..208R
2638:10.1248/bpb.31.1321
2583:2013PLoSO...867983S
2115:Parkinson's disease
2102:Parkinson's disease
2018:Parkinson's disease
1518:guanine deglycation
1513:glucose homeostasis
906:cuprous ion binding
861:mercury ion binding
618:islet of Langerhans
3743:, which is in the
3443:Movement Disorders
2128:has been shown to
2074:mutator phenotypes
1663:ENSMUSG00000028964
1041:Biological process
930:Cellular component
901:hydrolase activity
856:copper ion binding
791:peptidase activity
776:cupric ion binding
759:Molecular function
3735:
3734:
3455:10.1002/mds.10422
3214:978-3-211-28927-3
2737:(6347): 208–211.
2004:androgen receptor
1882:
1881:
1878:
1877:
1851:
1850:
1816:
1815:
1795:
1794:
1765:
1764:
1744:
1743:
1714:
1713:
1695:
1694:
1669:
1668:
1650:
1649:
1624:
1623:
1605:
1604:
1553:
1552:
1508:insulin secretion
962:neuron projection
743:
742:
739:
738:
708:
707:
695:
694:
633:
632:
531:
530:
430:
429:
325:PARK7 - orthologs
257:
256:
253:
252:
61:Ortholog search:
16:(Redirected from
3768:
3721:
3706:
3691:
3676:
3661:
3646:
3631:
3616:
3601:
3586:
3566:
3559:
3552:
3543:
3509:
3499:
3490:(38): 36588–95.
3474:
3437:
3399:
3389:
3380:(40): 37556–63.
3364:
3354:
3321:
3292:
3263:
3226:
3191:
3181:
3148:
3111:
3082:
3045:
3027:
2993:
2992:
2982:
2973:(40): 37556–63.
2958:
2952:
2951:
2930:
2924:
2923:
2913:
2903:
2879:
2873:
2872:
2827:
2821:
2820:
2810:
2800:
2776:
2765:
2764:
2754:
2722:
2711:
2710:
2700:
2690:
2681:(31): 22809–20.
2666:
2660:
2659:
2649:
2621:
2615:
2614:
2604:
2594:
2562:
2556:
2555:
2537:
2509:
2503:
2502:
2474:
2468:
2467:
2457:
2447:
2423:
2417:
2416:
2406:
2382:
2376:
2375:
2365:
2341:
2335:
2334:
2324:
2300:
2294:
2293:
2286:
2280:
2279:
2272:
2259:
2258:
2247:
2241:
2240:
2229:
2223:
2213:
2202:
2192:
2000:oxidative stress
1976:Escherichia coli
1926:, also known as
1887:, also known as
1864:
1863:
1835:
1828:
1811:
1799:
1790:
1774:
1770:RefSeq (protein)
1760:
1748:
1739:
1723:
1699:
1680:
1654:
1635:
1609:
1590:
1559:
911:cadherin binding
771:cytokine binding
749:
735:
726:
719:
704:
644:
642:Top expressed in
637:
598:endothelial cell
582:
580:Top expressed in
575:
554:
537:
527:
514:
503:
488:
481:
475:
464:
452:
436:
426:
413:
402:
387:
380:
374:
363:
349:
333:
327:
278:
271:
248:
75:
69:
48:
47:
42:
30:
21:
3776:
3775:
3771:
3770:
3769:
3767:
3766:
3765:
3751:
3750:
3736:
3731:
3728:
3722:
3713:
3707:
3698:
3692:
3683:
3677:
3668:
3662:
3653:
3647:
3638:
3632:
3623:
3617:
3608:
3602:
3593:
3587:
3573:
3570:
3517:
3512:
3477:
3440:
3402:
3367:
3324:
3304:(1–2): 285–92.
3295:
3266:
3229:
3215:
3194:
3151:
3114:
3085:
3048:
3005:
3001:
2999:Further reading
2996:
2960:
2959:
2955:
2932:
2931:
2927:
2881:
2880:
2876:
2839:(5604): 256–9.
2831:parkinsonism".
2829:
2828:
2824:
2778:
2777:
2768:
2724:
2723:
2714:
2668:
2667:
2663:
2623:
2622:
2618:
2564:
2563:
2559:
2511:
2510:
2506:
2476:
2475:
2471:
2425:
2424:
2420:
2397:(45): 44552–9.
2384:
2383:
2379:
2356:(33): 31372–9.
2343:
2342:
2338:
2315:(33): 31380–4.
2302:
2301:
2297:
2288:
2287:
2283:
2274:
2273:
2262:
2249:
2248:
2244:
2231:
2230:
2226:
2214:
2205:
2193:
2184:
2180:
2158:
2123:
2111:
2082:
1992:
1948:
1920:
1913:
1908:
1873:View/Edit Mouse
1868:View/Edit Human
1831:
1824:
1821:Location (UCSC)
1807:
1786:
1782:
1756:
1735:
1731:
1644:ENSG00000116288
1537:
1036:
997:plasma membrane
925:
841:protein binding
731:
700:
691:
686:
684:temporal muscle
682:
678:
674:
670:
666:
662:
658:
654:
640:
629:
626:right ventricle
624:
620:
616:
612:
608:
604:
600:
596:
592:
578:
522:
509:
501:
491:
490:
489:
482:
462:
439:Gene location (
421:
408:
400:
390:
389:
388:
381:
359:
336:Gene location (
287:
274:
267:
84:
62:
28:
23:
22:
15:
12:
11:
5:
3774:
3772:
3764:
3763:
3753:
3752:
3733:
3732:
3730:
3729:
3723:
3716:
3714:
3708:
3701:
3699:
3693:
3686:
3684:
3678:
3671:
3669:
3663:
3656:
3654:
3648:
3641:
3639:
3633:
3626:
3624:
3618:
3611:
3609:
3603:
3596:
3594:
3588:
3581:
3578:
3575:
3574:
3571:
3569:
3568:
3561:
3554:
3546:
3540:
3539:
3516:
3515:External links
3513:
3511:
3510:
3475:
3438:
3400:
3365:
3343:10.1086/322996
3322:
3293:
3264:
3227:
3213:
3192:
3149:
3112:
3083:
3046:
3002:
3000:
2997:
2995:
2994:
2953:
2925:
2874:
2822:
2766:
2712:
2661:
2616:
2557:
2504:
2485:(4): 1036–48.
2469:
2418:
2377:
2336:
2295:
2281:
2260:
2242:
2224:
2203:
2181:
2179:
2176:
2175:
2174:
2172:Stress granule
2169:
2164:
2157:
2154:
2153:
2152:
2146:
2140:
2122:
2119:
2110:
2107:
2081:
2078:
1991:
1988:
1947:
1944:
1912:
1909:
1907:
1904:
1880:
1879:
1876:
1875:
1870:
1860:
1859:
1853:
1852:
1849:
1848:
1846:
1844:
1837:
1836:
1829:
1822:
1818:
1817:
1814:
1813:
1803:
1802:
1796:
1793:
1792:
1778:
1777:
1771:
1767:
1766:
1763:
1762:
1752:
1751:
1745:
1742:
1741:
1727:
1726:
1720:
1716:
1715:
1712:
1711:
1703:
1702:
1696:
1693:
1692:
1684:
1683:
1677:
1671:
1670:
1667:
1666:
1658:
1657:
1651:
1648:
1647:
1639:
1638:
1632:
1626:
1625:
1622:
1621:
1613:
1612:
1606:
1603:
1602:
1594:
1593:
1587:
1581:
1580:
1575:
1570:
1566:
1565:
1555:
1554:
1551:
1550:
1539:
1538:
1536:
1535:
1530:
1525:
1520:
1515:
1510:
1505:
1500:
1495:
1490:
1485:
1480:
1475:
1470:
1465:
1460:
1455:
1450:
1445:
1440:
1435:
1430:
1425:
1420:
1415:
1410:
1405:
1400:
1395:
1390:
1385:
1380:
1375:
1370:
1365:
1360:
1355:
1350:
1345:
1340:
1335:
1330:
1325:
1320:
1315:
1310:
1305:
1300:
1295:
1290:
1285:
1280:
1275:
1270:
1265:
1260:
1255:
1250:
1245:
1240:
1235:
1230:
1225:
1220:
1215:
1210:
1205:
1200:
1195:
1190:
1185:
1180:
1175:
1170:
1165:
1160:
1155:
1150:
1145:
1140:
1135:
1130:
1125:
1120:
1115:
1110:
1105:
1100:
1095:
1090:
1085:
1080:
1075:
1070:
1065:
1060:
1055:
1050:
1044:
1042:
1038:
1037:
1035:
1034:
1029:
1024:
1019:
1014:
1009:
1004:
999:
994:
989:
984:
979:
974:
969:
964:
959:
954:
949:
944:
939:
933:
931:
927:
926:
924:
923:
918:
913:
908:
903:
898:
893:
888:
883:
878:
873:
868:
863:
858:
853:
848:
843:
838:
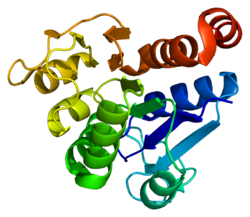
833:
828:
823:
818:
813:
808:
806:enzyme binding
803:
798:
796:kinase binding
793:
788:
783:
778:
773:
768:
762:
760:
756:
755:
745:
744:
741:
740:
737:
736:
728:
727:
716:
710:
709:
706:
705:
697:
696:
693:
692:
690:
689:
685:
681:
677:
673:
669:
665:
661:
657:
653:
649:
646:
645:
634:
631:
630:
628:
627:
623:
619:
615:
611:
607:
603:
599:
595:
594:deltoid muscle
591:
587:
584:
583:
571:
570:
562:
551:
545:
544:
541:RNA expression
533:
532:
529:
528:
520:
516:
515:
507:
504:
499:
493:
492:
483:
476:
470:
466:
465:
460:
454:
453:
445:
444:
432:
431:
428:
427:
419:
415:
414:
406:
403:
398:
392:
391:
382:
375:
369:
365:
364:
357:
351:
350:
342:
341:
329:
328:
285:
281:
280:
272:
264:
263:
259:
258:
255:
254:
251:
250:
80:
79:
71:
70:
59:
53:
52:
44:
43:
35:
34:
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
3773:
3762:
3759:
3758:
3756:
3749:
3748:
3746:
3745:public domain
3742:
3726:
3720:
3715:
3711:
3705:
3700:
3696:
3690:
3685:
3681:
3675:
3670:
3666:
3660:
3655:
3651:
3645:
3640:
3636:
3630:
3625:
3621:
3615:
3610:
3606:
3600:
3595:
3591:
3585:
3580:
3576:
3567:
3562:
3560:
3555:
3553:
3548:
3547:
3544:
3537:
3533:
3532:
3527:
3523:
3519:
3518:
3514:
3507:
3503:
3498:
3493:
3489:
3485:
3481:
3476:
3472:
3468:
3464:
3460:
3456:
3452:
3448:
3444:
3439:
3435:
3431:
3427:
3423:
3419:
3415:
3411:
3407:
3401:
3397:
3393:
3388:
3383:
3379:
3375:
3371:
3366:
3362:
3358:
3353:
3348:
3344:
3340:
3337:(3): 629–34.
3336:
3332:
3328:
3323:
3319:
3315:
3311:
3307:
3303:
3299:
3294:
3290:
3286:
3282:
3278:
3275:(2): 509–13.
3274:
3270:
3265:
3261:
3257:
3253:
3249:
3245:
3241:
3238:(3): 215–25.
3237:
3233:
3228:
3224:
3220:
3216:
3210:
3206:
3202:
3198:
3193:
3189:
3185:
3180:
3175:
3171:
3167:
3164:(2): 235–42.
3163:
3159:
3155:
3150:
3146:
3142:
3138:
3134:
3130:
3126:
3122:
3118:
3113:
3109:
3105:
3101:
3097:
3093:
3089:
3084:
3080:
3076:
3072:
3068:
3064:
3060:
3057:(3): 163–74.
3056:
3052:
3047:
3043:
3039:
3035:
3031:
3026:
3021:
3017:
3013:
3009:
3004:
3003:
2998:
2990:
2986:
2981:
2976:
2972:
2968:
2964:
2957:
2954:
2949:
2945:
2942:(4): 247–61.
2941:
2937:
2929:
2926:
2921:
2917:
2912:
2907:
2902:
2897:
2893:
2889:
2885:
2878:
2875:
2870:
2866:
2862:
2858:
2854:
2850:
2846:
2842:
2838:
2834:
2826:
2823:
2818:
2814:
2809:
2804:
2799:
2794:
2790:
2786:
2785:Int J Mol Sci
2782:
2775:
2773:
2771:
2767:
2762:
2758:
2753:
2748:
2744:
2740:
2736:
2732:
2728:
2721:
2719:
2717:
2713:
2708:
2704:
2699:
2694:
2689:
2684:
2680:
2676:
2672:
2665:
2662:
2657:
2653:
2648:
2643:
2639:
2635:
2632:(7): 1321–6.
2631:
2627:
2620:
2617:
2612:
2608:
2603:
2598:
2593:
2588:
2584:
2580:
2577:(7): e67983.
2576:
2572:
2568:
2561:
2558:
2553:
2549:
2545:
2541:
2536:
2531:
2527:
2523:
2519:
2515:
2514:Neurosci Lett
2508:
2505:
2500:
2496:
2492:
2488:
2484:
2480:
2473:
2470:
2465:
2461:
2456:
2451:
2446:
2441:
2437:
2433:
2429:
2422:
2419:
2414:
2410:
2405:
2400:
2396:
2392:
2388:
2381:
2378:
2373:
2369:
2364:
2359:
2355:
2351:
2347:
2340:
2337:
2332:
2328:
2323:
2318:
2314:
2310:
2306:
2299:
2296:
2291:
2285:
2282:
2277:
2271:
2269:
2267:
2265:
2261:
2256:
2252:
2246:
2243:
2238:
2234:
2228:
2225:
2221:
2217:
2212:
2210:
2208:
2204:
2200:
2196:
2191:
2189:
2187:
2183:
2177:
2173:
2170:
2168:
2165:
2163:
2160:
2159:
2155:
2150:
2147:
2144:
2141:
2138:
2135:
2134:
2133:
2131:
2127:
2120:
2118:
2116:
2108:
2106:
2103:
2099:
2095:
2091:
2087:
2079:
2077:
2075:
2071:
2067:
2063:
2059:
2055:
2051:
2048:
2045:
2041:
2040:methylglyoxal
2037:
2032:
2030:
2026:
2021:
2019:
2015:
2011:
2009:
2005:
2001:
1997:
1989:
1987:
1985:
1981:
1978:
1977:
1971:
1969:
1965:
1961:
1957:
1953:
1945:
1943:
1941:
1937:
1933:
1932:peptidase C56
1929:
1925:
1918:
1915:For C56, see
1910:
1905:
1903:
1901:
1898:
1894:
1890:
1886:
1874:
1869:
1865:
1861:
1858:
1854:
1847:
1845:
1842:
1838:
1834:
1830:
1827:
1823:
1819:
1812:
1810:
1804:
1800:
1797:
1791:
1789:
1785:
1779:
1775:
1772:
1768:
1761:
1759:
1753:
1749:
1746:
1740:
1738:
1734:
1728:
1724:
1721:
1719:RefSeq (mRNA)
1717:
1710:
1709:
1704:
1700:
1697:
1691:
1690:
1685:
1681:
1678:
1676:
1672:
1665:
1664:
1659:
1655:
1652:
1646:
1645:
1640:
1636:
1633:
1631:
1627:
1620:
1619:
1614:
1610:
1607:
1601:
1600:
1595:
1591:
1588:
1586:
1582:
1579:
1576:
1574:
1571:
1567:
1564:
1560:
1556:
1549:
1545:
1540:
1534:
1531:
1529:
1526:
1524:
1521:
1519:
1516:
1514:
1511:
1509:
1506:
1504:
1501:
1499:
1496:
1494:
1491:
1489:
1486:
1484:
1481:
1479:
1476:
1474:
1471:
1469:
1466:
1464:
1461:
1459:
1456:
1454:
1451:
1449:
1446:
1444:
1441:
1439:
1436:
1434:
1431:
1429:
1426:
1424:
1421:
1419:
1416:
1414:
1411:
1409:
1406:
1404:
1401:
1399:
1396:
1394:
1391:
1389:
1386:
1384:
1381:
1379:
1376:
1374:
1371:
1369:
1366:
1364:
1361:
1359:
1356:
1354:
1351:
1349:
1346:
1344:
1341:
1339:
1336:
1334:
1331:
1329:
1326:
1324:
1321:
1319:
1316:
1314:
1311:
1309:
1306:
1304:
1301:
1299:
1296:
1294:
1291:
1289:
1286:
1284:
1281:
1279:
1276:
1274:
1271:
1269:
1266:
1264:
1261:
1259:
1256:
1254:
1251:
1249:
1246:
1244:
1241:
1239:
1236:
1234:
1231:
1229:
1226:
1224:
1221:
1219:
1216:
1214:
1211:
1209:
1206:
1204:
1201:
1199:
1196:
1194:
1191:
1189:
1186:
1184:
1181:
1179:
1176:
1174:
1171:
1169:
1166:
1164:
1161:
1159:
1156:
1154:
1151:
1149:
1146:
1144:
1141:
1139:
1136:
1134:
1131:
1129:
1126:
1124:
1121:
1119:
1116:
1114:
1111:
1109:
1106:
1104:
1101:
1099:
1096:
1094:
1091:
1089:
1086:
1084:
1081:
1079:
1076:
1074:
1071:
1069:
1066:
1064:
1061:
1059:
1056:
1054:
1051:
1049:
1046:
1045:
1043:
1040:
1039:
1033:
1030:
1028:
1025:
1023:
1020:
1018:
1015:
1013:
1010:
1008:
1005:
1003:
1000:
998:
995:
993:
990:
988:
985:
983:
982:membrane raft
980:
978:
975:
973:
970:
968:
965:
963:
960:
958:
957:mitochondrion
955:
953:
950:
948:
945:
943:
940:
938:
935:
934:
932:
929:
928:
922:
919:
917:
914:
912:
909:
907:
904:
902:
899:
897:
894:
892:
889:
887:
884:
882:
879:
877:
874:
872:
869:
867:
864:
862:
859:
857:
854:
852:
849:
847:
844:
842:
839:
837:
834:
832:
829:
827:
824:
822:
819:
817:
814:
812:
809:
807:
804:
802:
799:
797:
794:
792:
789:
787:
784:
782:
779:
777:
774:
772:
769:
767:
764:
763:
761:
758:
757:
754:
753:Gene ontology
750:
746:
734:
729:
725:
720:
717:
715:
711:
703:
698:
687:
683:
679:
675:
671:
667:
663:
659:
655:
651:
650:
647:
643:
638:
635:
625:
621:
617:
613:
609:
605:
601:
597:
593:
589:
588:
585:
581:
576:
573:
572:
569:
567:
563:
561:
560:
556:
555:
552:
550:
546:
542:
538:
534:
526:
521:
517:
513:
508:
498:
494:
487:
480:
474:
467:
459:
455:
451:
446:
442:
437:
433:
425:
420:
416:
412:
407:
397:
393:
386:
379:
373:
366:
362:
356:
352:
348:
343:
339:
334:
330:
326:
322:
318:
314:
310:
306:
302:
298:
294:
290:
282:
277:
270:
265:
260:
249:
247:
243:
239:
235:
231:
227:
223:
219:
215:
211:
207:
203:
199:
195:
191:
187:
183:
179:
175:
171:
167:
163:
159:
155:
151:
147:
143:
139:
135:
131:
127:
123:
119:
115:
111:
107:
103:
99:
95:
91:
87:
81:
76:
73:
72:
68:
65:
58:
54:
49:
45:
41:
36:
31:
19:
3738:
3737:
3724:
3709:
3694:
3679:
3664:
3649:
3634:
3619:
3604:
3589:
3529:
3487:
3483:
3449:(7): 751–7.
3446:
3442:
3409:
3405:
3377:
3373:
3334:
3330:
3301:
3297:
3272:
3268:
3235:
3231:
3196:
3161:
3157:
3123:(1): 185–8.
3120:
3116:
3094:(1): 79–84.
3091:
3087:
3054:
3050:
3015:
3011:
2970:
2966:
2956:
2939:
2935:
2928:
2891:
2887:
2877:
2836:
2832:
2825:
2788:
2784:
2734:
2730:
2678:
2674:
2664:
2629:
2625:
2619:
2574:
2570:
2560:
2517:
2513:
2507:
2482:
2478:
2472:
2438:(11): e362.
2435:
2432:PLOS Biology
2431:
2421:
2394:
2390:
2380:
2353:
2349:
2339:
2312:
2308:
2298:
2284:
2254:
2245:
2236:
2227:
2125:
2124:
2121:Interactions
2112:
2083:
2069:
2058:nucleic acid
2035:
2033:
2022:
2012:
1993:
1974:
1972:
1949:
1935:
1927:
1923:
1921:
1896:
1888:
1884:
1883:
1806:
1784:NP_001116849
1781:
1755:
1733:NM_001123377
1730:
1706:
1687:
1661:
1642:
1616:
1597:
1577:
1572:
811:mRNA binding
564:
557:
523:150,998,894
510:150,981,590
284:External IDs
83:
3572:PDB gallery
3018:(1): 7–10.
2535:10045/94474
2050:nucleotides
1996:α-synuclein
1253:proteolysis
1032:nucleoplasm
886:RNA binding
680:Paneth cell
502:4|4 E2
262:Identifiers
2647:2115/53726
2520:: 134361.
2222:, May 2017
2201:, May 2017
2178:References
2086:DNA damage
2084:DJ-1 is a
2080:DNA repair
1498:DNA repair
1017:presynapse
568:(ortholog)
422:7,985,505
409:7,954,291
305:HomoloGene
2552:195813073
1980:chaperone
1968:peptidase
1960:α-helices
1958:and nine
1956:β-strands
1922:The gene
1906:Structure
1809:NP_065594
1788:NP_009193
1758:NM_020569
1737:NM_007262
1563:Orthologs
1363:autophagy
1007:cell body
987:chromatin
937:cytoplasm
313:GeneCards
3755:Category
3506:12851414
3471:44253517
3463:12815653
3434:13625056
3426:12548343
3396:11477070
3361:11462174
3318:11223268
3260:85481215
3252:17085780
3223:17017532
3188:15717024
3137:15503154
3108:15018843
3079:32685319
3071:14712351
3042:14513509
3034:12526767
2989:11477070
2948:12612053
2920:25071438
2869:27186691
2861:12446870
2817:37239999
2808:10218208
2761:28596309
2707:23792957
2656:18591768
2611:23844142
2571:PLOS ONE
2544:31276729
2499:16403519
2464:15502874
2413:12939276
2372:12761214
2331:12796482
2218:–
2197:–
2156:See also
2130:interact
2047:glycated
1990:Function
1857:Wikidata
1542:Sources:
1012:PML body
947:membrane
3536:PDBe-KB
3526:UniProt
3352:1235491
3289:9070310
3158:NeuroRx
3145:9453283
2911:4075472
2841:Bibcode
2833:Science
2739:Bibcode
2731:Science
2698:3829365
2602:3699467
2579:Bibcode
2220:Ensembl
2199:Ensembl
2054:Guanine
2044:glyoxal
2029:mercury
1984:Archaea
1946:Protein
1893:protein
1891:, is a
1675:UniProt
1630:Ensembl
1569:Species
1548:QuickGO
967:nucleus
942:cytosol
543:pattern
401:1p36.23
301:2135637
269:Aliases
3531:Q99497
3504:
3469:
3461:
3432:
3424:
3394:
3359:
3349:
3316:
3287:
3258:
3250:
3221:
3211:
3186:
3179:534935
3176:
3143:
3135:
3106:
3077:
3069:
3040:
3032:
3012:Neuron
2987:
2946:
2918:
2908:
2894:: 58.
2867:
2859:
2815:
2805:
2791:(10).
2759:
2705:
2695:
2654:
2609:
2599:
2550:
2542:
2497:
2462:
2455:521177
2452:
2411:
2370:
2329:
2143:EFCAB6
2132:with:
2025:copper
1938:has 8
1917:MEROPS
1843:search
1841:PubMed
1708:Q99LX0
1689:Q99497
1585:Entrez
714:BioGPS
293:602533
3467:S2CID
3430:S2CID
3256:S2CID
3141:S2CID
3075:S2CID
3038:S2CID
2865:S2CID
2548:S2CID
2149:PIAS2
2145:, and
2126:PARK7
2070:PARK7
2036:PARK7
2034:DJ-1/
1964:dimer
1940:exons
1936:PARK7
1924:PARK7
1897:PARK7
1618:57320
1599:11315
1578:Mouse
1573:Human
1544:Amigo
590:tibia
566:Mouse
559:Human
506:Start
441:Mouse
405:Start
338:Human
317:PARK7
309:38295
276:PARK7
33:PARK7
18:PARK7
3725:2or3
3710:1ucf
3695:1soa
3680:1q2u
3665:1ps4
3650:1pe0
3635:1pdw
3620:1pdv
3605:1p5f
3590:1j42
3524:for
3502:PMID
3459:PMID
3422:PMID
3392:PMID
3357:PMID
3314:PMID
3298:Gene
3285:PMID
3248:PMID
3219:PMID
3209:ISBN
3184:PMID
3133:PMID
3104:PMID
3067:PMID
3030:PMID
2985:PMID
2944:PMID
2916:PMID
2857:PMID
2813:PMID
2757:PMID
2703:PMID
2652:PMID
2607:PMID
2540:PMID
2495:PMID
2460:PMID
2409:PMID
2368:PMID
2327:PMID
2137:CASK
2096:and
2042:and
2027:and
1928:DJ-1
1911:Gene
1900:gene
1027:axon
549:Bgee
497:Band
458:Chr.
396:Band
355:Chr.
289:OMIM
246:4S0Z
242:4RKY
238:4RKW
234:4ZGG
230:4P36
226:4P35
222:4P34
218:4P2G
214:4OQ4
210:4OGF
206:4N12
202:4N0M
198:4MTC
194:4MNT
190:4BTE
186:3SF8
182:3F71
178:3EZG
174:3CZA
170:3CZ9
166:3CYF
162:3CY6
158:3BWE
154:3B3A
150:3B38
146:3B36
142:2RK6
138:2RK4
134:2RK3
130:2R1V
126:2R1U
122:2R1T
118:2OR3
114:1UCF
110:1SOA
106:1Q2U
102:1PE0
98:1PDW
94:1PDV
90:1P5F
86:1J42
67:RCSB
64:PDBe
3522:PDB
3492:doi
3488:278
3451:doi
3414:doi
3382:doi
3378:276
3347:PMC
3339:doi
3306:doi
3302:263
3277:doi
3273:231
3240:doi
3201:doi
3174:PMC
3166:doi
3125:doi
3121:318
3096:doi
3059:doi
3020:doi
2975:doi
2971:276
2906:PMC
2896:doi
2849:doi
2837:299
2803:PMC
2793:doi
2747:doi
2735:357
2693:PMC
2683:doi
2679:288
2642:hdl
2634:doi
2597:PMC
2587:doi
2530:hdl
2522:doi
2518:708
2487:doi
2483:356
2450:PMC
2440:doi
2399:doi
2395:278
2358:doi
2354:278
2317:doi
2313:278
2117:7.
2066:RNA
2064:or
2062:DNA
2052:.
1952:kDa
519:End
418:End
321:OMA
297:MGI
57:PDB
3757::
3528::
3500:.
3486:.
3482:.
3465:.
3457:.
3447:18
3445:.
3428:.
3420:.
3410:23
3408:.
3390:.
3376:.
3372:.
3355:.
3345:.
3335:69
3333:.
3329:.
3312:.
3300:.
3283:.
3271:.
3254:.
3246:.
3236:29
3234:.
3217:.
3207:.
3182:.
3172:.
3160:.
3156:.
3139:.
3131:.
3119:.
3102:.
3090:.
3073:.
3065:.
3055:82
3053:.
3036:.
3028:.
3016:37
3014:.
3010:.
2983:.
2969:.
2965:.
2938:.
2914:.
2904:.
2890:.
2886:.
2863:.
2855:.
2847:.
2835:.
2811:.
2801:.
2789:24
2787:.
2783:.
2769:^
2755:.
2745:.
2733:.
2729:.
2715:^
2701:.
2691:.
2677:.
2673:.
2650:.
2640:.
2630:31
2628:.
2605:.
2595:.
2585:.
2573:.
2569:.
2546:.
2538:.
2528:.
2516:.
2493:.
2481:.
2458:.
2448:.
2434:.
2430:.
2407:.
2393:.
2389:.
2366:.
2352:.
2348:.
2325:.
2311:.
2307:.
2263:^
2253:.
2235:.
2206:^
2185:^
2076:.
2031:.
2020:.
1902:.
1546:/
525:bp
512:bp
424:bp
411:bp
319:;
315::
311:;
307::
303:;
299::
295:;
291::
244:,
240:,
236:,
232:,
228:,
224:,
220:,
216:,
212:,
208:,
204:,
200:,
196:,
192:,
188:,
184:,
180:,
176:,
172:,
168:,
164:,
160:,
156:,
152:,
148:,
144:,
140:,
136:,
132:,
128:,
124:,
120:,
116:,
112:,
108:,
104:,
100:,
96:,
92:,
88:,
3747:.
3565:e
3558:t
3551:v
3538:.
3508:.
3494::
3473:.
3453::
3436:.
3416::
3398:.
3384::
3363:.
3341::
3320:.
3308::
3291:.
3279::
3262:.
3242::
3225:.
3203::
3190:.
3168::
3162:1
3147:.
3127::
3110:.
3098::
3092:4
3081:.
3061::
3044:.
3022::
2991:.
2977::
2950:.
2940:1
2922:.
2898::
2892:7
2871:.
2851::
2843::
2819:.
2795::
2763:.
2749::
2741::
2709:.
2685::
2658:.
2644::
2636::
2613:.
2589::
2581::
2575:8
2554:.
2532::
2524::
2501:.
2489::
2466:.
2442::
2436:2
2415:.
2401::
2374:.
2360::
2333:.
2319::
2292:.
2278:.
2257:.
2239:.
2151:.
2139:,
2060:(
1919:.
443:)
340:)
323::
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.