2078:
structural and functional mitochondrial alterations). These findings provided explanation for previous and subsequent reports demonstrating tissue protective effects of PARP inhibitors and the PARP1 knockout phenotypes in various models of ischemia-reperfusion injury (e.g. in stroke, in the heart and in the gut) where oxidative stress-induced cell death is a central cellular event. Later, apoptosis inducing factor (AIF; a misnomer) was identified as a key mediator of the PARP1-mediated regulated necrotic cell death pathway termed parthanatos.
477:
454:
5251:
351:
376:
5221:
5236:
5311:
5296:
1955:, in stark contrast to cells with at least one good copy of both BRCA1 and BRCA2. Many breast cancers have defects in the BRCA1/BRCA2 HR repair pathway due to mutations in either BRCA1 or BRCA2, or other essential genes in the pathway (the latter termed cancers with "BRCAness"). Tumors with BRCAness are hypothesized to be highly sensitive to PARP1 inhibitors, and it has been demonstrated in mice that these inhibitors can both prevent BRCA1/2-deficient
5266:
5281:
728:
483:
382:
40:
2090:(and presumably other plants), PARP2 plays more significant roles than PARP1 in protective responses to DNA damage and bacterial pathogenesis. The plant PARP2 carries PARP regulatory and catalytic domains with only intermediate similarity to PARP1, and carries N-terminal SAP DNA binding motifs rather than the Zn-finger DNA binding motifs of plant and animal PARP1 proteins.
2008:. These findings indicate a linkage between longevity and PARP-mediated DNA repair capability. Furthermore, PARP can also act against production of reactive oxygen species, which may contribute to longevity by inhibiting oxidative damage to DNA and proteins. These observations suggest that PARP activity contributes to mammalian longevity, consistent with the
1974:. It is hypothesized that PARP1 inhibitors may prove highly effective therapies for cancers with BRCAness, due to the high sensitivity of the tumors to the inhibitor and the lack of deleterious effects on the remaining healthy cells with functioning BRCA HR pathway. This is in contrast to conventional
2046:
expression level of both PARP1 and PARP2 genes reduced by half, compared to their levels in young adult humans (19 to 26 years old). However, centenarians (humans aged 100 to 107 years of age) have constitutive expression of PARP1 at levels similar to those of young individuals. This high level of
2077:
Following severe DNA damage, excessive activation of PARP1 can lead to cell death. Initially, overactivation of the enzyme was linked to apoptotic cell death but later, PARP1-mediated cell death turned out to show characteristics of necrotic cell death (i.e. early plasma membrane disruption,
66:
2030:
transcription factor which interacts with multiple ETS1 binding sites in the promoter region of PARP1. The degree to which the ETS1 transcription factor can bind to its binding sites on the PARP1 promoter depends on the methylation status of the
4110:
3840:
2560:
1902:
PARP1 is also over-expressed in a number of other cancers, including neuroblastoma, HPV infected oropharyngeal carcinoma, testicular and other germ cell tumors, Ewing's sarcoma, malignant lymphoma, breast cancer, and colon cancer.
4316:
Erdélyi K, Bakondi E, Gergely P, Szabó C, Virág L (April 2005). "Pathophysiologic role of oxidative stress-induced poly(ADP-ribose) polymerase-1 activation: focus on cell death and transcriptional regulation".
1950:
are at least partially necessary for the HR pathway to function. Cells that are deficient in BRCA1 or BRCA2 have been shown to be highly sensitive to PARP1 inhibition or knock-down, resulting in cell death by
2019:'s primary functional target through its interaction with the tyrosyl tRNA synthetase (TyrRS). Tyrosyl tRNA synthetase translocates to the nucleus under stress conditions stimulating NAD-dependent auto-poly-
5250:
1990:
blood cells of thirteen mammalian species (rat, guinea pig, rabbit, marmoset, sheep, pig, cattle, pigmy chimpanzee, horse, donkey, gorilla elephant and man) correlates with maximum lifespan of the species.
1995:
cell lines established from blood samples of humans who were centenarians (100 years old or older) have significantly higher PARP activity than cell lines from younger (20 to 70 years old) individuals. The
1884:(MMEJ). MMEJ is associated with frequent chromosome abnormalities such as deletions, translocations, inversions and other complex rearrangements. When PARP1 is up-regulated, MMEJ is increased, causing
2086:
Plants have a PARP1 with substantial similarity to animal PARP1, and roles of poly(ADP-ribosyl)ation in plant responses to DNA damage, infection and other stresses have been studied. Intriguingly, in
1791:(HR) repair, a potentially error-free repair mechanism. For this reason, cells lacking PARP1 show a hyper-recombinagenic phenotype (e.g., an increased frequency of HR), which has also been observed
2069:
and PARP1 have a roughly equal affinity for the NAD+ that both enzymes require for activity. But DNA damage can increase levels of PARP1 more than 100-fold, leaving little NAD+ for SIRT1.
5220:
1914:
of a DNA repair gene is less usual in cancer. For instance, at least 36 DNA repair enzymes, when mutationally defective in germ line cells, cause increased risk of cancer (hereditary
3728:
Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. (April 2005). "Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase".
4571:
Yu SW, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, et al. (July 2002). "Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor".
3309:"Promoter hypomethylation, especially around the E26 transformation-specific motif, and increased expression of poly (ADP-ribose) polymerase 1 in BRCA-mutated serous ovarian cancer"
3942:"Genetic cooperation between the Werner syndrome protein and poly(ADP-ribose) polymerase-1 in preventing chromatid breaks, complex chromosomal rearrangements, and cancer in mice"
5235:
3491:
Newman RE, Soldatenkov VA, Dritschilo A, Notario V (2002). "Poly(ADP-ribose) polymerase turnover alterations do not contribute to PARP overexpression in Ewing's sarcoma cells".
3258:"Poly (ADP-ribose) polymerase 1 transcriptional regulation: a novel crosstalk between histone modification H3K9ac and ETS1 motif hypomethylation in BRCA1-mutated ovarian cancer"
2441:
Nilov, DK; Pushkarev, SV; Gushchina, IV; Manasaryan, GA; Kirsanov, KI; Švedas, VK (2020). "Modeling of the enzyme-substrate complexes of human poly(ADP-ribose) polymerase 1".
5095:
5310:
3781:
Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. (April 2005). "Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy".
3405:"Subjugation of TGFβ Signaling by Human Papilloma Virus in Head and Neck Squamous Cell Carcinoma Shifts DNA Repair from Homologous Recombination to Alternative End Joining"
5295:
1642:
family of enzymes, accounting for 90% of the NAD+ used by the family. PARP1 is mostly present in cell nucleus, but cytosolic fraction of this protein was also reported.
490:
389:
4215:"Oxidative DNA damage repair and parp 1 and parp 2 expression in Epstein-Barr virus-immortalized B lymphocyte cells from young subjects, old subjects, and centenarians"
3528:
Tomoda T, Kurashige T, Moriki T, Yamamoto H, Fujimoto S, Taniguchi T (August 1991). "Enhanced expression of poly(ADP-ribose) synthetase gene in malignant lymphoma".
4993:"Functional association of poly(ADP-ribose) polymerase with DNA polymerase alpha-primase complex: a link between DNA strand break detection and DNA replication"
3993:"PARP-1 inhibition with or without ionizing radiation confers reactive oxygen species-mediated cytotoxicity preferentially to cancer cells with mutant TP53"
5265:
5280:
3895:
Muiras ML, Müller M, Schächter F, Bürkle A (April 1998). "Increased poly(ADP-ribose) polymerase activity in lymphoblastoid cell lines from centenarians".
1351:
2385:"Quantitative analysis of biochemical processes in living cells at a single-molecule level: a case of olaparib–PARP1 (DNA repair protein) interactions"
312:
2493:
1332:
1779:
or inhibiting PARP1 activity with small molecules reduces repair of ssDNA breaks. In the absence of PARP1, when these breaks are encountered during
2338:
2023:
of PARP1, thereby altering the functions of PARP1 from a chromatin architectural protein to a DNA damage responder and transcription regulator.
1899:
hypomethylated, and this contributes to progression to endometrial cancer, BRCA-mutated ovarian cancer, and BRCA-mutated serous ovarian cancer.
3146:"c-MYC Generates Repair Errors via Increased Transcription of Alternative-NHEJ Factors, LIG3 and PARP1, in Tyrosine Kinase-Activated Leukemias"
1997:
5200:
2266:
63:
5339:
4482:
4362:"Inhibition and down-regulation of poly(ADP-ribose) polymerase results in a marked resistance of HL-60 cells to various apoptosis-inducers"
4263:
2248:
1926:.) Ordinarily, deficient expression of a DNA repair enzyme results in increased un-repaired DNA damage which, through replication errors (
3039:"Multifaceted Role of PARP-1 in DNA Repair and Inflammation: Pathological and Therapeutic Implications in Cancer and Non-Cancer Diseases"
4951:
4908:
4865:
4822:
1922:.) Similarly, at least 12 DNA repair genes have frequently been found to be epigenetically repressed in one or more cancers. (See also
1553:
476:
4393:"Intact cell evidence for the early synthesis, and subsequent late apopain-mediated suppression, of poly(ADP-ribose) during apoptosis"
3446:
Mego M, Cierna Z, Svetlovska D, Macak D, Machalekova K, Miskovska V, et al. (July 2013). "PARP expression in germ cell tumours".
2127:
1931:
1881:
1761:
1560:
3946:
2062:). The high constitutive levels of PARP1 in centenarians were thought to be due to altered epigenetic control of PARP1 expression.
1655:
3836:"Poly(ADP-ribose) polymerase activity in mononuclear leukocytes of 13 mammalian species correlates with species-specific life span"
453:
2685:"PARP inhibition versus PARP-1 silencing: different outcomes in terms of single-strand break repair and radiation susceptibility"
1729:
2540:
5349:
2235:
2214:
5044:"XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage"
5344:
5093:
Ku MC, Stewart S, Hata A (November 2003). "Poly(ADP-ribose) polymerase 1 interacts with OAZ and regulates BMP-target genes".
5048:
3530:
1919:
2231:
375:
350:
3897:
3448:
3206:
1978:, which are highly toxic to all cells and can induce DNA damage in healthy cells, leading to secondary cancer generation.
2210:
1934:
repair is highly inaccurate, so in this case, over-expression, rather than under-expression, apparently leads to cancer.
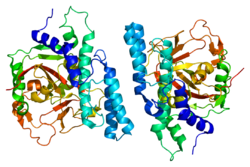
5259:: Crystal structure of catalytic domain of human poly(ADP-ribose) polymerase complexed with a quinoxaline-type inhibitor
1659:
292:
4818:"Regulation of protein synthesis in heart muscle. II. Effect of amino acid levels and insulin on ribosomal aggregation"
2787:"Stimulation of intrachromosomal homologous recombination in mammalian cells by an inhibitor of poly(ADP-ribosylation)"
2384:
2035:
in the ETS1 binding sites in the PARP1 promoter. If these CpG islands in ETS1 binding sites of the PARP1 promoter are
3202:"Hypomethylation of ETS transcription factor binding sites and upregulation of PARP1 expression in endometrial cancer"
2988:"Macrophage Immunometabolism and Inflammaging: Roles of Mitochondrial Dysfunction, Cellular Senescence, CD38, and NAD"
2099:
489:
388:
4434:"Peroxynitrite-induced thymocyte apoptosis: the role of caspases and poly (ADP-ribose) synthetase (PARS) activation"
2182:
2155:
2059:
2009:
1757:
1753:
1674:
1639:
482:
381:
5193:
3366:
3150:
1775:
PARP1 has a role in repair of single-stranded DNA (ssDNA) breaks. Knocking down intracellular PARP1 levels with
2992:
2004:, a human premature aging disorder. PARP1 and Wrn proteins are part of a complex involved in the processing of
1677:
family act on single strands; or, when BRCA fails, PARP takes over those jobs as well (in a DNA repair context).
4779:
3409:
1788:
1765:
1396:
300:
283:, ADPRT, ADPRT 1, ADPRT1, ARTD1, PARP, PARP-1, PPOL, pADPRT-1, poly(ADP-ribose) polymerase 1, Poly-PARP, PARS
4624:
3096:
2939:
2286:
1819:
1377:
4997:
4528:
Virág L, Szabó C (September 2002). "The therapeutic potential of poly(ADP-ribose) polymerase inhibitors".
2893:
2842:
2791:
2740:
2689:
2026:
The messenger RNA level and protein level of PARP1 is controlled, in part, by the expression level of the
1752:
factors. PARP1 is implicated in the regulation of several DNA repair processes including the pathways of
4483:"Poly(ADP-ribose) synthetase activation mediates mitochondrial injury during oxidant-induced cell death"
4219:
1835:
1827:
4904:"The enzymatic and DNA binding activity of PARP-1 are not required for NF-kappa B coactivator function"
3575:"Nuclear PARP-1 protein overexpression is associated with poor overall survival in early breast cancer"
2489:"Pleiotropic cellular functions of PARP1 in longevity and aging: genome maintenance meets inflammation"
3144:
Muvarak N, Kelley S, Robert C, Baer MR, Perrotti D, Gambacorti-Passerini C, et al. (April 2015).
1959:
from becoming tumors and eradicate tumors having previously formed from BRCA1/2-deficient xenografts.
5186:
5139:
4717:"PARP2 Is the Predominant Poly(ADP-Ribose) Polymerase in Arabidopsis DNA Damage and Immune Responses"
4580:
4119:
4106:"Automodification switches PARP-1 function from chromatin architectural protein to histone chaperone"
4060:
3849:
3792:
3739:
3631:
2638:
2569:
2399:
364:
3616:
Dziaman T, Ludwiczak H, Ciesla JM, Banaszkiewicz Z, Winczura A, Chmielarczyk M, et al. (2014).
2889:"Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease"
5229:: Crystal structure of catalytic domain of human poly(ADP-ribose) polymerase with a novel inhibitor
1971:
1846:
1769:
1712:
279:
4391:
Rosenthal DS, Ding R, Simbulan-Rosenthal CM, Vaillancourt JP, Nicholson DW, Smulson M (May 1997).
1517:
4604:
4553:
4510:
4342:
4298:
4171:
3997:
3922:
3816:
3763:
3579:
3573:
Rojo F, García-Parra J, Zazo S, Tusquets I, Ferrer-Lozano J, Menendez S, et al. (May 2012).
3555:
3473:
2683:
Godon C, Cordelières FP, Biard D, Giocanti N, Mégnin-Chanet F, Hall J, Favaudon V (August 2008).
2466:
2423:
2311:
1885:
1663:
324:
2556:"Activation of the abundant nuclear factor poly(ADP-ribose) polymerase-1 by Helicobacter pylori"
1513:
1487:
1466:
5042:
Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, de Murcia G (June 1998).
1796:
5166:
5112:
5075:
5024:
4970:
4927:
4884:
4841:
4798:
4750:
4694:
4641:
4596:
4545:
4502:
4463:
4414:
4373:
4334:
4290:
4236:
4190:
4167:"Regulation of the human poly(ADP-ribose) polymerase promoter by the ETS transcription factor"
4147:
4086:
4024:
3973:
3914:
3877:
3808:
3755:
3710:
3659:
3598:
3547:
3510:
3465:
3428:
3385:
3342:
3289:
3235:
3177:
3123:
3072:
3019:
2968:
2912:
2869:
2818:
2767:
2716:
2665:
2597:
2554:
Nossa CW, Jain P, Tamilselvam B, Gupta VR, Chen LF, Schreiber V, et al. (November 2009).
2522:
2458:
2415:
2365:
2303:
2058:
sublethal oxidative DNA damage. Higher DNA repair is thought to contribute to longevity (see
2048:
1787:
stalls, and double-strand DNA (dsDNA) breaks accumulate. These dsDNA breaks are repaired via
272:
56:
4213:
Chevanne M, Calia C, Zampieri M, Cecchinelli B, Caldini R, Monti D, et al. (June 2007).
5156:
5148:
5104:
5065:
5057:
5014:
5006:
4960:
4917:
4874:
4831:
4788:
4740:
4730:
4684:
4674:
4633:
4588:
4537:
4494:
4453:
4445:
4404:
4326:
4280:
4272:
4228:
4180:
4137:
4127:
4076:
4068:
4051:
4014:
4006:
3963:
3955:
3906:
3867:
3857:
3800:
3783:
3747:
3730:
3700:
3692:
3649:
3639:
3588:
3539:
3502:
3493:
3457:
3418:
3375:
3332:
3322:
3279:
3271:
3225:
3215:
3167:
3159:
3113:
3105:
3062:
3052:
3009:
3001:
2958:
2948:
2902:
2859:
2851:
2808:
2800:
2757:
2749:
2706:
2698:
2655:
2647:
2587:
2577:
2512:
2502:
2450:
2407:
2355:
2347:
2295:
2160:
2020:
1784:
1737:
1667:
569:
500:
444:
399:
3403:
Liu Q, Ma L, Jones T, Palomero L, Pujana MA, Martinez-Ruiz H, et al. (December 2018).
2887:
Wang ZQ, Auer B, Stingl L, Berghammer H, Haidacher D, Schweiger M, Wagner EF (March 1995).
727:
320:
5244:: Crystal structure of human poly(ADP-ribose) polymerase complexed with a potent inhibitor
3683:
2043:
2001:
1915:
1780:
1701:
544:
2736:"Poly(ADP-ribose) polymerase (PARP-1) has a controlling role in homologous recombination"
1803:(cells without functioning PARP1) do not show an unhealthy phenotype, and in fact, PARP1
4947:"Poly(ADP-ribose) binds to specific domains of p53 and alters its DNA binding functions"
4584:
4285:
4258:
4123:
4104:
Muthurajan UM, Hepler MR, Hieb AR, Clark NJ, Kramer M, Yao T, Luger K (September 2014).
4064:
3991:
Liu Q, Gheorghiu L, Drumm M, Clayman R, Eidelman A, Wszolek MF, et al. (May 2018).
3853:
3796:
3743:
3635:
3618:"PARP-1 expression is increased in colon adenoma and carcinoma and correlates with OGG1"
3362:"Alternative NHEJ Pathway Components Are Therapeutic Targets in High-Risk Neuroblastoma"
3092:"Homology and enzymatic requirements of microhomology-dependent alternative end joining"
2573:
2403:
5161:
5134:
4745:
4716:
4689:
4665:
4660:
4458:
4433:
4142:
4105:
4081:
4046:
4019:
3992:
3968:
3941:
3705:
3678:
3654:
3617:
3337:
3308:
3284:
3257:
3230:
3201:
3172:
3145:
3118:
3091:
3067:
3043:
3038:
3014:
2987:
2963:
2934:
2864:
2837:
2711:
2684:
2660:
2633:
2592:
2555:
2517:
2488:
2360:
2333:
2284:
Ha HC, Snyder SH (August 2000). "Poly(ADP-ribose) polymerase-1 in the nervous system".
2171:
1992:
1986:
PARP activity (which is mainly due to PARP1) measured in the permeabilized mononuclear
1975:
1967:
1888:. PARP1 is up-regulated and MMEJ is increased in tyrosine kinase-activated leukemias.
1869:
1716:
1692:
39:
5319:: Solution structure of the first zf-PARP domain of human Poly(ADP-ribose)polymerase-1
5070:
5043:
5019:
4992:
4836:
4817:
4047:"A human tRNA synthetase is a potent PARP1-activating effector target for resveratrol"
3959:
2813:
2786:
2762:
2735:
1266:
1261:
1256:
1251:
1246:
1241:
1236:
1231:
1226:
1221:
1216:
1211:
1206:
1201:
1196:
1191:
1186:
1181:
1176:
1171:
1166:
1161:
1156:
1151:
1146:
1141:
1136:
1131:
1126:
1121:
1116:
1111:
1106:
1101:
1096:
1091:
1086:
1081:
1076:
1071:
1066:
1061:
1056:
1051:
1046:
1041:
1036:
1031:
1026:
1021:
1016:
1011:
1006:
1001:
996:
991:
986:
981:
976:
971:
955:
950:
945:
940:
935:
930:
925:
920:
915:
910:
905:
900:
895:
879:
874:
869:
864:
859:
854:
849:
844:
839:
834:
829:
824:
819:
814:
809:
804:
799:
794:
789:
784:
779:
774:
769:
5333:
4991:
Dantzer F, Nasheuer HP, Vonesch JL, de Murcia G, Ménissier-de Murcia J (April 1998).
4773:
Gueven N, Becherel OJ, Kijas AW, Chen P, Howe O, Rudolph JH, et al. (May 2004).
4721:
4449:
3872:
3835:
2470:
2427:
1861:
1831:
1804:
756:
5304:: Solution structure of the second Zn-finger domain of poly(ADP-ribose) polymerase-1
4608:
4557:
4346:
4302:
3926:
3559:
3090:
Sharma S, Javadekar SM, Pandey M, Srivastava M, Kumari R, Raghavan SC (March 2015).
2315:
304:
4514:
4214:
4165:
Soldatenkov VA, Albor A, Patel BK, Dreszer R, Dritschilo A, Notario V (July 1999).
3820:
3767:
1880:
PARP1 is one of six enzymes required for the highly error-prone DNA repair pathway
1823:
1102:
regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway
562:
341:
4637:
4259:"Nicotinamide is an inhibitor of SIRT1 in vitro, but can be a stimulator in cells"
3477:
3461:
3423:
3404:
3380:
3361:
3163:
4735:
4679:
3644:
3005:
2651:
328:
4498:
4360:
Tanaka Y, Yoshihara K, Tohno Y, Kojima K, Kameoka M, Kamiya T (September 1995).
2271:
National Center for
Biotechnology Information, U.S. National Library of Medicine
2253:
National Center for
Biotechnology Information, U.S. National Library of Medicine
2177:
2036:
2016:
1896:
1800:
5108:
4111:
Proceedings of the
National Academy of Sciences of the United States of America
3841:
Proceedings of the
National Academy of Sciences of the United States of America
3696:
2561:
Proceedings of the
National Academy of Sciences of the United States of America
2351:
870:
RNA polymerase II transcription regulatory region sequence-specific DNA binding
645:
4330:
4276:
4010:
3313:
3262:
2454:
2187:
2005:
1927:
1923:
1865:
1839:
1749:
1733:
1267:
positive regulation of double-strand break repair via homologous recombination
461:
358:
308:
4965:
4946:
4879:
4860:
3862:
3275:
2953:
2804:
2047:
PARP1 expression in centenarians was shown to allow more efficient repair of
5010:
4592:
4132:
3593:
3574:
3327:
2609:
2582:
2107:
2066:
1987:
1956:
1952:
1850:
1745:
1296:
705:
583:
528:
515:
427:
414:
316:
5170:
5116:
4931:
4922:
4903:
4888:
4802:
4754:
4698:
4645:
4600:
4549:
4432:
Virág L, Scott GS, Cuzzocrea S, Marmer D, Salzman AL, Szabó C (July 1998).
4409:
4392:
4361:
4338:
4294:
4240:
4194:
4185:
4166:
4151:
4090:
4028:
3977:
3812:
3759:
3714:
3663:
3602:
3543:
3514:
3469:
3432:
3389:
3346:
3293:
3239:
3181:
3127:
3076:
3023:
2972:
2873:
2771:
2720:
2669:
2601:
2526:
2462:
2419:
2369:
2307:
2299:
5079:
5061:
5028:
4974:
4845:
4506:
4467:
4418:
4377:
4232:
3918:
3910:
3881:
3551:
3360:
Newman EA, Lu F, Bashllari D, Wang L, Opipari AW, Castle VP (March 2015).
3220:
3109:
2916:
2822:
2507:
1807:
show no negative phenotype and no increased incidence of tumor formation.
1600:
1595:
4793:
4774:
3622:
3057:
2855:
2753:
2702:
2334:"NAD + metabolism: pathophysiologic mechanisms and therapeutic potential"
2165:
2032:
1816:
1584:
1441:
1422:
4661:"Protein ADP-Ribosylation Takes Control in Plant-Bacterium Interactions"
4541:
4072:
3804:
3751:
2907:
2888:
2610:"Team finds link between stomach-cancer bug and cancer-promoting factor"
1137:
positive regulation of intracellular estrogen receptor signaling pathway
875:
DNA-binding transcription activator activity, RNA polymerase II-specific
3679:"Therapeutic Potential of NAD-Boosting Molecules: The In Vivo Evidence"
3506:
2935:"Poly(ADP-Ribose)Polymerase-1 in Lung Inflammatory Disorders: A Review"
2411:
1792:
1741:
1673:
In conjunction with BRCA, which acts on double strands; members of the
1408:
1363:
5152:
5135:"Beyond DNA repair, the immunological role of PARP-1 and its siblings"
1860:
PARP1 facilitates recruitment and function of immune cells, including
249:
245:
241:
237:
233:
229:
225:
221:
217:
213:
209:
205:
201:
197:
193:
189:
185:
181:
177:
173:
169:
165:
161:
157:
153:
149:
145:
141:
137:
133:
129:
125:
121:
117:
113:
109:
105:
101:
97:
93:
89:
85:
17:
2142:
1857:
1628:
1568:
1318:
5274:: Solution structure of BRCT domain of poly(ADP-ribose) polymerase-1
4622:
Briggs AG, Bent AF (July 2011). "Poly(ADP-ribosyl)ation in plants".
1062:
negative regulation of telomere maintenance via telomere lengthening
5289:: Solution structure of WGR domain of poly(ADP-ribose) polymerase-1
4775:"Aprataxin, a novel protein that protects against genotoxic stress"
4902:
Hassa PO, Covic M, Hasan S, Imhof R, Hottiger MO (December 2001).
3940:
Lebel M, Lavoie J, Gaudreault I, Bronsard M, Drouin R (May 2003).
2838:"PARP1 suppresses homologous recombination events in mice in vivo"
2541:"Entrez Gene: PARP1 poly (ADP-ribose) polymerase family, member 1"
2137:
2132:
2112:
1947:
1943:
1843:
1776:
972:
positive regulation of transcription regulatory region DNA binding
3256:
Li D, Bi FF, Cao JM, Cao C, Li CY, Liu B, Yang Q (January 2014).
2734:
Schultz N, Lopez E, Saleh-Gohari N, Helleday T (September 2003).
1281:
1277:
2117:
2027:
1892:
1842:
that increase with age in many tissues. ADP-riboyslation of the
1635:
296:
5182:
4945:
Malanga M, Pleschke JM, Kleczkowska HE, Althaus FR (May 1998).
5178:
4816:
Morgan HE, Jefferson LS, Wolpert EB, Rannels DE (April 1971).
2122:
736:
1192:
nucleotide-excision repair, preincision complex stabilization
1152:
cellular response to transforming growth factor beta stimulus
1007:
signal transduction involved in regulation of gene expression
2634:"The comings and goings of PARP-1 in response to DNA damage"
1748:
structure, and by interacting with and modifying multiple
1187:
transforming growth factor beta receptor signaling pathway
2039:
hypomethylated, PARP1 is expressed at an elevated level.
1930:), lead to mutations and cancer. However, PARP1 mediated
1232:
positive regulation of transcription by RNA polymerase II
1017:
negative regulation of transcription by RNA polymerase II
5133:
Rosado MM, Bennici E, Novelli F, Pioli C (August 2013).
2836:
Claybon A, Karia B, Bruce C, Bishop AJ (November 2010).
1685:
Differentiation, proliferation, and tumor transformation
1097:
nucleotide-excision repair, preincision complex assembly
552:
2042:
Cells from older humans (69 to 75 years of age) have a
1092:
positive regulation of SMAD protein signal transduction
1047:
double-strand break repair via homologous recombination
1891:
PARP1 is also over-expressed when its promoter region
1147:
nucleotide-excision repair, DNA incision, 5'-to lesion
1142:
positive regulation of protein localization to nucleus
1132:
nucleotide-excision repair, DNA incision, 3'-to lesion
717:
4861:"Poly(ADP-ribose) polymerase is a B-MYB coactivator"
1838:. PARP1 activity contributes to the proinflammatory
1736:
pathway. PARP1 contributes to repair efficiency by
1167:
positive regulation of myofibroblast differentiation
5096:
1910:in expression of one or more DNA repair genes, but
1531:
1506:
1480:
1459:
1127:
positive regulation of mitochondrial depolarization
2227:
2225:
2223:
2206:
2204:
2202:
997:nucleotide-excision repair, DNA damage recognition
4715:Song J, Keppler BD, Wise RR, Bent AF (May 2015).
1242:positive regulation of single strand break repair
1037:positive regulation of cardiac muscle hypertrophy
499:
398:
1799:. Thus, if the HR pathway is functioning, PARP1
1172:nucleotide-excision repair, DNA duplex unwinding
4768:
4766:
4764:
4252:
4250:
3251:
3249:
3195:
3193:
3191:
2928:
2926:
1212:negative regulation of ATP biosynthetic process
4986:
4984:
2482:
2480:
2332:Xie N, Zhang L, Gao W, Huang C, Zou B (2020).
2327:
2325:
2232:GRCm38: Ensembl release 89: ENSMUSG00000026496
5194:
4659:Feng B, Liu C, Shan L, He P (December 2016).
4481:Virág L, Salzman AL, Szabó C (October 1998).
2627:
2625:
1728:PARP1 acts as a first responder that detects
8:
1924:Epigenetically reduced DNA repair and cancer
1247:peptidyl-glutamic acid poly-ADP-ribosylation
4710:
4708:
4208:
4206:
4204:
2211:GRCh38: Ensembl release 89: ENSG00000143799
1688:Normal or abnormal recovery from DNA damage
1112:regulation of SMAD protein complex assembly
5201:
5187:
5179:
3139:
3137:
2174:class of investigational anti-cancer drugs
1292:
982:regulation of transcription, DNA-templated
752:
540:
439:
336:
74:
5160:
5069:
5018:
4964:
4921:
4878:
4835:
4792:
4744:
4734:
4688:
4678:
4457:
4408:
4284:
4184:
4141:
4131:
4080:
4040:
4038:
4018:
3967:
3871:
3861:
3704:
3677:Rajman L, Chwalek K, Sinclair DA (2018).
3653:
3643:
3592:
3422:
3379:
3336:
3326:
3283:
3229:
3219:
3171:
3117:
3066:
3056:
3013:
2962:
2952:
2906:
2863:
2812:
2761:
2710:
2659:
2591:
2581:
2516:
2506:
2494:Oxidative Medicine and Cellular Longevity
2359:
2785:Waldman AS, Waldman BC (November 1991).
2339:Signal Transduction and Targeted Therapy
1853:cells, thereby sustaining inflammation.
1715:in the development and proliferation of
1082:nucleotide-excision repair, DNA incision
1042:global genome nucleotide-excision repair
1032:cellular response to DNA damage stimulus
5216:
2986:Yarbro JR, Emmons RS, Pence BD (2020).
2198:
880:NAD DNA ADP-ribosyltransferase activity
27:Mammalian protein found in Homo sapiens
2933:Sethi GS, Dharwal V, Naura AS (2017).
1970:are being tested for effectiveness in
29:
3307:Bi FF, Li D, Yang Q (February 2013).
2000:protein is deficient in persons with
1849:protein by PARP1 inhibits removal of
1207:ATP generation from poly-ADP-D-ribose
1072:cellular response to oxidative stress
1057:cellular response to insulin stimulus
504:
465:
460:
403:
362:
357:
7:
4859:Cervellera MN, Sala A (April 2000).
4319:Cellular and Molecular Life Sciences
4264:Cellular and Molecular Life Sciences
860:NAD+ ADP-ribosyltransferase activity
4952:The Journal of Biological Chemistry
4909:The Journal of Biological Chemistry
4866:The Journal of Biological Chemistry
4823:The Journal of Biological Chemistry
4045:Sajish M, Schimmel P (March 2015).
3834:Grube K, Bürkle A (December 1992).
1177:positive regulation of neuron death
1077:DNA ligation involved in DNA repair
987:mitochondrial DNA metabolic process
1882:microhomology-mediated end joining
1762:microhomology-mediated end joining
1528:
1503:
1477:
1456:
1432:
1413:
1387:
1368:
1342:
1323:
1022:transcription by RNA polymerase II
722:
640:
578:
557:
25:
3947:The American Journal of Pathology
1638:. It is the most abundant of the
1631:that in humans is encoded by the
1117:cellular response to amyloid-beta
5309:
5294:
5279:
5264:
5249:
5234:
5219:
4450:10.1046/j.1365-2567.1998.00534.x
1938:Interaction with BRCA1 and BRCA2
1227:peptidyl-serine ADP-ribosylation
1202:regulation of catalytic activity
726:
488:
481:
475:
452:
387:
380:
374:
349:
38:
1732:and then facilitates choice of
1691:May be the site of mutation in
906:transcription regulator complex
865:protein ADP-ribosylase activity
5049:Molecular and Cellular Biology
4366:Cellular and Molecular Biology
3531:American Journal of Hematology
2487:Mangerich A, Bürkle A (2012).
1920:DNA repair-deficiency disorder
737:More reference expression data
706:More reference expression data
1:
4837:10.1016/S0021-9258(19)77203-2
4638:10.1016/j.tplants.2011.03.008
3960:10.1016/S0002-9440(10)64290-3
3898:Journal of Molecular Medicine
3462:10.1136/jclinpath-2012-201088
3449:Journal of Clinical Pathology
3424:10.1158/1078-0432.CCR-18-1346
3381:10.1158/1541-7786.MCR-14-0337
3207:BioMed Research International
3164:10.1158/1541-7786.MCR-14-0422
3037:Pazzaglia S, Pioli C (2019).
1963:Application to cancer therapy
1262:protein auto-ADP-ribosylation
1122:regulation of DNA methylation
1107:cellular response to zinc ion
1067:protein poly-ADP-ribosylation
473:
372:
4736:10.1371/journal.pgen.1005200
4680:10.1371/journal.ppat.1005941
3645:10.1371/journal.pone.0115558
3200:Bi FF, Li D, Yang Q (2013).
3006:10.20900/immunometab20200026
2652:10.1016/j.dnarep.2018.08.022
1621:NAD ADP-ribosyltransferase 1
1162:transcription, DNA-templated
1052:protein modification process
810:transcription factor binding
795:glycosyltransferase activity
5340:Genes on human chromosome 1
4499:10.4049/jimmunol.161.7.3753
2632:Pascal JM (November 2018).
1744:leading to decompaction of
1662:(PAR) and transferring PAR
1197:response to gamma radiation
946:site of double-strand break
815:histone deacetylase binding
5366:
5109:10.1016/j.bbrc.2003.10.053
4397:Experimental Cell Research
4257:Hwang ES, Song SB (2017).
3697:10.1016/j.cmet.2018.02.011
2352:10.1038/s41392-020-00311-7
2183:Poly ADP ribose polymerase
2156:DNA damage theory of aging
2060:DNA damage theory of aging
2010:DNA damage theory of aging
1766:homologous recombinational
1758:non-homologous end joining
1754:nucleotide excision repair
1697:Induction of inflammation.
1157:double-strand break repair
1027:macrophage differentiation
1002:mitochondrion organization
941:protein-containing complex
790:protein N-terminus binding
5214:
4331:10.1007/s00018-004-4506-0
4277:10.1007/s00018-017-2527-8
4011:10.1038/s41388-018-0130-6
3367:Molecular Cancer Research
3151:Molecular Cancer Research
2455:10.1134/S0006297920010095
2383:Karpińska, Aneta (2021).
2267:"Mouse PubMed Reference:"
2249:"Human PubMed Reference:"
1876:Over-expression in cancer
1724:Role in DNA damage repair
1599:
1594:
1590:
1583:
1567:
1554:Chr 1: 226.36 – 226.41 Mb
1548:
1535:
1510:
1499:
1484:
1463:
1452:
1439:
1435:
1420:
1416:
1407:
1394:
1390:
1375:
1371:
1362:
1349:
1345:
1330:
1326:
1317:
1302:
1295:
1291:
1275:
977:lagging strand elongation
840:estrogen receptor binding
830:identical protein binding
785:DNA ligase (ATP) activity
755:
751:
734:
725:
716:
703:
652:
643:
590:
581:
551:
543:
539:
522:
509:
472:
451:
442:
438:
421:
408:
371:
348:
339:
335:
290:
287:
277:
270:
265:
82:
77:
60:
55:
50:
46:
37:
32:
4966:10.1074/jbc.273.19.11839
4880:10.1074/jbc.275.14.10692
4780:Human Molecular Genetics
3863:10.1073/pnas.89.24.11759
3410:Clinical Cancer Research
3276:10.18632/oncotarget.1549
3097:Cell Death & Disease
2954:10.3389/fimmu.2017.01172
2098:PARP1 has been shown to
1789:homologous recombination
1561:Chr 1: 180.4 – 180.43 Mb
1222:protein ADP-ribosylation
992:mitochondrial DNA repair
684:Rostral migratory stream
4625:Trends in Plant Science
4593:10.1126/science.1072221
4530:Pharmacological Reviews
4133:10.1073/pnas.1405005111
3328:10.1186/1471-2407-13-90
2940:Frontiers in Immunology
2894:Genes & Development
2583:10.1073/pnas.0906753106
2287:Neurobiology of Disease
1906:Cancers are very often
1708:PARP1 is activated by:
1700:The pathophysiology of
1257:cellular response to UV
1182:response to aldosterone
688:external carotid artery
5350:Aging-related proteins
5175:Review of the subject.
4998:Nucleic Acids Research
4923:10.1074/jbc.M106528200
4410:10.1006/excr.1997.3536
4186:10.1038/sj.onc.1202778
3544:10.1002/ajh.2830370402
2843:Nucleic Acids Research
2805:10.1093/nar/19.21.5943
2792:Nucleic Acids Research
2741:Nucleic Acids Research
2690:Nucleic Acids Research
2300:10.1006/nbdi.2000.0324
1815:PARP1 is required for
1681:PARP1 is involved in:
1012:protein autoprocessing
835:protein kinase binding
5345:Aging-related enzymes
5062:10.1128/MCB.18.6.3563
5011:10.1093/nar/26.8.1891
4487:Journal of Immunology
4233:10.1089/rej.2006.0514
4220:Rejuvenation Research
3911:10.1007/s001090050226
3594:10.1093/annonc/mdr361
3110:10.1038/cddis.2015.58
2443:Biochemistry (Moscow)
1928:translesion synthesis
1836:nitric oxide synthase
1828:tumor necrosis factor
3058:10.3390/cells9010041
2088:Arabidopsis thaliana
2015:PARP1 appears to be
1811:Role in inflammation
1252:DNA ADP-ribosylation
1217:telomere maintenance
770:transferase activity
614:middle frontal gyrus
506:1 H4|1 84.44 cM
467:Chromosome 1 (mouse)
365:Chromosome 1 (human)
78:List of PDB id codes
51:Available structures
4585:2002Sci...297..259Y
4542:10.1124/pr.54.3.375
4124:2014PNAS..11112752M
4073:10.1038/nature14028
4065:2015Natur.519..370S
3854:1992PNAS...8911759G
3805:10.1038/nature03445
3797:2005Natur.434..917F
3752:10.1038/nature03443
3744:2005Natur.434..913B
3636:2014PLoSO...9k5558D
3221:10.1155/2013/946268
2908:10.1101/gad.9.5.509
2574:2009PNAS..10619998N
2568:(47): 19998–20003.
2508:10.1155/2012/321653
2404:2021Ana...146.7131K
1847:high-mobility group
1770:DNA mismatch repair
1713:Helicobacter pylori
1658:to synthesize poly
931:protein-DNA complex
598:ganglionic eminence
4794:10.1093/hmg/ddh122
3580:Annals of Oncology
3507:10.3892/or.9.3.529
2856:10.1093/nar/gkq624
2754:10.1093/nar/gkg703
2703:10.1093/nar/gkn403
2616:. January 6, 2010.
2412:10.1039/D1AN01769A
2168:– a PARP inhibitor
2073:Role in cell death
1886:genome instability
1826:mediators such as
1795:in mice using the
1613:Poly polymerase 1
1397:ENSMUSG00000026496
965:Biological process
951:site of DNA damage
889:Cellular component
763:Molecular function
5327:
5326:
5153:10.1111/imm.12099
4579:(5579): 259–263.
4271:(18): 3347–3362.
4005:(21): 2793–2805.
3417:(23): 6001–6014.
2398:(23): 7131–7143.
1610:
1609:
1606:
1605:
1579:
1578:
1544:
1543:
1525:
1524:
1495:
1494:
1474:
1473:
1448:
1447:
1429:
1428:
1403:
1402:
1384:
1383:
1358:
1357:
1339:
1338:
1287:
1286:
1237:apoptotic process
820:metal ion binding
747:
746:
743:
742:
712:
711:
699:
698:
637:
636:
535:
534:
434:
433:
329:PARP1 - orthologs
261:
260:
257:
256:
61:Ortholog search:
16:(Redirected from
5357:
5313:
5298:
5283:
5268:
5253:
5238:
5223:
5203:
5196:
5189:
5180:
5174:
5164:
5121:
5120:
5090:
5084:
5083:
5073:
5039:
5033:
5032:
5022:
4988:
4979:
4978:
4968:
4959:(19): 11839–43.
4942:
4936:
4935:
4925:
4916:(49): 45588–97.
4899:
4893:
4892:
4882:
4856:
4850:
4849:
4839:
4813:
4807:
4806:
4796:
4770:
4759:
4758:
4748:
4738:
4712:
4703:
4702:
4692:
4682:
4673:(12): e1005941.
4656:
4650:
4649:
4619:
4613:
4612:
4568:
4562:
4561:
4525:
4519:
4518:
4493:(7): 3753–3759.
4478:
4472:
4471:
4461:
4429:
4423:
4422:
4412:
4388:
4382:
4381:
4357:
4351:
4350:
4325:(7–8): 751–759.
4313:
4307:
4306:
4288:
4254:
4245:
4244:
4210:
4199:
4198:
4188:
4162:
4156:
4155:
4145:
4135:
4101:
4095:
4094:
4084:
4042:
4033:
4032:
4022:
3988:
3982:
3981:
3971:
3937:
3931:
3930:
3892:
3886:
3885:
3875:
3865:
3848:(24): 11759–63.
3831:
3825:
3824:
3791:(7035): 917–21.
3778:
3772:
3771:
3725:
3719:
3718:
3708:
3674:
3668:
3667:
3657:
3647:
3613:
3607:
3606:
3596:
3570:
3564:
3563:
3525:
3519:
3518:
3494:Oncology Reports
3488:
3482:
3481:
3443:
3437:
3436:
3426:
3400:
3394:
3393:
3383:
3357:
3351:
3350:
3340:
3330:
3304:
3298:
3297:
3287:
3253:
3244:
3243:
3233:
3223:
3197:
3186:
3185:
3175:
3141:
3132:
3131:
3121:
3087:
3081:
3080:
3070:
3060:
3034:
3028:
3027:
3017:
2993:Immunometabolism
2983:
2977:
2976:
2966:
2956:
2930:
2921:
2920:
2910:
2884:
2878:
2877:
2867:
2833:
2827:
2826:
2816:
2782:
2776:
2775:
2765:
2731:
2725:
2724:
2714:
2680:
2674:
2673:
2663:
2629:
2620:
2617:
2605:
2595:
2585:
2551:
2545:
2544:
2537:
2531:
2530:
2520:
2510:
2484:
2475:
2474:
2438:
2432:
2431:
2389:
2380:
2374:
2373:
2363:
2329:
2320:
2319:
2281:
2275:
2274:
2263:
2257:
2256:
2245:
2239:
2229:
2218:
2208:
2161:Maximum lifespan
2021:ADP-ribosylation
1968:PARP1 inhibitors
1916:cancer syndromes
1834:, and inducible
1785:replication fork
1738:ADP-ribosylation
1668:ADP-ribosylation
1619:) also known as
1592:
1591:
1563:
1556:
1539:
1529:
1520:
1504:
1500:RefSeq (protein)
1490:
1478:
1469:
1457:
1433:
1414:
1388:
1369:
1343:
1324:
1293:
896:nuclear envelope
805:zinc ion binding
753:
739:
730:
723:
708:
648:
646:Top expressed in
641:
594:ventricular zone
586:
584:Top expressed in
579:
558:
541:
531:
518:
507:
492:
485:
479:
468:
456:
440:
430:
417:
406:
391:
384:
378:
367:
353:
337:
331:
282:
275:
252:
75:
69:
48:
47:
42:
30:
21:
5365:
5364:
5360:
5359:
5358:
5356:
5355:
5354:
5330:
5329:
5328:
5323:
5320:
5314:
5305:
5299:
5290:
5284:
5275:
5269:
5260:
5254:
5245:
5239:
5230:
5224:
5210:
5207:
5132:
5129:
5127:Further reading
5124:
5092:
5091:
5087:
5041:
5040:
5036:
4990:
4989:
4982:
4944:
4943:
4939:
4901:
4900:
4896:
4873:(14): 10692–6.
4858:
4857:
4853:
4815:
4814:
4810:
4787:(10): 1081–93.
4772:
4771:
4762:
4729:(5): e1005200.
4714:
4713:
4706:
4658:
4657:
4653:
4621:
4620:
4616:
4570:
4569:
4565:
4527:
4526:
4522:
4480:
4479:
4475:
4431:
4430:
4426:
4390:
4389:
4385:
4359:
4358:
4354:
4315:
4314:
4310:
4256:
4255:
4248:
4212:
4211:
4202:
4179:(27): 3954–62.
4164:
4163:
4159:
4118:(35): 12752–7.
4103:
4102:
4098:
4059:(7543): 370–3.
4044:
4043:
4036:
3990:
3989:
3985:
3939:
3938:
3934:
3894:
3893:
3889:
3833:
3832:
3828:
3780:
3779:
3775:
3738:(7035): 913–7.
3727:
3726:
3722:
3684:Cell Metabolism
3676:
3675:
3671:
3630:(12): e115558.
3615:
3614:
3610:
3572:
3571:
3567:
3527:
3526:
3522:
3490:
3489:
3485:
3445:
3444:
3440:
3402:
3401:
3397:
3359:
3358:
3354:
3306:
3305:
3301:
3255:
3254:
3247:
3199:
3198:
3189:
3143:
3142:
3135:
3089:
3088:
3084:
3036:
3035:
3031:
2985:
2984:
2980:
2932:
2931:
2924:
2886:
2885:
2881:
2850:(21): 7538–45.
2835:
2834:
2830:
2784:
2783:
2779:
2748:(17): 4959–64.
2733:
2732:
2728:
2697:(13): 4454–64.
2682:
2681:
2677:
2631:
2630:
2623:
2608:
2553:
2552:
2548:
2539:
2538:
2534:
2486:
2485:
2478:
2440:
2439:
2435:
2387:
2382:
2381:
2377:
2331:
2330:
2323:
2283:
2282:
2278:
2265:
2264:
2260:
2247:
2246:
2242:
2230:
2221:
2209:
2200:
2196:
2152:
2147:
2096:
2084:
2075:
2056:
2052:
2002:Werner syndrome
1984:
1965:
1940:
1912:over-expression
1878:
1870:dendritic cells
1824:proinflammatory
1813:
1781:DNA replication
1726:
1702:type I diabetes
1648:
1625:poly synthase 1
1601:View/Edit Mouse
1596:View/Edit Human
1559:
1552:
1549:Location (UCSC)
1537:
1516:
1512:
1486:
1465:
1378:ENSG00000143799
1271:
960:
884:
825:protein binding
735:
704:
695:
690:
686:
682:
678:
674:
670:
666:
662:
658:
644:
633:
628:
626:corpus callosum
624:
620:
616:
612:
608:
604:
600:
596:
582:
526:
513:
505:
495:
494:
493:
486:
466:
443:Gene location (
425:
412:
404:
394:
393:
392:
385:
363:
340:Gene location (
291:
278:
271:
84:
62:
28:
23:
22:
15:
12:
11:
5:
5363:
5361:
5353:
5352:
5347:
5342:
5332:
5331:
5325:
5324:
5322:
5321:
5315:
5308:
5306:
5300:
5293:
5291:
5285:
5278:
5276:
5270:
5263:
5261:
5255:
5248:
5246:
5240:
5233:
5231:
5225:
5218:
5215:
5212:
5211:
5208:
5206:
5205:
5198:
5191:
5183:
5177:
5176:
5128:
5125:
5123:
5122:
5085:
5056:(6): 3563–71.
5034:
4980:
4937:
4894:
4851:
4830:(7): 2163–70.
4808:
4760:
4704:
4666:PLOS Pathogens
4651:
4614:
4563:
4536:(3): 375–429.
4520:
4473:
4444:(3): 345–355.
4424:
4403:(2): 313–321.
4383:
4372:(6): 771–781.
4352:
4308:
4246:
4227:(2): 191–204.
4200:
4157:
4096:
4034:
3983:
3954:(5): 1559–69.
3932:
3887:
3826:
3773:
3720:
3691:(3): 529–547.
3669:
3608:
3587:(5): 1156–64.
3565:
3520:
3483:
3438:
3395:
3352:
3299:
3245:
3187:
3158:(4): 699–712.
3133:
3082:
3029:
3000:(3): e200026.
2978:
2922:
2879:
2828:
2799:(21): 5943–7.
2777:
2726:
2675:
2621:
2619:
2618:
2614:Medical Xpress
2546:
2532:
2476:
2433:
2375:
2321:
2276:
2258:
2240:
2219:
2197:
2195:
2192:
2191:
2190:
2185:
2180:
2175:
2172:PARP inhibitor
2169:
2163:
2158:
2151:
2148:
2146:
2145:
2140:
2135:
2130:
2125:
2120:
2115:
2110:
2104:
2095:
2092:
2083:
2080:
2074:
2071:
2054:
2050:
2037:epigenetically
1993:Lymphoblastoid
1983:
1980:
1976:chemotherapies
1972:cancer therapy
1964:
1961:
1939:
1936:
1918:). (Also see
1897:epigenetically
1877:
1874:
1812:
1809:
1725:
1722:
1721:
1720:
1717:gastric cancer
1706:
1705:
1698:
1695:
1693:Fanconi anemia
1689:
1686:
1679:
1678:
1671:
1647:
1644:
1608:
1607:
1604:
1603:
1598:
1588:
1587:
1581:
1580:
1577:
1576:
1574:
1572:
1565:
1564:
1557:
1550:
1546:
1545:
1542:
1541:
1533:
1532:
1526:
1523:
1522:
1508:
1507:
1501:
1497:
1496:
1493:
1492:
1482:
1481:
1475:
1472:
1471:
1461:
1460:
1454:
1450:
1449:
1446:
1445:
1437:
1436:
1430:
1427:
1426:
1418:
1417:
1411:
1405:
1404:
1401:
1400:
1392:
1391:
1385:
1382:
1381:
1373:
1372:
1366:
1360:
1359:
1356:
1355:
1347:
1346:
1340:
1337:
1336:
1328:
1327:
1321:
1315:
1314:
1309:
1304:
1300:
1299:
1289:
1288:
1285:
1284:
1273:
1272:
1270:
1269:
1264:
1259:
1254:
1249:
1244:
1239:
1234:
1229:
1224:
1219:
1214:
1209:
1204:
1199:
1194:
1189:
1184:
1179:
1174:
1169:
1164:
1159:
1154:
1149:
1144:
1139:
1134:
1129:
1124:
1119:
1114:
1109:
1104:
1099:
1094:
1089:
1084:
1079:
1074:
1069:
1064:
1059:
1054:
1049:
1044:
1039:
1034:
1029:
1024:
1019:
1014:
1009:
1004:
999:
994:
989:
984:
979:
974:
968:
966:
962:
961:
959:
958:
953:
948:
943:
938:
933:
928:
923:
918:
913:
908:
903:
898:
892:
890:
886:
885:
883:
882:
877:
872:
867:
862:
857:
852:
847:
845:enzyme binding
842:
837:
832:
827:
822:
817:
812:
807:
802:
797:
792:
787:
782:
780:R-SMAD binding
777:
772:
766:
764:
760:
759:
749:
748:
745:
744:
741:
740:
732:
731:
720:
714:
713:
710:
709:
701:
700:
697:
696:
694:
693:
689:
685:
681:
677:
673:
672:abdominal wall
669:
668:tail of embryo
665:
661:
657:
653:
650:
649:
638:
635:
634:
632:
631:
627:
623:
619:
615:
611:
607:
603:
599:
595:
591:
588:
587:
575:
574:
566:
555:
549:
548:
545:RNA expression
537:
536:
533:
532:
524:
520:
519:
511:
508:
503:
497:
496:
487:
480:
474:
470:
469:
464:
458:
457:
449:
448:
436:
435:
432:
431:
423:
419:
418:
410:
407:
402:
396:
395:
386:
379:
373:
369:
368:
361:
355:
354:
346:
345:
333:
332:
289:
285:
284:
276:
268:
267:
263:
262:
259:
258:
255:
254:
80:
79:
71:
70:
59:
53:
52:
44:
43:
35:
34:
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
5362:
5351:
5348:
5346:
5343:
5341:
5338:
5337:
5335:
5318:
5312:
5307:
5303:
5297:
5292:
5288:
5282:
5277:
5273:
5267:
5262:
5258:
5252:
5247:
5243:
5237:
5232:
5228:
5222:
5217:
5213:
5204:
5199:
5197:
5192:
5190:
5185:
5184:
5181:
5172:
5168:
5163:
5158:
5154:
5150:
5147:(4): 428–37.
5146:
5142:
5141:
5136:
5131:
5130:
5126:
5118:
5114:
5110:
5106:
5102:
5098:
5097:
5089:
5086:
5081:
5077:
5072:
5067:
5063:
5059:
5055:
5051:
5050:
5045:
5038:
5035:
5030:
5026:
5021:
5016:
5012:
5008:
5005:(8): 1891–8.
5004:
5000:
4999:
4994:
4987:
4985:
4981:
4976:
4972:
4967:
4962:
4958:
4954:
4953:
4948:
4941:
4938:
4933:
4929:
4924:
4919:
4915:
4911:
4910:
4905:
4898:
4895:
4890:
4886:
4881:
4876:
4872:
4868:
4867:
4862:
4855:
4852:
4847:
4843:
4838:
4833:
4829:
4825:
4824:
4819:
4812:
4809:
4804:
4800:
4795:
4790:
4786:
4782:
4781:
4776:
4769:
4767:
4765:
4761:
4756:
4752:
4747:
4742:
4737:
4732:
4728:
4724:
4723:
4722:PLOS Genetics
4718:
4711:
4709:
4705:
4700:
4696:
4691:
4686:
4681:
4676:
4672:
4668:
4667:
4662:
4655:
4652:
4647:
4643:
4639:
4635:
4632:(7): 372–80.
4631:
4627:
4626:
4618:
4615:
4610:
4606:
4602:
4598:
4594:
4590:
4586:
4582:
4578:
4574:
4567:
4564:
4559:
4555:
4551:
4547:
4543:
4539:
4535:
4531:
4524:
4521:
4516:
4512:
4508:
4504:
4500:
4496:
4492:
4488:
4484:
4477:
4474:
4469:
4465:
4460:
4455:
4451:
4447:
4443:
4439:
4435:
4428:
4425:
4420:
4416:
4411:
4406:
4402:
4398:
4394:
4387:
4384:
4379:
4375:
4371:
4367:
4363:
4356:
4353:
4348:
4344:
4340:
4336:
4332:
4328:
4324:
4320:
4312:
4309:
4304:
4300:
4296:
4292:
4287:
4282:
4278:
4274:
4270:
4266:
4265:
4260:
4253:
4251:
4247:
4242:
4238:
4234:
4230:
4226:
4222:
4221:
4216:
4209:
4207:
4205:
4201:
4196:
4192:
4187:
4182:
4178:
4174:
4173:
4168:
4161:
4158:
4153:
4149:
4144:
4139:
4134:
4129:
4125:
4121:
4117:
4113:
4112:
4107:
4100:
4097:
4092:
4088:
4083:
4078:
4074:
4070:
4066:
4062:
4058:
4054:
4053:
4048:
4041:
4039:
4035:
4030:
4026:
4021:
4016:
4012:
4008:
4004:
4000:
3999:
3994:
3987:
3984:
3979:
3975:
3970:
3965:
3961:
3957:
3953:
3949:
3948:
3943:
3936:
3933:
3928:
3924:
3920:
3916:
3912:
3908:
3905:(5): 346–54.
3904:
3900:
3899:
3891:
3888:
3883:
3879:
3874:
3869:
3864:
3859:
3855:
3851:
3847:
3843:
3842:
3837:
3830:
3827:
3822:
3818:
3814:
3810:
3806:
3802:
3798:
3794:
3790:
3786:
3785:
3777:
3774:
3769:
3765:
3761:
3757:
3753:
3749:
3745:
3741:
3737:
3733:
3732:
3724:
3721:
3716:
3712:
3707:
3702:
3698:
3694:
3690:
3686:
3685:
3680:
3673:
3670:
3665:
3661:
3656:
3651:
3646:
3641:
3637:
3633:
3629:
3625:
3624:
3619:
3612:
3609:
3604:
3600:
3595:
3590:
3586:
3582:
3581:
3576:
3569:
3566:
3561:
3557:
3553:
3549:
3545:
3541:
3537:
3533:
3532:
3524:
3521:
3516:
3512:
3508:
3504:
3501:(3): 529–32.
3500:
3496:
3495:
3487:
3484:
3479:
3475:
3471:
3467:
3463:
3459:
3456:(7): 607–12.
3455:
3451:
3450:
3442:
3439:
3434:
3430:
3425:
3420:
3416:
3412:
3411:
3406:
3399:
3396:
3391:
3387:
3382:
3377:
3374:(3): 470–82.
3373:
3369:
3368:
3363:
3356:
3353:
3348:
3344:
3339:
3334:
3329:
3324:
3320:
3316:
3315:
3310:
3303:
3300:
3295:
3291:
3286:
3281:
3277:
3273:
3269:
3265:
3264:
3259:
3252:
3250:
3246:
3241:
3237:
3232:
3227:
3222:
3217:
3213:
3209:
3208:
3203:
3196:
3194:
3192:
3188:
3183:
3179:
3174:
3169:
3165:
3161:
3157:
3153:
3152:
3147:
3140:
3138:
3134:
3129:
3125:
3120:
3115:
3111:
3107:
3103:
3099:
3098:
3093:
3086:
3083:
3078:
3074:
3069:
3064:
3059:
3054:
3050:
3046:
3045:
3040:
3033:
3030:
3025:
3021:
3016:
3011:
3007:
3003:
2999:
2995:
2994:
2989:
2982:
2979:
2974:
2970:
2965:
2960:
2955:
2950:
2946:
2942:
2941:
2936:
2929:
2927:
2923:
2918:
2914:
2909:
2904:
2901:(5): 509–20.
2900:
2896:
2895:
2890:
2883:
2880:
2875:
2871:
2866:
2861:
2857:
2853:
2849:
2845:
2844:
2839:
2832:
2829:
2824:
2820:
2815:
2810:
2806:
2802:
2798:
2794:
2793:
2788:
2781:
2778:
2773:
2769:
2764:
2759:
2755:
2751:
2747:
2743:
2742:
2737:
2730:
2727:
2722:
2718:
2713:
2708:
2704:
2700:
2696:
2692:
2691:
2686:
2679:
2676:
2671:
2667:
2662:
2657:
2653:
2649:
2645:
2641:
2640:
2635:
2628:
2626:
2622:
2615:
2611:
2607:
2606:
2603:
2599:
2594:
2589:
2584:
2579:
2575:
2571:
2567:
2563:
2562:
2557:
2550:
2547:
2542:
2536:
2533:
2528:
2524:
2519:
2514:
2509:
2504:
2500:
2496:
2495:
2490:
2483:
2481:
2477:
2472:
2468:
2464:
2460:
2456:
2452:
2449:(1): 99–107.
2448:
2444:
2437:
2434:
2429:
2425:
2421:
2417:
2413:
2409:
2405:
2401:
2397:
2393:
2386:
2379:
2376:
2371:
2367:
2362:
2357:
2353:
2349:
2345:
2341:
2340:
2335:
2328:
2326:
2322:
2317:
2313:
2309:
2305:
2301:
2297:
2294:(4): 225–39.
2293:
2289:
2288:
2280:
2277:
2272:
2268:
2262:
2259:
2254:
2250:
2244:
2241:
2237:
2233:
2228:
2226:
2224:
2220:
2216:
2212:
2207:
2205:
2203:
2199:
2193:
2189:
2186:
2184:
2181:
2179:
2176:
2173:
2170:
2167:
2164:
2162:
2159:
2157:
2154:
2153:
2149:
2144:
2141:
2139:
2136:
2134:
2131:
2129:
2126:
2124:
2121:
2119:
2116:
2114:
2111:
2109:
2106:
2105:
2103:
2101:
2093:
2091:
2089:
2081:
2079:
2072:
2070:
2068:
2063:
2061:
2057:
2045:
2040:
2038:
2034:
2029:
2024:
2022:
2018:
2013:
2011:
2007:
2003:
1999:
1994:
1989:
1981:
1979:
1977:
1973:
1969:
1962:
1960:
1958:
1954:
1949:
1945:
1937:
1935:
1933:
1929:
1925:
1921:
1917:
1913:
1909:
1904:
1900:
1898:
1894:
1889:
1887:
1883:
1875:
1873:
1871:
1867:
1863:
1859:
1854:
1852:
1848:
1845:
1841:
1837:
1833:
1832:interleukin 6
1829:
1825:
1821:
1820:transcription
1818:
1810:
1808:
1806:
1805:knockout mice
1802:
1798:
1794:
1790:
1786:
1782:
1778:
1773:
1771:
1767:
1763:
1759:
1755:
1751:
1747:
1743:
1739:
1735:
1731:
1723:
1718:
1714:
1711:
1710:
1709:
1703:
1699:
1696:
1694:
1690:
1687:
1684:
1683:
1682:
1676:
1672:
1669:
1666:to proteins (
1665:
1661:
1657:
1653:
1652:
1651:
1650:PARP1 works:
1645:
1643:
1641:
1637:
1634:
1630:
1626:
1622:
1618:
1614:
1602:
1597:
1593:
1589:
1586:
1582:
1575:
1573:
1570:
1566:
1562:
1558:
1555:
1551:
1547:
1540:
1534:
1530:
1527:
1521:
1519:
1515:
1509:
1505:
1502:
1498:
1491:
1489:
1483:
1479:
1476:
1470:
1468:
1462:
1458:
1455:
1453:RefSeq (mRNA)
1451:
1444:
1443:
1438:
1434:
1431:
1425:
1424:
1419:
1415:
1412:
1410:
1406:
1399:
1398:
1393:
1389:
1386:
1380:
1379:
1374:
1370:
1367:
1365:
1361:
1354:
1353:
1348:
1344:
1341:
1335:
1334:
1329:
1325:
1322:
1320:
1316:
1313:
1310:
1308:
1305:
1301:
1298:
1294:
1290:
1283:
1279:
1274:
1268:
1265:
1263:
1260:
1258:
1255:
1253:
1250:
1248:
1245:
1243:
1240:
1238:
1235:
1233:
1230:
1228:
1225:
1223:
1220:
1218:
1215:
1213:
1210:
1208:
1205:
1203:
1200:
1198:
1195:
1193:
1190:
1188:
1185:
1183:
1180:
1178:
1175:
1173:
1170:
1168:
1165:
1163:
1160:
1158:
1155:
1153:
1150:
1148:
1145:
1143:
1140:
1138:
1135:
1133:
1130:
1128:
1125:
1123:
1120:
1118:
1115:
1113:
1110:
1108:
1105:
1103:
1100:
1098:
1095:
1093:
1090:
1088:
1085:
1083:
1080:
1078:
1075:
1073:
1070:
1068:
1065:
1063:
1060:
1058:
1055:
1053:
1050:
1048:
1045:
1043:
1040:
1038:
1035:
1033:
1030:
1028:
1025:
1023:
1020:
1018:
1015:
1013:
1010:
1008:
1005:
1003:
1000:
998:
995:
993:
990:
988:
985:
983:
980:
978:
975:
973:
970:
969:
967:
964:
963:
957:
954:
952:
949:
947:
944:
942:
939:
937:
934:
932:
929:
927:
924:
922:
921:mitochondrion
919:
917:
914:
912:
909:
907:
904:
902:
899:
897:
894:
893:
891:
888:
887:
881:
878:
876:
873:
871:
868:
866:
863:
861:
858:
856:
853:
851:
848:
846:
843:
841:
838:
836:
833:
831:
828:
826:
823:
821:
818:
816:
813:
811:
808:
806:
803:
801:
798:
796:
793:
791:
788:
786:
783:
781:
778:
776:
773:
771:
768:
767:
765:
762:
761:
758:
757:Gene ontology
754:
750:
738:
733:
729:
724:
721:
719:
715:
707:
702:
691:
687:
683:
679:
676:Gonadal ridge
675:
671:
667:
663:
659:
655:
654:
651:
647:
642:
639:
630:apex of heart
629:
625:
621:
617:
613:
609:
605:
601:
597:
593:
592:
589:
585:
580:
577:
576:
573:
571:
567:
565:
564:
560:
559:
556:
554:
550:
546:
542:
538:
530:
525:
521:
517:
512:
502:
498:
491:
484:
478:
471:
463:
459:
455:
450:
446:
441:
437:
429:
424:
420:
416:
411:
401:
397:
390:
383:
377:
370:
366:
360:
356:
352:
347:
343:
338:
334:
330:
326:
322:
318:
314:
310:
306:
302:
298:
294:
286:
281:
274:
269:
264:
253:
251:
247:
243:
239:
235:
231:
227:
223:
219:
215:
211:
207:
203:
199:
195:
191:
187:
183:
179:
175:
171:
167:
163:
159:
155:
151:
147:
143:
139:
135:
131:
127:
123:
119:
115:
111:
107:
103:
99:
95:
91:
87:
81:
76:
73:
72:
68:
65:
58:
54:
49:
45:
41:
36:
31:
19:
5316:
5301:
5286:
5271:
5256:
5241:
5226:
5144:
5138:
5103:(3): 702–7.
5100:
5094:
5088:
5053:
5047:
5037:
5002:
4996:
4956:
4950:
4940:
4913:
4907:
4897:
4870:
4864:
4854:
4827:
4821:
4811:
4784:
4778:
4726:
4720:
4670:
4664:
4654:
4629:
4623:
4617:
4576:
4572:
4566:
4533:
4529:
4523:
4490:
4486:
4476:
4441:
4437:
4427:
4400:
4396:
4386:
4369:
4365:
4355:
4322:
4318:
4311:
4268:
4262:
4224:
4218:
4176:
4170:
4160:
4115:
4109:
4099:
4056:
4050:
4002:
3996:
3986:
3951:
3945:
3935:
3902:
3896:
3890:
3845:
3839:
3829:
3788:
3782:
3776:
3735:
3729:
3723:
3688:
3682:
3672:
3627:
3621:
3611:
3584:
3578:
3568:
3538:(4): 223–7.
3535:
3529:
3523:
3498:
3492:
3486:
3453:
3447:
3441:
3414:
3408:
3398:
3371:
3365:
3355:
3318:
3312:
3302:
3270:(1): 291–7.
3267:
3261:
3211:
3205:
3155:
3149:
3104:(3): e1697.
3101:
3095:
3085:
3048:
3042:
3032:
2997:
2991:
2981:
2944:
2938:
2898:
2892:
2882:
2847:
2841:
2831:
2796:
2790:
2780:
2745:
2739:
2729:
2694:
2688:
2678:
2643:
2637:
2613:
2565:
2559:
2549:
2535:
2498:
2492:
2446:
2442:
2436:
2395:
2391:
2378:
2343:
2337:
2291:
2285:
2279:
2270:
2261:
2252:
2243:
2097:
2094:Interactions
2087:
2085:
2076:
2064:
2044:constitutive
2041:
2025:
2014:
1985:
1966:
1941:
1911:
1907:
1905:
1901:
1890:
1879:
1862:CD4+ T-cells
1855:
1814:
1801:null mutants
1774:
1768:repair, and
1727:
1707:
1680:
1649:
1632:
1624:
1620:
1616:
1612:
1611:
1536:
1511:
1485:
1464:
1440:
1421:
1395:
1376:
1350:
1331:
1311:
1306:
850:SMAD binding
664:otic vesicle
656:otic placode
568:
561:
527:180,428,819
514:180,396,489
426:226,408,154
413:226,360,210
288:External IDs
83:
5209:PDB gallery
2646:: 177–182.
2178:Parthanatos
2082:Plant PARP1
2033:CpG islands
2017:resveratrol
1866:eosinophils
1840:macrophages
1518:NP_001609.2
911:nucleoplasm
855:RNA binding
800:NAD binding
775:DNA binding
266:Identifiers
5334:Categories
5140:Immunology
4438:Immunology
3314:BMC Cancer
3263:Oncotarget
3214:: 946268.
2639:DNA Repair
2501:: 321653.
2346:(1): 227.
2238:, May 2017
2217:, May 2017
2194:References
2188:Senescence
2006:DNA breaks
1957:xenografts
1750:DNA repair
1730:DNA damage
1660:ADP ribose
1087:DNA repair
956:chromosome
610:lymph node
606:C1 segment
572:(ortholog)
309:HomoloGene
3051:(1): 41.
2471:211028760
2428:240110114
2067:sirtuin 1
1988:leukocyte
1953:apoptosis
1908:deficient
1851:apoptotic
1797:pun assay
1746:chromatin
1654:By using
1514:NP_001609
1488:NM_007415
1467:NM_001618
1297:Orthologs
936:cytoplasm
916:nucleolus
317:GeneCards
5171:23489378
5117:14623329
4932:11590148
4889:10744766
4803:15044383
4755:25950582
4699:27907213
4646:21482174
4609:22991897
4601:12114629
4558:27100634
4550:12223530
4347:43817844
4339:15868400
4303:25896400
4295:28417163
4286:11107671
4241:17518695
4195:10435618
4172:Oncogene
4152:25136112
4091:25533949
4029:29511347
3998:Oncogene
3978:12707040
3927:24616650
3813:15829967
3760:15829966
3715:29514064
3664:25526641
3623:PLOS ONE
3603:21908496
3560:26905918
3515:11956622
3470:23486608
3433:30087144
3390:25563294
3347:23442605
3294:24448423
3240:23762867
3182:25828893
3128:25789972
3077:31877876
3024:32774895
2973:28974953
2947:: 1172.
2874:20660013
2772:12930944
2721:18603595
2670:30177435
2602:19897724
2527:23050038
2463:32079521
2420:34726203
2370:33028824
2316:41201067
2308:10964595
2234:–
2213:–
2166:Olaparib
2150:See also
2100:interact
1895:site is
1742:histones
1664:moieties
1646:Function
1585:Wikidata
1276:Sources:
901:membrane
692:yolk sac
680:epiblast
622:testicle
618:appendix
5162:3719060
5080:9584196
5029:9518481
4975:9565608
4846:5555565
4746:4423837
4690:5131896
4581:Bibcode
4573:Science
4515:5734113
4507:9759901
4468:9767416
4459:1364252
4419:9168807
4378:8535170
4143:4156740
4120:Bibcode
4082:4368482
4061:Bibcode
4020:5970015
3969:1851180
3919:9587069
3882:1465394
3850:Bibcode
3821:4364706
3793:Bibcode
3768:4391043
3740:Bibcode
3706:6342515
3655:4272268
3632:Bibcode
3552:1907096
3338:3599366
3285:3960209
3231:3666359
3173:4398615
3119:4385936
3068:7017201
3015:7409778
2964:5610677
2917:7698643
2865:2995050
2823:1945881
2712:2490739
2661:6637744
2593:2785281
2570:Bibcode
2518:3459245
2400:Bibcode
2392:Analyst
2361:7539288
2236:Ensembl
2215:Ensembl
1793:in vivo
1409:UniProt
1364:Ensembl
1303:Species
1282:QuickGO
926:nucleus
660:saccule
547:pattern
405:1q42.12
305:1340806
273:Aliases
5169:
5159:
5115:
5078:
5071:108937
5068:
5027:
5020:147507
5017:
4973:
4930:
4887:
4844:
4801:
4753:
4743:
4697:
4687:
4644:
4607:
4599:
4556:
4548:
4513:
4505:
4466:
4456:
4417:
4376:
4345:
4337:
4301:
4293:
4283:
4239:
4193:
4150:
4140:
4089:
4079:
4052:Nature
4027:
4017:
3976:
3966:
3925:
3917:
3880:
3870:
3819:
3811:
3784:Nature
3766:
3758:
3731:Nature
3713:
3703:
3662:
3652:
3601:
3558:
3550:
3513:
3478:535704
3476:
3468:
3431:
3388:
3345:
3335:
3321:: 90.
3292:
3282:
3238:
3228:
3180:
3170:
3126:
3116:
3075:
3065:
3022:
3012:
2971:
2961:
2915:
2872:
2862:
2821:
2814:329051
2811:
2770:
2763:212803
2760:
2719:
2709:
2668:
2658:
2600:
2590:
2525:
2515:
2469:
2461:
2426:
2418:
2368:
2358:
2314:
2306:
2143:ZNF423
2102:with:
1868:, and
1858:asthma
1783:, the
1734:repair
1629:enzyme
1627:is an
1617:PARP-1
1571:search
1569:PubMed
1442:P11103
1423:P09874
1319:Entrez
718:BioGPS
297:173870
18:PARP-1
4605:S2CID
4554:S2CID
4511:S2CID
4343:S2CID
4299:S2CID
3923:S2CID
3873:50636
3817:S2CID
3764:S2CID
3556:S2CID
3474:S2CID
3044:Cells
2467:S2CID
2424:S2CID
2388:(PDF)
2312:S2CID
2138:XRCC1
2133:POLA2
2128:POLA1
2113:MYBL2
2065:Both
1982:Aging
1948:BRCA2
1944:BRCA1
1942:Both
1844:HMGB1
1817:NF-κB
1777:siRNA
1633:PARP1
1352:11545
1312:Mouse
1307:Human
1278:Amigo
602:gonad
570:Mouse
563:Human
510:Start
445:Mouse
409:Start
342:Human
321:PARP1
280:PARP1
33:PARP1
5317:2dmj
5302:2cs2
5287:2cr9
5272:2cok
5257:1wok
5242:1uk1
5227:1uk0
5167:PMID
5113:PMID
5076:PMID
5025:PMID
4971:PMID
4928:PMID
4885:PMID
4842:PMID
4799:PMID
4751:PMID
4695:PMID
4642:PMID
4597:PMID
4546:PMID
4503:PMID
4464:PMID
4415:PMID
4374:PMID
4335:PMID
4291:PMID
4237:PMID
4191:PMID
4148:PMID
4087:PMID
4025:PMID
3974:PMID
3915:PMID
3878:PMID
3809:PMID
3756:PMID
3711:PMID
3660:PMID
3599:PMID
3548:PMID
3511:PMID
3466:PMID
3429:PMID
3386:PMID
3343:PMID
3290:PMID
3236:PMID
3212:2013
3178:PMID
3124:PMID
3073:PMID
3020:PMID
2969:PMID
2913:PMID
2870:PMID
2819:PMID
2768:PMID
2717:PMID
2666:PMID
2598:PMID
2523:PMID
2499:2012
2459:PMID
2416:PMID
2366:PMID
2304:PMID
2118:RELA
2108:APTX
2028:ETS1
1946:and
1932:MMEJ
1675:PARP
1656:NAD+
1640:PARP
1636:gene
553:Bgee
501:Band
462:Chr.
400:Band
359:Chr.
313:1222
293:OMIM
250:5DS3
246:5HA9
242:4RV6
238:4XHU
234:2N8A
230:4UXB
226:4R6E
222:4R5W
218:5A00
214:4ZZZ
210:4UND
206:4PJT
202:4OQB
198:4OQA
194:4OPX
190:4L6S
186:4HHZ
182:4HHY
178:4GV7
174:4DQY
170:4AV1
166:3ODE
162:3ODC
158:3ODA
154:3OD8
150:3L3M
146:3L3L
142:3GN7
138:3GJW
134:2RIQ
130:2RD6
126:2RCW
122:2L31
118:2L30
114:2JVN
110:2DMJ
106:2CS2
102:2CR9
98:2COK
94:1WOK
90:1UK1
86:1UK0
67:RCSB
64:PDBe
5157:PMC
5149:doi
5145:139
5105:doi
5101:311
5066:PMC
5058:doi
5015:PMC
5007:doi
4961:doi
4957:273
4918:doi
4914:276
4875:doi
4871:275
4832:doi
4828:246
4789:doi
4741:PMC
4731:doi
4685:PMC
4675:doi
4634:doi
4589:doi
4577:297
4538:doi
4495:doi
4491:161
4454:PMC
4446:doi
4405:doi
4401:232
4327:doi
4281:PMC
4273:doi
4229:doi
4181:doi
4138:PMC
4128:doi
4116:111
4077:PMC
4069:doi
4057:519
4015:PMC
4007:doi
3964:PMC
3956:doi
3952:162
3907:doi
3868:PMC
3858:doi
3801:doi
3789:434
3748:doi
3736:434
3701:PMC
3693:doi
3650:PMC
3640:doi
3589:doi
3540:doi
3503:doi
3458:doi
3419:doi
3376:doi
3333:PMC
3323:doi
3280:PMC
3272:doi
3226:PMC
3216:doi
3168:PMC
3160:doi
3114:PMC
3106:doi
3063:PMC
3053:doi
3010:PMC
3002:doi
2959:PMC
2949:doi
2903:doi
2860:PMC
2852:doi
2809:PMC
2801:doi
2758:PMC
2750:doi
2707:PMC
2699:doi
2656:PMC
2648:doi
2588:PMC
2578:doi
2566:106
2513:PMC
2503:doi
2451:doi
2408:doi
2396:146
2356:PMC
2348:doi
2296:doi
2123:P53
1998:Wrn
1893:ETS
1856:In
1822:of
1740:of
1623:or
1538:n/a
1333:142
523:End
422:End
325:OMA
301:MGI
57:PDB
5336::
5165:.
5155:.
5143:.
5137:.
5111:.
5099:.
5074:.
5064:.
5054:18
5052:.
5046:.
5023:.
5013:.
5003:26
5001:.
4995:.
4983:^
4969:.
4955:.
4949:.
4926:.
4912:.
4906:.
4883:.
4869:.
4863:.
4840:.
4826:.
4820:.
4797:.
4785:13
4783:.
4777:.
4763:^
4749:.
4739:.
4727:11
4725:.
4719:.
4707:^
4693:.
4683:.
4671:12
4669:.
4663:.
4640:.
4630:16
4628:.
4603:.
4595:.
4587:.
4575:.
4552:.
4544:.
4534:54
4532:.
4509:.
4501:.
4489:.
4485:.
4462:.
4452:.
4442:94
4440:.
4436:.
4413:.
4399:.
4395:.
4370:41
4368:.
4364:.
4341:.
4333:.
4323:62
4321:.
4297:.
4289:.
4279:.
4269:74
4267:.
4261:.
4249:^
4235:.
4225:10
4223:.
4217:.
4203:^
4189:.
4177:18
4175:.
4169:.
4146:.
4136:.
4126:.
4114:.
4108:.
4085:.
4075:.
4067:.
4055:.
4049:.
4037:^
4023:.
4013:.
4003:37
4001:.
3995:.
3972:.
3962:.
3950:.
3944:.
3921:.
3913:.
3903:76
3901:.
3876:.
3866:.
3856:.
3846:89
3844:.
3838:.
3815:.
3807:.
3799:.
3787:.
3762:.
3754:.
3746:.
3734:.
3709:.
3699:.
3689:27
3687:.
3681:.
3658:.
3648:.
3638:.
3626:.
3620:.
3597:.
3585:23
3583:.
3577:.
3554:.
3546:.
3536:37
3534:.
3509:.
3497:.
3472:.
3464:.
3454:66
3452:.
3427:.
3415:24
3413:.
3407:.
3384:.
3372:13
3370:.
3364:.
3341:.
3331:.
3319:13
3317:.
3311:.
3288:.
3278:.
3266:.
3260:.
3248:^
3234:.
3224:.
3210:.
3204:.
3190:^
3176:.
3166:.
3156:13
3154:.
3148:.
3136:^
3122:.
3112:.
3100:.
3094:.
3071:.
3061:.
3047:.
3041:.
3018:.
3008:.
2996:.
2990:.
2967:.
2957:.
2943:.
2937:.
2925:^
2911:.
2897:.
2891:.
2868:.
2858:.
2848:38
2846:.
2840:.
2817:.
2807:.
2797:19
2795:.
2789:.
2766:.
2756:.
2746:31
2744:.
2738:.
2715:.
2705:.
2695:36
2693:.
2687:.
2664:.
2654:.
2644:71
2642:.
2636:.
2624:^
2612:.
2596:.
2586:.
2576:.
2564:.
2558:.
2521:.
2511:.
2497:.
2491:.
2479:^
2465:.
2457:.
2447:85
2445:.
2422:.
2414:.
2406:.
2394:.
2390:.
2364:.
2354:.
2342:.
2336:.
2324:^
2310:.
2302:.
2290:.
2269:.
2251:.
2222:^
2201:^
2012:.
1872:.
1864:,
1830:,
1772:.
1764:,
1760:,
1756:,
1670:).
1280:/
529:bp
516:bp
428:bp
415:bp
323:;
319::
315:;
311::
307:;
303::
299:;
295::
248:,
244:,
240:,
236:,
232:,
228:,
224:,
220:,
216:,
212:,
208:,
204:,
200:,
196:,
192:,
188:,
184:,
180:,
176:,
172:,
168:,
164:,
160:,
156:,
152:,
148:,
144:,
140:,
136:,
132:,
128:,
124:,
120:,
116:,
112:,
108:,
104:,
100:,
96:,
92:,
88:,
5202:e
5195:t
5188:v
5173:.
5151::
5119:.
5107::
5082:.
5060::
5031:.
5009::
4977:.
4963::
4934:.
4920::
4891:.
4877::
4848:.
4834::
4805:.
4791::
4757:.
4733::
4701:.
4677::
4648:.
4636::
4611:.
4591::
4583::
4560:.
4540::
4517:.
4497::
4470:.
4448::
4421:.
4407::
4380:.
4349:.
4329::
4305:.
4275::
4243:.
4231::
4197:.
4183::
4154:.
4130::
4122::
4093:.
4071::
4063::
4031:.
4009::
3980:.
3958::
3929:.
3909::
3884:.
3860::
3852::
3823:.
3803::
3795::
3770:.
3750::
3742::
3717:.
3695::
3666:.
3642::
3634::
3628:9
3605:.
3591::
3562:.
3542::
3517:.
3505::
3499:9
3480:.
3460::
3435:.
3421::
3392:.
3378::
3349:.
3325::
3296:.
3274::
3268:5
3242:.
3218::
3184:.
3162::
3130:.
3108::
3102:6
3079:.
3055::
3049:9
3026:.
3004::
2998:2
2975:.
2951::
2945:8
2919:.
2905::
2899:9
2876:.
2854::
2825:.
2803::
2774:.
2752::
2723:.
2701::
2672:.
2650::
2604:.
2580::
2572::
2543:.
2529:.
2505::
2473:.
2453::
2430:.
2410::
2402::
2372:.
2350::
2344:5
2318:.
2298::
2292:7
2273:.
2255:.
2055:2
2053:O
2051:2
2049:H
1719:.
1704:.
1615:(
447:)
344:)
327::
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.