247:-dependent protein kinase) can reduce the activity of PP1. The glycogen binding region, GM, becomes phosphorylated, which causes its dissociation from the catalytic PP1 unit. This separation of the catalytic PP1 unit, glycogen, and other substrates causes a significant decrease in dephosphorylation. Also, when other substrates become phosphorylated by protein kinase A, they can bind to the catalytic subunit of PP1 and directly inhibit it. In the end, glycogen phosphorylase is kept in its active form and glycogen synthase in its inactive form. Separately from inhibition of PP1,
37:
3048:
134:
317:-associated protein inhibits the assembly of microtubules in neurons. Researchers at the New York State Institute for Basic Research in Developmental Disabilities showed that there is significantly lower type 1 phosphatase activity in both gray and white matters in Alzheimer disease brains. This suggests that dysfunctional phosphatases play a role in Alzheimer's disease.
397:
1075:
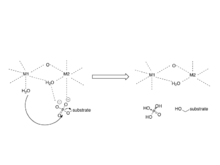
As described earlier, a catalytic subunit is always paired with one or more regulatory subunits. The core sequence motif for binding to the catalytic subunit is "RVxF", but additional motifs allow for extra sites to be used. Some complexes with two regulatory subunits attached have been reported in
336:
pathogenesis by dephosphorylating the viral transcription activator VP30, allowing it to produce viral mRNAs. Inhibition of PP1 prevents VP30 dephosphorylation, thus preventing manufacture of viral mRNA, and thus viral protein. The viral L polymerase is, however, still capable of replicating viral
129:
X-ray crystallographic structural data is available for PP1 catalytic subunit. The catalytic subunit of PP1 forms an α/β fold with a central β-sandwich arranged between two α-helical domains. The interaction of the three β-sheets of the β-sandwich creates a channel for catalytic activity, as it is
274:, indirectly activating glycogen synthase and triggering glycogen synthesis. Although it has been known that PP1 is one of the most important phosphatases involved in insulin action since the late 1990s, the precise mechanisms by which insulin regulates PP1 has only been uncovered more recently.
372:
so by shutting down eIF-2A, the cell prevents the virus from hijacking its own protein-making machinery. Herpesviruses in turn evolved ICP34.5 to defeat the defense; ICP34.5 activates protein phosphatase-1A which dephosphorylates eIF-2A, allowing translation to occur again. ICP34.5 shares the
192:
levels in the liver and glycogen metabolism. PP1 is important to the reciprocal regulation of glycogen metabolism by ensuring the opposite regulation of glycogen breakdown and glycogen synthesis. A key regulator of PP1 is
170:. Microcystin is a liver toxin produced by blue-green algae and contains a cyclic heptapeptide structure that interacts with three distinct regions of the surface of the catalytic subunit of PP1. The structure of
117:
only encodes one catalytic subunit, mammals have four isozymes encoded by three genes, each attracting a different set of regulatory subunits. Regulation of these different processes is performed by distinct PP1
1414:
Zhang Y, Zhang M, Zhang Y (March 2011). "Crystal structure of Ssu72, an essential eukaryotic phosphatase specific for the C-terminal domain of RNA polymerase II, in complex with a transition state analogue".
107:, thus suggesting a common catalytic mechanism. The catalytic subunit can form complexes with various regulatory subunits. These regulatory subunits play an important role in substrate specificity as well as
328:
by
Protein Phosphatase 1 (PP1). It has been recognized that protein phosphatase-1 (PP1) serves as an important regulator of HIV-1 transcription. Researchers at Howard University showed that
130:
the site of coordination of metal ions. These metal ions have been identified as Mn and Fe and their coordination is provided by three histidines, two aspartic acids, and one asparagine.
111:. Some common regulatory subunits include GM (PPP1R3A) and GL (PPP1R3B), which are named after their locations of action within the body (muscle and liver respectively), While the yeast
103:
subunit. The catalytic subunit consists of a 30-kD single-domain protein that can form complexes with other regulatory subunits. The catalytic subunit is highly conserved among all
1185:
Goldberg J, Huang HB, Kwon YG, Greengard P, Nairn AC, Kuriyan J (August 1995). "Three-dimensional structure of the catalytic subunit of protein serine/threonine phosphatase-1".
297:, which then binds to the PP1 complex, activating its phosphatase activity. The study demonstrated that phosphorylated PPP1R3G was also able to bind phosphorylated
1768:
Ilinykh PA, Tigabu B, Ivanov A, Ammosova T, Obukhov Y, Garron T, Kumari N, Kovalskyy D, Platonov MO, Naumchik VS, Freiberg AN, Nekhai S, Bukreyev A (August 2014).
786:
625:
464:
286:
174:
does not change when complexed with PP1, but the catalytic subunit of PP1 does in order to avoid steric effects of Tyr 276 of PP1 and Mdha side chain of MCLR.
1284:
Armstrong CG, Browne GJ, Cohen P, Cohen PT (November 1997). "PPP1R6, a novel member of the family of glycogen-targetting subunits of protein phosphatase 1".
212:
prevents any phosphatase activity of PP1 and maintains the glycogen phosphorylase in its active phosphorylated configuration. Therefore, there phosphorylase
1593:
2122:
2369:
3078:
3068:
1836:
1576:
1770:"Role of protein phosphatase 1 in dephosphorylation of Ebola virus VP30 protein and its targeting for the inhibition of viral transcription"
3073:
2283:
239:
When the muscles of the body signal the need for glycogen degradation and an increase in blood glucose, PP1 will be regulated accordingly.
60:
216:
will accelerate glycogen breakdown until adequate levels of glucose are achieved. When glucose concentrations get too high, phosphorylase
301:(p-GS) independently and recruit p-GS towards PP1, allowing PP1 to dephosphorylate and thereby activate glycogen synthase independent of
2384:
2378:
2179:
1929:
810:
649:
488:
332:
protein targets PP1 to the nucleus and the consequent interaction is important for HIV-1 transcription. The protein also contributes to
1497:"Cyanobacterial microcystin-LR is a potent and specific inhibitor of protein phosphatases 1 and 2A from both mammals and higher plants"
2227:
2767:
2338:
2127:
1379:
Barford D, Das AK, Egloff MP (1998). "The structure and mechanism of protein phosphatases: insights into catalysis and regulation".
1682:
Gong CX, Singh TJ, Grundke-Iqbal I, Iqbal K (September 1993). "Phosphoprotein phosphatase activities in
Alzheimer disease brain".
798:
637:
476:
2445:
2333:
1725:
Nekhai S, Jerebtsova M, Jackson A, Southerland W (January 2007). "Regulation of HIV-1 transcription by protein phosphatase 1".
791:
630:
469:
2254:
2164:
2117:
256:
244:
2923:
224:
to its T state, PP1 dissociates from the complex. This dissociation activates glycogen synthase and converts phosphorylase
2676:
2617:
3038:
1643:"The Protein Phosphatase 1 Complex Is a Direct Target of AKT that Links Insulin Signaling to Hepatic Glycogen Deposition"
2721:
2288:
2278:
2681:
2573:
2198:
2267:
2263:
2259:
2175:
2008:
411:
345:
2908:
3024:
3011:
2998:
2985:
2972:
2959:
2946:
2529:
2475:
2435:
2394:
2298:
2137:
2105:
1991:
1955:
40:
PP1 plays an instrumental role in glycogen metabolism through its responsibility for the interconversion between
2918:
2872:
2815:
2183:
2032:
1946:
1901:
1329:"Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1"
851:
690:
529:
349:
325:
2820:
2052:
1922:
2608:
2247:
194:
803:
642:
481:
2841:
2760:
2543:
2440:
2232:
2193:
2057:
1979:
868:
546:
369:
2913:
2726:
2561:
2556:
2489:
2149:
1974:
1194:
1143:"Identification of binding sites on protein targeting to glycogen for enzymes of glycogen metabolism"
707:
341:
252:
108:
30:
1094:"Distinct roles of PP1 and PP2A-like phosphatases in control of microtubule dynamics during mitosis"
408:
about approximate functions of the reg subunits: not all inhibitory, and does merit some explaining.
2877:
2581:
2551:
2355:
2350:
2274:
2210:
1996:
357:
278:
122:
that facilitate the complexation of the PP1 catalytic subunit to various regulatory subunits. and
2810:
2714:
2566:
2047:
2037:
1915:
1897:
1750:
1707:
1526:
1309:
1218:
72:
1544:
36:
3083:
2515:
2460:
2428:
2303:
2022:
1877:
1842:
1832:
1801:
1742:
1699:
1664:
1616:
1572:
1518:
1477:
1432:
1396:
1358:
1301:
1266:
1210:
1164:
1123:
1069:
1065:
1059:
1049:
1025:
298:
1641:
Li Q, Zhao Q, Zhang J, Linkang L, Wenhao W, Chua B, Chen Y, Xu L, Li P (September 24, 2019).
973:
969:
965:
957:
941:
933:
2856:
2851:
2825:
2753:
2591:
2171:
2076:
2071:
2027:
1869:
1824:
1819:
Agarwalla PK, Aghi MK (2012). "Oncolytic herpes simplex virus engineering and preparation".
1791:
1781:
1734:
1691:
1654:
1608:
1508:
1467:
1459:
1424:
1388:
1348:
1340:
1293:
1256:
1202:
1154:
1113:
1105:
863:
702:
541:
353:
282:
240:
177:
159:
856:
695:
534:
2903:
2887:
2800:
2693:
2507:
2423:
2418:
2413:
2326:
2321:
2081:
1959:
892:
872:
731:
570:
550:
361:
711:
1198:
410:
Please expand the section to include this information. Further details may exist on the
3052:
2941:
2882:
2666:
2661:
2656:
2042:
2017:
2013:
1986:
1969:
1796:
1769:
1695:
1472:
1451:
1353:
1328:
1118:
1093:
1297:
914:
Protein phosphatase 1 is a multimeric enzyme that may contain the following subunits:
146:
The mechanism involves two metal ions binding and activating water, which initiates a
3062:
2846:
2805:
2407:
2316:
2066:
1513:
1496:
1392:
189:
113:
41:
1754:
1711:
1530:
1313:
1092:
Tournebize R, Andersen SS, Verde F, Dorée M, Karsenti E, Hyman AA (September 1997).
63:. This type of phosphatase includes metal-dependent protein phosphatases (PPMs) and
2795:
2534:
2465:
2188:
2109:
1222:
827:
666:
505:
163:
147:
56:
1659:
1642:
1261:
1244:
379:
289:
demonstrated in both cell culture experiments and in PPP1R3G-knockdown mice that
3019:
2954:
2790:
2709:
2480:
2374:
2237:
2203:
2141:
1907:
1828:
902:
741:
580:
314:
167:
119:
3047:
1860:
Cohen PT (January 2002). "Protein phosphatase 1--targeted in many directions".
1738:
1109:
2686:
1344:
1327:
Egloff MP, Johnson DF, Moorhead G, Cohen PT, Cohen P, Barford D (April 1997).
1141:
Fong NM, Jensen TC, Shah AS, Parekh NN, Saltiel AR, Brady MJ (November 2000).
834:
673:
512:
333:
201:
133:
104:
100:
2993:
2967:
2599:
2398:
2311:
1938:
1786:
329:
267:
96:
84:
64:
1881:
1846:
1805:
1746:
1668:
1620:
1436:
1270:
1168:
1159:
1142:
396:
208:
in its active R state has PP1 bound tightly. This binding to phosphorylase
1873:
1703:
1522:
1481:
1400:
1362:
1305:
1214:
1127:
137:
The PP1 mechanism involves the use of a di-metal ion and activating water.
67:-based phosphatases. PP1 has been found to be important in the control of
2644:
2639:
2634:
2455:
2090:
1942:
1055:
1045:
1041:
1037:
1031:
1021:
1017:
839:
678:
517:
374:
271:
248:
68:
1612:
87:, protein synthesis, and regulation of membrane receptors and channels.
2651:
2629:
2624:
2242:
1428:
1011:
1005:
999:
995:
977:
961:
953:
937:
294:
263:
123:
80:
1463:
3006:
2776:
2671:
2604:
2497:
2095:
2003:
1206:
989:
983:
947:
927:
923:
919:
822:
771:
756:
661:
610:
595:
500:
449:
434:
1245:"From promiscuity to precision: protein phosphatases get a makeover"
17:
2980:
2613:
2364:
2360:
132:
1495:
MacKintosh C, Beattie KA, Klumpp S, Cohen P, Codd GA (May 1990).
220:
is converted to its inactive, T state. By shifting phosphorylase
2345:
2222:
2215:
2159:
2154:
815:
654:
493:
365:
302:
171:
2749:
1911:
1823:. Methods in Molecular Biology. Vol. 797. pp. 1–19.
390:
321:
290:
76:
368:(eIF-2A), which inactivates eIF-2A. EIF-2A is required for
166:, a diarrhetic shellfish poison, strong tumor promoter, and
620:
PP1, PP1b, PP1beta, PP-1B; PPP1CD; MGC3672; PP1beta; PPP1CB
2745:
348:
also activates protein phosphatase 1, which overcomes the
162:
include a variety of naturally occurring toxins including
757:
protein phosphatase 1, catalytic subunit, gamma isozyme
435:
protein phosphatase 1, catalytic subunit, alpha isozyme
1381:
Annual Review of
Biophysics and Biomolecular Structure
596:
protein phosphatase 1, catalytic subunit, beta isozyme
3036:
1594:"The role of protein phosphatase-1 in insulin action"
295:
Protein phosphatase 1 regulatory subunit 3G (PPP1R3G)
75:, cell progression, neuronal activities, splicing of
459:
PP1, PP1a, MGC15877, MGC1674, PP-1A, PP1alpha, PPP1A
236:
does not bind PP1 allowing PP1 to remain activated.
2932:
2896:
2865:
2834:
2783:
2702:
2590:
2542:
2528:
2506:
2488:
2474:
2454:
2393:
2297:
2136:
2104:
1954:
898:
888:
883:
862:
850:
845:
833:
821:
809:
797:
785:
777:
767:
762:
755:
737:
727:
722:
701:
689:
684:
672:
660:
648:
636:
624:
616:
606:
601:
594:
576:
566:
561:
540:
528:
523:
511:
499:
487:
475:
463:
455:
445:
440:
433:
305:(which is already known to be inhibited by Akt).
259:, thereby keeping glycogen phosphorylase active.
2233:Fructose 6-P,2-kinase:fructose 2,6-bisphosphatase
337:genomes without VP30 dephosphorylation by PP1.
1238:
1236:
1234:
1232:
188:PP1 plays a crucial role in the regulation of
2761:
1923:
1567:Berg JM, Stryer L, Tymoczko JL (2010-12-24).
287:University of the Chinese Academy of Sciences
204:. When glucose levels are low, phosphorylase
8:
383:) with protein phosphatase 1 subunit 15A/B.
313:In Alzheimer's, hyperphosphorylation of the
293:(protein kinase B) directly phosphorylates
2768:
2754:
2746:
2539:
2485:
2471:
1930:
1916:
1908:
1180:
1178:
880:
719:
558:
29:"PP1" redirects here. For other uses, see
1900:at the U.S. National Library of Medicine
1795:
1785:
1658:
1512:
1471:
1374:
1372:
1352:
1260:
1158:
1117:
1571:(7th ed.). New York: W.H. Freeman.
1545:"Cantharidic Acid | CAS 28874-45-5"
35:
3043:
2123:Ubiquitin carboxy-terminal hydrolase L1
1562:
1560:
1558:
1556:
1554:
1452:"Serine/threonine protein phosphatases"
1243:Virshup DM, Shenolikar S (March 2009).
1084:
1592:Brady M, Saltiel A (January 1, 2001).
752:
591:
430:
200:, which serves as a glucose sensor in
2703:either deoxy- or ribo-
61:protein serine/threonine phosphatases
7:
2284:Protein serine/threonine phosphatase
1450:Wera S, Hemmings BA (October 1995).
2385:Cyclic nucleotide phosphodiesterase
2379:Clostridium perfringens alpha toxin
2180:Tartrate-resistant acid phosphatase
1774:The Journal of Biological Chemistry
1147:The Journal of Biological Chemistry
2228:Pyruvate dehydrogenase phosphatase
1696:10.1111/j.1471-4159.1993.tb03603.x
184:Biological function and regulation
25:
2128:4-hydroxybenzoyl-CoA thioesterase
3046:
1393:10.1146/annurev.biophys.27.1.133
395:
95:Each PP1 enzyme contains both a
55:) belongs to a certain class of
2446:N-acetylglucosamine-6-sulfatase
2334:Sphingomyelin phosphodiesterase
781:PP1gamma, PP1y, PP1gamma, PPP1G
366:eukaryotic initiation factor-2A
277:A 2019 study by researchers at
150:attack on the phosphorus atom.
2255:Inositol-phosphate phosphatase
2118:Palmitoyl protein thioesterase
373:C-terminal regulatory domain (
1:
2618:RNA-induced silencing complex
1298:10.1016/S0014-5793(97)01385-9
180:is also an inhibitor of PP1.
3079:Genes on human chromosome 12
3069:Genes on human chromosome 11
2722:Serratia marcescens nuclease
2289:Dual-specificity phosphatase
2279:Protein tyrosine phosphatase
1660:10.1016/j.celrep.2019.08.066
1514:10.1016/0014-5793(90)80245-E
1262:10.1016/j.molcel.2009.02.015
360:, and protein kinase R then
3074:Genes on human chromosome 2
2199:Fructose 1,6-bisphosphatase
1829:10.1007/978-1-61779-340-0_1
356:is activated by the virus'
3100:
1739:10.2174/157016207779316279
1458:. 311 ( Pt 1) (1): 17–29.
262:When blood sugar is high,
28:
2924:Michaelis–Menten kinetics
2436:Galactosamine-6 sulfatase
1992:6-phosphogluconolactonase
1684:Journal of Neurochemistry
879:
718:
557:
99:subunit and at least one
2816:Diffusion-limited enzyme
2184:Purple acid phosphatases
1902:Medical Subject Headings
1110:10.1093/emboj/16.18.5537
350:cellular stress response
1862:Journal of Cell Science
1787:10.1074/jbc.M114.575050
1456:The Biochemical Journal
1417:The Biochemical Journal
1345:10.1093/emboj/16.8.1876
1064:regulatory subunit 16:
1054:regulatory subunit 15:
1036:regulatory subunit 14:
1030:regulatory subunit 13:
1016:regulatory subunit 12:
1010:regulatory subunit 11:
1004:regulatory subunit 10:
195:glycogen phosphorylase
2609:Microprocessor complex
2248:Beta-propeller phytase
1160:10.1074/jbc.M005541200
994:regulatory subunit 9:
988:regulatory subunit 8:
982:regulatory subunit 7:
952:regulatory subunit 3:
946:regulatory subunit 2:
932:regulatory subunit 1:
406:is missing information
138:
45:
2909:Eadie–Hofstee diagram
2842:Allosteric regulation
2544:Endodeoxyribonuclease
2441:Iduronate-2-sulfatase
2194:Glucose 6-phosphatase
1980:Butyrylcholinesterase
1898:Protein+Phosphatase+1
1874:10.1242/jcs.115.2.241
154:Exogeneous inhibitors
136:
49:Protein phosphatase 1
39:
2919:Lineweaver–Burk plot
2727:Micrococcal nuclease
2562:Deoxyribonuclease IV
2557:Deoxyribonuclease II
2490:Exodeoxyribonuclease
2150:Alkaline phosphatase
1975:Acetylcholinesterase
1727:Current HIV Research
1601:Recent Prog Horm Res
352:to viral infection;
342:herpes simplex virus
266:will be secreted by
253:phosphorylase kinase
109:compartmentalization
2582:UvrABC endonuclease
2552:Deoxyribonuclease I
2275:Protein phosphatase
2211:Protein phosphatase
2009:Bile salt-dependent
1997:PAF acetylhydrolase
1613:10.1210/rp.56.1.157
1199:1995Natur.376..745G
918:catalytic subunit:
358:double-stranded RNA
142:Catalytic mechanism
2878:Enzyme superfamily
2811:Enzyme promiscuity
2715:Mung bean nuclease
2574:Restriction enzyme
2567:Restriction enzyme
1429:10.1042/BJ20101471
309:Clinical relevance
139:
73:muscle contraction
46:
3034:
3033:
2743:
2742:
2739:
2738:
2735:
2734:
2524:
2523:
2516:Oligonucleotidase
2461:deoxyribonuclease
2429:Steroid sulfatase
2304:Phosphodiesterase
2033:Hormone-sensitive
1838:978-1-61779-339-4
1821:Oncolytic Viruses
1653:(13): 3406–3422.
1578:978-1-4292-2936-4
1464:10.1042/bj3110017
912:
911:
908:
907:
751:
750:
747:
746:
590:
589:
586:
585:
429:
428:
364:a protein called
299:glycogen synthase
228:to phosphorylase
83:, cell division,
16:(Redirected from
3091:
3051:
3050:
3042:
2914:Hanes–Woolf plot
2857:Enzyme activator
2852:Enzyme inhibitor
2826:Enzyme catalysis
2770:
2763:
2756:
2747:
2592:Endoribonuclease
2578:
2572:
2540:
2486:
2472:
2172:Acid phosphatase
2053:Monoacylglycerol
1963:ester hydrolases
1932:
1925:
1918:
1909:
1886:
1885:
1868:(Pt 2): 241–56.
1857:
1851:
1850:
1816:
1810:
1809:
1799:
1789:
1780:(33): 22723–38.
1765:
1759:
1758:
1722:
1716:
1715:
1679:
1673:
1672:
1662:
1638:
1632:
1631:
1629:
1627:
1598:
1589:
1583:
1582:
1564:
1549:
1548:
1541:
1535:
1534:
1516:
1492:
1486:
1485:
1475:
1447:
1441:
1440:
1411:
1405:
1404:
1376:
1367:
1366:
1356:
1333:The EMBO Journal
1324:
1318:
1317:
1281:
1275:
1274:
1264:
1240:
1227:
1226:
1207:10.1038/376745a0
1193:(6543): 745–53.
1182:
1173:
1172:
1162:
1138:
1132:
1131:
1121:
1098:The EMBO Journal
1089:
881:
753:
720:
592:
559:
431:
424:
421:
415:
399:
391:
354:protein kinase R
241:Protein kinase A
232:. Phosphorylase
178:Cantharidic acid
21:
3099:
3098:
3094:
3093:
3092:
3090:
3089:
3088:
3059:
3058:
3057:
3045:
3037:
3035:
3030:
2942:Oxidoreductases
2928:
2904:Enzyme kinetics
2892:
2888:List of enzymes
2861:
2830:
2801:Catalytic triad
2779:
2774:
2744:
2731:
2698:
2586:
2576:
2570:
2533:
2520:
2508:Exoribonuclease
2502:
2479:
2463:
2459:
2450:
2424:Arylsulfatase L
2419:Arylsulfatase B
2414:Arylsulfatase A
2389:
2302:
2293:
2132:
2100:
1962:
1950:
1936:
1894:
1889:
1859:
1858:
1854:
1839:
1818:
1817:
1813:
1767:
1766:
1762:
1724:
1723:
1719:
1681:
1680:
1676:
1640:
1639:
1635:
1625:
1623:
1596:
1591:
1590:
1586:
1579:
1566:
1565:
1552:
1543:
1542:
1538:
1494:
1493:
1489:
1449:
1448:
1444:
1413:
1412:
1408:
1378:
1377:
1370:
1326:
1325:
1321:
1283:
1282:
1278:
1242:
1241:
1230:
1184:
1183:
1176:
1153:(45): 35034–9.
1140:
1139:
1135:
1104:(18): 5537–49.
1091:
1090:
1086:
1082:
1076:2002 and 2007.
425:
419:
416:
409:
400:
389:
311:
251:will also keep
186:
156:
144:
93:
34:
23:
22:
15:
12:
11:
5:
3097:
3095:
3087:
3086:
3081:
3076:
3071:
3061:
3060:
3056:
3055:
3032:
3031:
3029:
3028:
3015:
3002:
2989:
2976:
2963:
2950:
2936:
2934:
2930:
2929:
2927:
2926:
2921:
2916:
2911:
2906:
2900:
2898:
2894:
2893:
2891:
2890:
2885:
2880:
2875:
2869:
2867:
2866:Classification
2863:
2862:
2860:
2859:
2854:
2849:
2844:
2838:
2836:
2832:
2831:
2829:
2828:
2823:
2818:
2813:
2808:
2803:
2798:
2793:
2787:
2785:
2781:
2780:
2775:
2773:
2772:
2765:
2758:
2750:
2741:
2740:
2737:
2736:
2733:
2732:
2730:
2729:
2724:
2719:
2718:
2717:
2706:
2704:
2700:
2699:
2697:
2696:
2691:
2690:
2689:
2684:
2679:
2674:
2664:
2659:
2654:
2649:
2648:
2647:
2642:
2637:
2632:
2622:
2621:
2620:
2611:
2596:
2594:
2588:
2587:
2585:
2584:
2579:
2564:
2559:
2554:
2548:
2546:
2537:
2526:
2525:
2522:
2521:
2519:
2518:
2512:
2510:
2504:
2503:
2501:
2500:
2494:
2492:
2483:
2469:
2452:
2451:
2449:
2448:
2443:
2438:
2433:
2432:
2431:
2426:
2421:
2416:
2403:
2401:
2391:
2390:
2388:
2387:
2382:
2372:
2367:
2358:
2353:
2348:
2343:
2342:
2341:
2331:
2330:
2329:
2324:
2314:
2308:
2306:
2295:
2294:
2292:
2291:
2286:
2281:
2272:
2271:
2270:
2252:
2251:
2250:
2240:
2235:
2230:
2225:
2220:
2219:
2218:
2208:
2207:
2206:
2196:
2191:
2186:
2169:
2168:
2167:
2162:
2157:
2146:
2144:
2134:
2133:
2131:
2130:
2125:
2120:
2114:
2112:
2102:
2101:
2099:
2098:
2093:
2087:
2086:
2085:
2084:
2079:
2074:
2063:
2062:
2061:
2060:
2058:Diacylglycerol
2055:
2050:
2045:
2040:
2035:
2030:
2025:
2020:
2011:
2000:
1999:
1994:
1989:
1987:Pectinesterase
1984:
1983:
1982:
1977:
1970:Cholinesterase
1966:
1964:
1952:
1951:
1937:
1935:
1934:
1927:
1920:
1912:
1906:
1905:
1893:
1892:External links
1890:
1888:
1887:
1852:
1837:
1811:
1760:
1717:
1674:
1633:
1584:
1577:
1550:
1536:
1487:
1442:
1406:
1368:
1339:(8): 1876–87.
1319:
1292:(1–2): 210–4.
1276:
1249:Molecular Cell
1228:
1174:
1133:
1083:
1081:
1078:
1073:
1072:
1062:
1052:
1034:
1028:
1014:
1008:
1002:
992:
986:
980:
950:
944:
930:
910:
909:
906:
905:
900:
896:
895:
890:
886:
885:
877:
876:
866:
860:
859:
854:
848:
847:
843:
842:
837:
831:
830:
825:
819:
818:
813:
807:
806:
801:
795:
794:
789:
783:
782:
779:
775:
774:
769:
765:
764:
760:
759:
749:
748:
745:
744:
739:
735:
734:
729:
725:
724:
716:
715:
705:
699:
698:
693:
687:
686:
682:
681:
676:
670:
669:
664:
658:
657:
652:
646:
645:
640:
634:
633:
628:
622:
621:
618:
614:
613:
608:
604:
603:
599:
598:
588:
587:
584:
583:
578:
574:
573:
568:
564:
563:
555:
554:
544:
538:
537:
532:
526:
525:
521:
520:
515:
509:
508:
503:
497:
496:
491:
485:
484:
479:
473:
472:
467:
461:
460:
457:
453:
452:
447:
443:
442:
438:
437:
427:
426:
403:
401:
394:
388:
385:
362:phosphorylates
320:Regulation of
310:
307:
185:
182:
155:
152:
143:
140:
92:
89:
24:
14:
13:
10:
9:
6:
4:
3:
2:
3096:
3085:
3082:
3080:
3077:
3075:
3072:
3070:
3067:
3066:
3064:
3054:
3049:
3044:
3040:
3026:
3022:
3021:
3016:
3013:
3009:
3008:
3003:
3000:
2996:
2995:
2990:
2987:
2983:
2982:
2977:
2974:
2970:
2969:
2964:
2961:
2957:
2956:
2951:
2948:
2944:
2943:
2938:
2937:
2935:
2931:
2925:
2922:
2920:
2917:
2915:
2912:
2910:
2907:
2905:
2902:
2901:
2899:
2895:
2889:
2886:
2884:
2883:Enzyme family
2881:
2879:
2876:
2874:
2871:
2870:
2868:
2864:
2858:
2855:
2853:
2850:
2848:
2847:Cooperativity
2845:
2843:
2840:
2839:
2837:
2833:
2827:
2824:
2822:
2819:
2817:
2814:
2812:
2809:
2807:
2806:Oxyanion hole
2804:
2802:
2799:
2797:
2794:
2792:
2789:
2788:
2786:
2782:
2778:
2771:
2766:
2764:
2759:
2757:
2752:
2751:
2748:
2728:
2725:
2723:
2720:
2716:
2713:
2712:
2711:
2708:
2707:
2705:
2701:
2695:
2692:
2688:
2685:
2683:
2680:
2678:
2675:
2673:
2670:
2669:
2668:
2665:
2663:
2660:
2658:
2655:
2653:
2650:
2646:
2643:
2641:
2638:
2636:
2633:
2631:
2628:
2627:
2626:
2623:
2619:
2615:
2612:
2610:
2606:
2603:
2602:
2601:
2598:
2597:
2595:
2593:
2589:
2583:
2580:
2575:
2568:
2565:
2563:
2560:
2558:
2555:
2553:
2550:
2549:
2547:
2545:
2541:
2538:
2536:
2531:
2527:
2517:
2514:
2513:
2511:
2509:
2505:
2499:
2496:
2495:
2493:
2491:
2487:
2484:
2482:
2477:
2473:
2470:
2467:
2462:
2457:
2453:
2447:
2444:
2442:
2439:
2437:
2434:
2430:
2427:
2425:
2422:
2420:
2417:
2415:
2412:
2411:
2410:
2409:
2408:arylsulfatase
2405:
2404:
2402:
2400:
2396:
2392:
2386:
2383:
2380:
2376:
2373:
2371:
2368:
2366:
2362:
2359:
2357:
2354:
2352:
2349:
2347:
2344:
2340:
2337:
2336:
2335:
2332:
2328:
2325:
2323:
2320:
2319:
2318:
2317:Phospholipase
2315:
2313:
2310:
2309:
2307:
2305:
2300:
2296:
2290:
2287:
2285:
2282:
2280:
2276:
2273:
2269:
2265:
2261:
2258:
2257:
2256:
2253:
2249:
2246:
2245:
2244:
2241:
2239:
2236:
2234:
2231:
2229:
2226:
2224:
2221:
2217:
2214:
2213:
2212:
2209:
2205:
2202:
2201:
2200:
2197:
2195:
2192:
2190:
2187:
2185:
2181:
2177:
2173:
2170:
2166:
2163:
2161:
2158:
2156:
2153:
2152:
2151:
2148:
2147:
2145:
2143:
2139:
2135:
2129:
2126:
2124:
2121:
2119:
2116:
2115:
2113:
2111:
2107:
2103:
2097:
2094:
2092:
2089:
2088:
2083:
2080:
2078:
2075:
2073:
2070:
2069:
2068:
2067:Phospholipase
2065:
2064:
2059:
2056:
2054:
2051:
2049:
2046:
2044:
2041:
2039:
2036:
2034:
2031:
2029:
2026:
2024:
2021:
2019:
2015:
2012:
2010:
2007:
2006:
2005:
2002:
2001:
1998:
1995:
1993:
1990:
1988:
1985:
1981:
1978:
1976:
1973:
1972:
1971:
1968:
1967:
1965:
1961:
1957:
1953:
1948:
1944:
1940:
1933:
1928:
1926:
1921:
1919:
1914:
1913:
1910:
1903:
1899:
1896:
1895:
1891:
1883:
1879:
1875:
1871:
1867:
1863:
1856:
1853:
1848:
1844:
1840:
1834:
1830:
1826:
1822:
1815:
1812:
1807:
1803:
1798:
1793:
1788:
1783:
1779:
1775:
1771:
1764:
1761:
1756:
1752:
1748:
1744:
1740:
1736:
1732:
1728:
1721:
1718:
1713:
1709:
1705:
1701:
1697:
1693:
1689:
1685:
1678:
1675:
1670:
1666:
1661:
1656:
1652:
1648:
1644:
1637:
1634:
1622:
1618:
1614:
1610:
1606:
1602:
1595:
1588:
1585:
1580:
1574:
1570:
1563:
1561:
1559:
1557:
1555:
1551:
1546:
1540:
1537:
1532:
1528:
1524:
1520:
1515:
1510:
1507:(2): 187–92.
1506:
1502:
1498:
1491:
1488:
1483:
1479:
1474:
1469:
1465:
1461:
1457:
1453:
1446:
1443:
1438:
1434:
1430:
1426:
1423:(3): 435–44.
1422:
1418:
1410:
1407:
1402:
1398:
1394:
1390:
1386:
1382:
1375:
1373:
1369:
1364:
1360:
1355:
1350:
1346:
1342:
1338:
1334:
1330:
1323:
1320:
1315:
1311:
1307:
1303:
1299:
1295:
1291:
1287:
1280:
1277:
1272:
1268:
1263:
1258:
1255:(5): 537–45.
1254:
1250:
1246:
1239:
1237:
1235:
1233:
1229:
1224:
1220:
1216:
1212:
1208:
1204:
1200:
1196:
1192:
1188:
1181:
1179:
1175:
1170:
1166:
1161:
1156:
1152:
1148:
1144:
1137:
1134:
1129:
1125:
1120:
1115:
1111:
1107:
1103:
1099:
1095:
1088:
1085:
1079:
1077:
1071:
1067:
1063:
1061:
1057:
1053:
1051:
1047:
1043:
1039:
1035:
1033:
1029:
1027:
1023:
1019:
1015:
1013:
1009:
1007:
1003:
1001:
997:
993:
991:
987:
985:
981:
979:
975:
971:
967:
963:
959:
955:
951:
949:
945:
943:
939:
935:
931:
929:
925:
921:
917:
916:
915:
904:
901:
897:
894:
891:
887:
882:
878:
875:
874:
870:
867:
865:
861:
858:
855:
853:
849:
844:
841:
838:
836:
832:
829:
826:
824:
820:
817:
814:
812:
808:
805:
802:
800:
796:
793:
790:
788:
784:
780:
776:
773:
770:
766:
761:
758:
754:
743:
740:
736:
733:
730:
726:
721:
717:
714:
713:
709:
706:
704:
700:
697:
694:
692:
688:
683:
680:
677:
675:
671:
668:
665:
663:
659:
656:
653:
651:
647:
644:
641:
639:
635:
632:
629:
627:
623:
619:
615:
612:
609:
605:
600:
597:
593:
582:
579:
575:
572:
569:
565:
560:
556:
553:
552:
548:
545:
543:
539:
536:
533:
531:
527:
522:
519:
516:
514:
510:
507:
504:
502:
498:
495:
492:
490:
486:
483:
480:
478:
474:
471:
468:
466:
462:
458:
454:
451:
448:
444:
439:
436:
432:
423:
413:
407:
404:This section
402:
398:
393:
392:
386:
384:
382:
381:
376:
371:
367:
363:
359:
355:
351:
347:
343:
338:
335:
331:
327:
326:transcription
323:
318:
316:
308:
306:
304:
300:
296:
292:
288:
284:
280:
275:
273:
269:
265:
260:
258:
254:
250:
246:
242:
237:
235:
231:
227:
223:
219:
215:
211:
207:
203:
199:
198:
191:
190:blood glucose
183:
181:
179:
175:
173:
169:
165:
161:
153:
151:
149:
141:
135:
131:
127:
125:
121:
116:
115:
114:S. cerevisiae
110:
106:
102:
98:
90:
88:
86:
82:
78:
74:
70:
66:
62:
58:
54:
50:
43:
42:phosphorylase
38:
32:
27:
19:
3020:Translocases
3017:
3004:
2991:
2978:
2965:
2955:Transferases
2952:
2939:
2796:Binding site
2577:}}
2571:{{
2535:Endonuclease
2466:ribonuclease
2406:
2189:Nucleotidase
2110:Thioesterase
1865:
1861:
1855:
1820:
1814:
1777:
1773:
1763:
1730:
1726:
1720:
1690:(3): 921–7.
1687:
1683:
1677:
1650:
1647:Cell Reports
1646:
1636:
1626:December 15,
1624:. Retrieved
1604:
1600:
1587:
1569:Biochemistry
1568:
1539:
1504:
1501:FEBS Letters
1500:
1490:
1455:
1445:
1420:
1416:
1409:
1384:
1380:
1336:
1332:
1322:
1289:
1286:FEBS Letters
1285:
1279:
1252:
1248:
1190:
1186:
1150:
1146:
1136:
1101:
1097:
1087:
1074:
913:
871:
778:Alt. symbols
710:
617:Alt. symbols
549:
456:Alt. symbols
417:
405:
378:
339:
319:
312:
276:
261:
238:
233:
229:
225:
221:
217:
213:
209:
205:
196:
187:
176:
164:okadaic acid
157:
148:nucleophilic
145:
128:
112:
94:
71:metabolism,
57:phosphatases
52:
48:
47:
26:
2791:Active site
2710:Nuclease S1
2481:Exonuclease
2375:Lecithinase
2204:Calcineurin
2142:Phosphatase
2048:Lipoprotein
2038:Endothelial
1607:: 157–173.
893:Swiss-model
828:NP_002701.1
763:Identifiers
732:Swiss-model
667:NP_002700.1
602:Identifiers
571:Swiss-model
506:NP_002699.1
441:Identifiers
420:August 2019
370:translation
315:microtubule
255:active via
202:hepatocytes
168:microcystin
120:holoenzymes
3063:Categories
2994:Isomerases
2968:Hydrolases
2835:Regulation
2023:Pancreatic
1960:Carboxylic
1733:(1): 3–9.
1387:: 133–64.
1080:References
889:Structures
884:Search for
846:Other data
728:Structures
723:Search for
685:Other data
567:Structures
562:Search for
524:Other data
334:ebolavirus
268:beta cells
160:inhibitors
158:Potential
105:eukaryotes
101:regulatory
2873:EC number
2600:RNase III
2458:(includes
2399:Sulfatase
2312:Autotaxin
2176:Prostatic
2028:Lysosomal
1943:esterases
1939:Hydrolase
852:EC number
787:NCBI gene
691:EC number
626:NCBI gene
530:EC number
465:NCBI gene
412:talk page
380:IPR019523
97:catalytic
91:Structure
85:apoptosis
65:aspartate
59:known as
3084:EC 3.1.3
2897:Kinetics
2821:Cofactor
2784:Activity
2694:RNase T1
2456:Nuclease
2091:Cutinase
1882:11839776
1847:21948465
1806:24936058
1755:12105058
1747:17266553
1712:30225343
1669:31553910
1621:11237211
1531:27643473
1437:21204787
1314:21169749
1271:19285938
1169:10938087
1070:PPP1R16B
1066:PPP1R16A
1060:PPP1R15B
1056:PPP1R15A
1050:PPP1R14D
1046:PPP1R14C
1042:PPP1R14B
1038:PPP1R14A
1032:PPP1R13B
1026:PPP1R12C
1022:PPP1R12B
1018:PPP1R12A
903:InterPro
857:3.1.3.16
742:InterPro
696:3.1.3.16
581:InterPro
535:3.1.3.16
387:Subunits
375:InterPro
344:protein
285:and the
279:Tsinghua
272:pancreas
249:glucagon
69:glycogen
44:a and b.
3053:Biology
3007:Ligases
2777:Enzymes
2667:RNase E
2662:RNase Z
2657:RNase A
2652:RNase P
2625:RNase H
2243:Phytase
2043:Hepatic
2018:Lingual
2014:Gastric
1797:4132779
1704:8395566
1523:2162782
1482:7575450
1473:1136113
1401:9646865
1363:9155014
1354:1169791
1306:9414128
1223:4256743
1215:7651533
1195:Bibcode
1128:9312013
1119:1170186
1012:PPP1R11
1006:PPP1R10
1000:PPP1R9B
996:PPP1R9A
978:PPP1R3G
974:PPP1R3F
970:PPP1R3E
966:PPP1R3D
962:PPP1R3C
958:PPP1R3B
954:PPP1R3A
942:PPP1R1C
938:PPP1R1B
934:PPP1R1A
899:Domains
869:Chr. 12
835:UniProt
738:Domains
674:UniProt
577:Domains
547:Chr. 11
513:UniProt
377::
346:ICP34.5
270:of the
264:insulin
124:PPP1R3G
81:mitosis
3039:Portal
2981:Lyases
2605:Drosha
2530:3.1.21
2498:RecBCD
2476:3.1.11
2096:PETase
2004:Lipase
1904:(MeSH)
1880:
1845:
1835:
1804:
1794:
1753:
1745:
1710:
1702:
1667:
1619:
1575:
1529:
1521:
1480:
1470:
1435:
1399:
1361:
1351:
1312:
1304:
1269:
1221:
1213:
1187:Nature
1167:
1126:
1116:
990:PPP1R8
984:PPP1R7
948:PPP1R2
928:PPP1CC
924:PPP1CB
920:PPP1CA
840:P36873
823:RefSeq
816:176914
772:PPP1CC
768:Symbol
708:Chr. 2
679:P62140
662:RefSeq
655:600590
611:PPP1CB
607:Symbol
518:P62136
501:RefSeq
494:176875
450:PPP1CA
446:Symbol
2933:Types
2614:Dicer
2569:;see
2395:3.1.6
2365:PDE4B
2361:PDE4A
2299:3.1.4
2268:IMPA3
2264:IMPA2
2260:IMPA1
2138:3.1.3
2106:3.1.2
1956:3.1.1
1751:S2CID
1708:S2CID
1597:(PDF)
1527:S2CID
1310:S2CID
1219:S2CID
864:Locus
703:Locus
542:Locus
283:Fudan
3025:list
3018:EC7
3012:list
3005:EC6
2999:list
2992:EC5
2986:list
2979:EC4
2973:list
2966:EC3
2960:list
2953:EC2
2947:list
2940:EC1
2532:-31:
2478:-16:
2464:and
2370:PDE5
2356:PDE3
2351:PDE2
2346:PDE1
2238:PTEN
2223:OCRL
2216:PP2A
2165:ALPP
2160:ALPL
2155:ALPI
1949:3.1)
1878:PMID
1843:PMID
1833:ISBN
1802:PMID
1743:PMID
1700:PMID
1665:PMID
1628:2023
1617:PMID
1573:ISBN
1519:PMID
1478:PMID
1433:PMID
1397:PMID
1359:PMID
1302:PMID
1267:PMID
1211:PMID
1165:PMID
1124:PMID
811:OMIM
804:9283
799:HGNC
792:5501
650:OMIM
643:9282
638:HGNC
631:5500
489:OMIM
482:9281
477:HGNC
470:5499
340:The
303:GSK3
257:cAMP
245:cAMP
172:MCLR
2687:4/5
1870:doi
1866:115
1825:doi
1792:PMC
1782:doi
1778:289
1735:doi
1692:doi
1655:doi
1609:doi
1509:doi
1505:264
1468:PMC
1460:doi
1425:doi
1421:434
1389:doi
1349:PMC
1341:doi
1294:doi
1290:418
1257:doi
1203:doi
1191:376
1155:doi
1151:275
1114:PMC
1106:doi
873:q24
712:p23
551:q13
330:Tat
324:-1
322:HIV
291:Akt
77:RNA
53:PP1
31:Pp1
18:PP1
3065::
2645:2C
2640:2B
2635:2A
2616::
2607::
2397::
2277::
2266:,
2262:,
2178:)/
2140::
2108::
2077:A2
2072:A1
1958::
1947:EC
1941::
1876:.
1864:.
1841:.
1831:.
1800:.
1790:.
1776:.
1772:.
1749:.
1741:.
1729:.
1706:.
1698:.
1688:61
1686:.
1663:.
1651:28
1649:.
1645:.
1615:.
1605:56
1603:.
1599:.
1553:^
1525:.
1517:.
1503:.
1499:.
1476:.
1466:.
1454:.
1431:.
1419:.
1395:.
1385:27
1383:.
1371:^
1357:.
1347:.
1337:16
1335:.
1331:.
1308:.
1300:.
1288:.
1265:.
1253:33
1251:.
1247:.
1231:^
1217:.
1209:.
1201:.
1189:.
1177:^
1163:.
1149:.
1145:.
1122:.
1112:.
1102:16
1100:.
1096:.
1068:,
1058:,
1048:,
1044:,
1040:,
1024:,
1020:,
998:,
976:,
972:,
968:,
964:,
960:,
956:,
940:,
936:,
926:,
922:,
281:,
126:.
79:,
3041::
3027:)
3023:(
3014:)
3010:(
3001:)
2997:(
2988:)
2984:(
2975:)
2971:(
2962:)
2958:(
2949:)
2945:(
2769:e
2762:t
2755:v
2682:3
2677:2
2672:1
2630:1
2468:)
2381:)
2377:(
2363:/
2339:1
2327:D
2322:C
2301::
2182:/
2174:(
2082:B
2016:/
1945:(
1931:e
1924:t
1917:v
1884:.
1872::
1849:.
1827::
1808:.
1784::
1757:.
1737::
1731:5
1714:.
1694::
1671:.
1657::
1630:.
1611::
1581:.
1547:.
1533:.
1511::
1484:.
1462::
1439:.
1427::
1403:.
1391::
1365:.
1343::
1316:.
1296::
1273:.
1259::
1225:.
1205::
1197::
1171:.
1157::
1130:.
1108::
422:)
418:(
414:.
243:(
234:b
230:b
226:a
222:a
218:a
214:a
210:a
206:a
197:a
51:(
33:.
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.