323:
1230:
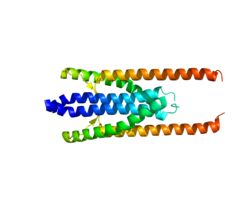
rod domain adopts multiple conformations, all affected by its C-terminal spectrin domain. A model has been suggested in which PRC1 is likely to be a flexible molecule both in solution and on single microtubules but becomes more rigid when the microtubule-binding domains are restricted with antiparallel microtubule filament crosslinking, seen at the spindle midzone. The overall structure of the PRC1 homodimer is reminiscent of actin-bundling proteins, and this process of microtubule filament crosslinking is similar to that of
300:
197:
222:
574:
329:
228:
1243:
by CDK1 mediated phosphorylation, preventing its dimerization. Upon anaphase onset and removal of inhibitory CDK1 phosphorylation, PRC1 dimers form. These homodimers specifically recognize antiparallel microtubule overlaps, found at the spindle midzone, and bind, allowing microtubule sliding, cross-linking of microtubule filaments, and assembly of central-spindle-mediating proteins, including but not limited to
Kinesin-4.
38:
1242:
PRC1’s role in midzone microtubule formation, essential to the cytokinetic machinery of mammals, is made possible through its collaboration with
Kinesin-4 in setting up a controlled zone of overlapping, antiparallel microtubules at the spindle midzone. PRC1 is normally inhibited until anaphase onset
1229:
microtubule-binding domain, an extended rod domain, and an N-terminal dimerization domain. Consisting of an intricate arrangement of α-helices, the rod domain, together with the dimerization-conducting N terminus cooperate to facilitate binding of other proteins, such as
Kinesin-4, to PRC1. PRC1’s
1246:
PRC1 dimers, required for the high-affinity interaction with
Kinesin-4, recruit Kinesin-4 to regions of antiparallel microtubule overlap, where Kinesin-4, a plus-end directed motor protein that inhibits microtubule dynamics, helps to form length-dependent end tags that help stabilize and regulate
2255:
Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P,
1841:
Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P,
64:
1197:, becomes associated with the mitotic spindle in a highly dynamic manner during anaphase, and localizes to the cell midbody during cytokinesis. PRC1 was first identified in 1998 using an ''in vitro'' phosphorylation screening method and shown to be a substrate of several
1247:
spindle microtubule assembly within cytokinesis. This PRC1-Kinesin-4 complex differentially identifies and regulates the spindle midzone microtubules during cell division. This regulation is crucial in order for cytokinesis to progress properly.
2256:
Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M (2005). "Towards a proteome-scale map of the human protein-protein interaction network".
1842:
Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M (2005). "Towards a proteome-scale map of the human protein-protein interaction network".
1204:
At least three alternatively spliced transcript variants encoding distinct isoforms of PRC1 have been observed. Additionally, PRC1 has sequence homology with Ase1 in yeasts, SPD-1 (spindle defective 1) in
1189:
PRC1 protein is expressed at relatively high levels during S and G2/M phases of the cell cycle before dropping dramatically after mitotic exit and entrance into G1 phase. PRC1 is located in the
2161:"Ablation of PRC1 by small interfering RNA demonstrates that cytokinetic abscission requires a central spindle bundle in mammalian cells, whereas completion of furrowing does not"
336:
235:
2407:
Beausoleil SA, Villén J, Gerber SA, Rush J, Gygi SP (2006). "A probability-based approach for high-throughput protein phosphorylation analysis and site localization".
1256:
PRC1 is a non-motor microtubule-associated protein (MAP) whose C-terminal spectrin domain (aa 341-640) binds microtubules with micromolar affinity (0.6 +/- 0.3uM)
1225:
The crystal structure of PRC1 has only recently been characterized in vitro. In 2013, PRC1 was illustrated as a lengthy molecule consisting of a C-terminal
832:
158:
813:
1270:
PRC1 interacts with
Kinesin-4 that plays an important role in crossing spindle microtubules and setting midzone length in mammalian cytokinesis.
1371:
2576:
1353:
322:
1100:
1107:
299:
1436:
1340:
1319:
1214:
221:
196:
1336:
61:
22:
1315:
138:
2204:"Cell cycle-dependent translocation of PRC1 on the spindle by Kif4 is essential for midzone formation and cytokinesis"
1260:
335:
234:
1201:(CDKs). Correspondingly, ablation of PRC1 has been shown to disrupt spindle midzone assembly in mammalian systems.
2126:"Identification of PRC1 as the p53 target gene uncovers a novel function of p53 in the regulation of cytokinesis"
328:
227:
1792:
Subramanian R, Wilson-Kubalek EM, Arthur CP, Bick MJ, Campbell EA, Darst SA, Milligan RA, Kapoor TM (Aug 2010).
2497:"Insights into antiparallel microtubule crosslinking by PRC1, a conserved nonmotor microtubule binding protein"
1794:"Insights into antiparallel microtubule crosslinking by PRC1, a conserved nonmotor microtubule binding protein"
1198:
877:
146:
2048:"Human mitotic spindle-associated protein PRC1 inhibits MgcRacGAP activity toward Cdc42 during the metaphase"
1944:"Direct interaction between centralspindlin and PRC1 reinforces mechanical resilience of the central spindle"
2495:
Subramanian R, Wilson-Kubalek EM, Arthur CP, Bick MJ, Campbell EA, Darst SA, Milligan RA, Kapoor TM (2010).
858:
2446:"Elevated expression of protein regulator of cytokinesis 1, involved in the growth of breast cancer cells"
1206:
210:
2005:"PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone"
2367:
2314:
2265:
2215:
2083:"Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation"
1955:
1851:
1601:
1487:"A Minimal Midzone Protein Module Controls Formation and Length of Antiparallel Microtubule Overlaps"
1294:
PRC1 binds directly to CYK-4 subunit of the centralspindlin complex to stabilise the central spindle.
125:
1083:
1079:
1075:
1071:
1067:
1057:
1053:
1045:
1041:
1018:
992:
984:
980:
947:
2483:
2432:
2289:
1875:
1774:
170:
1049:
1026:
1022:
988:
959:
955:
951:
1450:
Eggert US, Mitchison TJ, Field CM (2006). "Animal
Cytokinesis: From Parts List to Mechanisms".
2546:
2526:
2475:
2424:
2395:
2342:
2281:
2243:
2190:
2147:
2112:
2069:
2034:
1981:
1924:
1867:
1823:
1766:
1718:
1668:
1619:
1566:
1508:
1467:
1415:
118:
54:
1463:
2516:
2508:
2465:
2457:
2416:
2385:
2375:
2332:
2322:
2273:
2233:
2223:
2180:
2172:
2137:
2102:
2094:
2059:
2024:
2016:
1971:
1963:
1914:
1906:
1859:
1813:
1805:
1756:
1748:
1708:
1658:
1650:
1609:
1556:
1548:
1498:
1459:
1405:
573:
415:
346:
290:
245:
1895:"Plk1 negatively regulates PRC1 to prevent premature midzone formation before cytokinesis"
1284:
1210:
390:
166:
1394:"PRC1: a human mitotic spindle-associated CDK substrate protein required for cytokinesis"
2470:
2445:
2371:
2318:
2269:
2219:
1959:
1855:
1605:
2521:
2496:
2390:
2355:
2337:
2302:
2029:
2004:
1976:
1943:
1919:
1894:
1818:
1793:
1663:
1638:
1561:
1536:
2238:
2203:
2185:
2160:
2159:
Mollinari C, Kleman JP, Saoudi Y, Jablonski SA, Perard J, Yen TJ, Margolis RL (2005).
2107:
2082:
1584:
Vernì F, Somma MP, Gunsalus KC, Bonaccorsi S, Belloni G, Goldberg ML, Gatti M (2004).
1410:
1393:
747:
742:
737:
732:
727:
722:
717:
712:
696:
691:
686:
681:
676:
671:
666:
661:
656:
651:
635:
630:
625:
620:
615:
37:
2570:
2555:
2461:
602:
2436:
150:
2487:
2293:
1879:
1778:
1190:
408:
187:
1713:
1696:
174:
1376:
National Center for
Biotechnology Information, U.S. National Library of Medicine
1358:
National Center for
Biotechnology Information, U.S. National Library of Medicine
1178:
2512:
1809:
1552:
1503:
1486:
491:
1739:
Fededa JP, Gerlich DW (2012). "Molecular
Control of animal cell cytokinesis".
1614:
1585:
1288:
1194:
307:
204:
154:
2098:
1392:
Jiang W, Jimenez G, Wells NJ, Hope TJ, Wahl GM, Hunter T, Fukunaga R (1998).
2380:
2356:"Spatiotemporal control of spindle midzone formation by PRC1 in human cells"
2327:
2228:
2176:
1910:
1654:
777:
551:
429:
374:
361:
273:
260:
162:
2530:
2479:
2428:
2399:
2346:
2285:
2247:
2194:
2151:
2142:
2125:
2116:
2073:
2064:
2047:
2038:
1985:
1928:
1871:
1827:
1770:
1722:
1672:
1623:
1590:
Homolog of PRC1, Is
Required for Central-Spindle Formation and Cytokinesis"
1570:
1512:
1471:
1213:, and MAP65 in plants, all of which fall in a conserved family of nonmotor
2020:
2003:
Mollinari C, Kleman JP, Jiang W, Schoehn G, Hunter T, Margolis RL (2002).
1419:
1147:
1142:
1291:
of PRC1, only after dephosphorylation of PRC1 at an inhibitory CDK1 site.
1226:
1131:
922:
903:
2277:
1863:
2560:
2550:
2444:
Shimo A, Nishidate T, Ohta T, Fukuda M, Nakamura Y, Katagiri T (2007).
1967:
1761:
1167:
889:
844:
95:
91:
87:
83:
1264:
1115:
799:
2420:
1752:
2081:
Kurasawa Y, Earnshaw WC, Mochizuki Y, Dohmae N, Todokoro K (2005).
1231:
2354:
Zhu C, Lau E, Schwarzenbacher R, Bossy-Wetzel E, Jiang W (2006).
1537:"Marking and Measuring Single Microtubules by PRC1 and Kinesin-4"
762:
758:
1280:
1274:
1174:
142:
582:
2545:
Overview of all the structural information available in the
2301:
Nousiainen M, Silljé HH, Sauer G, Nigg EA, Körner R (2006).
1893:
Hu CK, Ozlü N, Coughlin M, Steen JJ, Mitchison TJ (2012).
1535:
Subramanian R, Ti SC, Tan L, Darst SA, Kapoor TM (2013).
1639:"Midbody assembly and its regulation during cytokinesis"
398:
2303:"Phosphoproteome analysis of the human mitotic spindle"
1437:"Entrez Gene: PRC1 protein regulator of cytokinesis 1"
1697:"Motors and MAPs Collaborate to Size Up Microtubules"
563:
738:
positive regulation of cell population proliferation
21:
For polycomb-group repressive complex 1 (PRC1), see
1273:PRC1 is negatively modulated by CDKs, particularly
1040:
1011:
973:
940:
1332:
1330:
1328:
1311:
1309:
1307:
1942:Lee KY, Esmaeili B, Zealley B, Mishima M (2015).
345:
244:
1734:
1732:
1690:
1688:
1686:
1684:
1682:
16:Protein-coding gene in the species Homo sapiens
1530:
1528:
1526:
1524:
1522:
1337:GRCm38: Ensembl release 89: ENSMUSG00000038943
1387:
1385:
8:
2559:(Protein regulator of cytokinesis 1) at the
2046:Ban R, Irino Y, Fukami K, Tanaka H (2004).
1316:GRCh38: Ensembl release 89: ENSG00000198901
1037:
773:
598:
386:
285:
182:
129:, ASE1, protein regulator of cytokinesis 1
72:
2520:
2469:
2389:
2379:
2336:
2326:
2237:
2227:
2184:
2141:
2106:
2063:
2028:
1975:
1918:
1817:
1760:
1712:
1662:
1613:
1560:
1502:
1409:
1637:Hu CK, Coughlin M, Mitchison TJ (2012).
1464:10.1146/annurev.biochem.74.082803.133425
1431:
1429:
1485:Bieling P, Telley IA, Surrey T (2010).
1303:
27:
728:microtubule cytoskeleton organization
350:
311:
306:
249:
208:
203:
7:
538:endothelial cell of lymphatic vessel
1283:negatively regulates PRC1 through
1160:Protein Regulator of cytokinesis 1
1008:
970:
937:
913:
894:
868:
849:
823:
804:
568:
486:
424:
403:
14:
1695:Bechstedt S, Brouhard GJ (2013).
1170:that in humans is encoded by the
2462:10.1111/j.1349-7006.2006.00381.x
572:
334:
327:
321:
298:
233:
226:
220:
195:
36:
1215:microtubule-associated proteins
583:More reference expression data
552:More reference expression data
1:
1411:10.1016/S1097-2765(00)80302-0
319:
218:
23:Polycomb repressive complex 1
2577:Genes on human chromosome 15
2360:Proc. Natl. Acad. Sci. U.S.A
2307:Proc. Natl. Acad. Sci. U.S.A
2208:Proc. Natl. Acad. Sci. U.S.A
1714:10.1016/j.devcel.2013.07.010
713:microtubule bundle formation
2124:Li C, Lin M, Liu J (2005).
468:stromal cell of endometrium
2593:
2513:10.1016/j.cell.2010.07.012
1810:10.1016/j.cell.2010.07.012
1553:10.1016/j.cell.2013.06.021
1504:10.1016/j.cell.2010.06.033
733:mitotic spindle elongation
472:mucosa of transverse colon
20:
1615:10.1016/j.cub.2004.08.054
1372:"Mouse PubMed Reference:"
1354:"Human PubMed Reference:"
1146:
1141:
1137:
1130:
1114:
1095:
1064:
1015:
1004:
977:
944:
933:
920:
916:
901:
897:
888:
875:
871:
856:
852:
843:
830:
826:
811:
807:
798:
783:
776:
772:
756:
748:regulation of cytokinesis
621:identical protein binding
601:
597:
580:
571:
562:
549:
498:
489:
436:
427:
397:
389:
385:
368:
355:
318:
297:
288:
284:
267:
254:
217:
194:
185:
181:
136:
133:
123:
116:
111:
80:
75:
58:
53:
48:
44:
35:
30:
2099:10.1038/sj.emboj.7600347
1199:cyclin-dependent kinases
2381:10.1073/pnas.0506926103
2328:10.1073/pnas.0507066103
2229:10.1073/pnas.0408438102
2202:Zhu C, Jiang W (2005).
2177:10.1091/mbc.E04-04-0346
1911:10.1091/mbc.E12-01-0058
1655:10.1091/mbc.E11-08-0721
1259:PRC1 has been shown to
1108:Chr 7: 80.29 – 80.32 Mb
2143:10.1038/sj.onc.1208114
2065:10.1074/jbc.M313257200
616:protein kinase binding
2021:10.1083/jcb.200111052
1948:Nature Communications
1287:at Thr-602, near the
1101:Chr 15: 90.97 – 91 Mb
211:Chromosome 15 (human)
352:7 D2|7 45.62 cM
313:Chromosome 7 (mouse)
76:List of PDB id codes
49:Available structures
2372:2006PNAS..103.6196Z
2319:2006PNAS..103.5391N
2278:10.1038/nature04209
2270:2005Natur.437.1173R
2220:2005PNAS..102..343Z
1960:2015NatCo...6.7290L
1864:10.1038/nature04209
1856:2005Natur.437.1173R
1606:2004CBio...14.1569V
1238:Role in cytokinesis
1177:and is involved in
672:spindle microtubule
631:microtubule binding
444:ganglionic eminence
1968:10.1038/ncomms8290
1452:Annu. Rev. Biochem
878:ENSMUSG00000038943
706:Biological process
645:Cellular component
609:Molecular function
2058:(16): 16394–402.
1157:
1156:
1153:
1152:
1126:
1125:
1091:
1090:
1034:
1033:
1000:
999:
967:
966:
929:
928:
910:
909:
884:
883:
865:
864:
839:
838:
820:
819:
768:
767:
593:
592:
589:
588:
558:
557:
545:
544:
483:
482:
476:bone marrow cells
381:
380:
280:
279:
107:
106:
103:
102:
59:Ortholog search:
2584:
2534:
2524:
2491:
2473:
2440:
2403:
2393:
2383:
2366:(16): 6196–201.
2350:
2340:
2330:
2297:
2264:(7062): 1173–8.
2251:
2241:
2231:
2198:
2188:
2155:
2145:
2120:
2110:
2077:
2067:
2042:
2032:
1990:
1989:
1979:
1939:
1933:
1932:
1922:
1890:
1884:
1883:
1850:(7062): 1173–8.
1838:
1832:
1831:
1821:
1789:
1783:
1782:
1764:
1736:
1727:
1726:
1716:
1692:
1677:
1676:
1666:
1634:
1628:
1627:
1617:
1581:
1575:
1574:
1564:
1532:
1517:
1516:
1506:
1482:
1476:
1475:
1447:
1441:
1440:
1433:
1424:
1423:
1413:
1389:
1380:
1379:
1368:
1362:
1361:
1350:
1344:
1334:
1323:
1313:
1139:
1138:
1110:
1103:
1086:
1038:
1029:
1009:
1005:RefSeq (protein)
995:
971:
962:
938:
914:
895:
869:
850:
824:
805:
774:
677:contractile ring
599:
585:
576:
569:
554:
526:genital tubercle
522:ventricular zone
506:secondary oocyte
494:
492:Top expressed in
487:
440:ventricular zone
432:
430:Top expressed in
425:
404:
387:
377:
364:
353:
338:
331:
325:
314:
302:
286:
276:
263:
252:
237:
230:
224:
213:
199:
183:
177:
175:PRC1 - orthologs
128:
121:
98:
73:
67:
46:
45:
40:
28:
2592:
2591:
2587:
2586:
2585:
2583:
2582:
2581:
2567:
2566:
2542:
2537:
2494:
2443:
2421:10.1038/nbt1240
2415:(10): 1285–92.
2409:Nat. Biotechnol
2406:
2353:
2300:
2254:
2201:
2165:Mol. Biol. Cell
2158:
2136:(58): 9336–47.
2123:
2093:(16): 3237–48.
2080:
2045:
2002:
1998:
1996:Further reading
1993:
1941:
1940:
1936:
1899:Mol. Biol. Cell
1892:
1891:
1887:
1840:
1839:
1835:
1791:
1790:
1786:
1753:10.1038/ncb2482
1738:
1737:
1730:
1694:
1693:
1680:
1643:Mol. Biol. Cell
1636:
1635:
1631:
1600:(17): 1569–75.
1583:
1582:
1578:
1534:
1533:
1520:
1484:
1483:
1479:
1449:
1448:
1444:
1435:
1434:
1427:
1391:
1390:
1383:
1370:
1369:
1365:
1352:
1351:
1347:
1335:
1326:
1314:
1305:
1301:
1285:phosphorylation
1253:
1240:
1223:
1211:D. melanogaster
1187:
1148:View/Edit Mouse
1143:View/Edit Human
1106:
1099:
1096:Location (UCSC)
1082:
1078:
1074:
1070:
1066:
1060:
1056:
1052:
1048:
1044:
1025:
1021:
1017:
991:
987:
983:
979:
958:
954:
950:
946:
859:ENSG00000198901
752:
701:
640:
636:protein binding
626:kinesin binding
581:
550:
541:
536:
532:
528:
524:
520:
516:
512:
508:
504:
490:
479:
474:
470:
466:
462:
458:
454:
450:
446:
442:
428:
372:
359:
351:
341:
340:
339:
332:
312:
289:Gene location (
271:
258:
250:
240:
239:
238:
231:
209:
186:Gene location (
137:
124:
117:
82:
60:
26:
17:
12:
11:
5:
2590:
2588:
2580:
2579:
2569:
2568:
2565:
2564:
2541:
2540:External links
2538:
2536:
2535:
2492:
2441:
2404:
2351:
2313:(14): 5391–6.
2298:
2252:
2199:
2171:(3): 1043–55.
2156:
2121:
2078:
2043:
2015:(7): 1175–86.
1999:
1997:
1994:
1992:
1991:
1934:
1905:(4): 2702–11.
1885:
1833:
1784:
1741:Nat. Cell Biol
1728:
1678:
1649:(6): 1024–34.
1629:
1576:
1518:
1477:
1442:
1425:
1381:
1363:
1345:
1324:
1302:
1300:
1297:
1296:
1295:
1292:
1278:
1271:
1268:
1257:
1252:
1249:
1239:
1236:
1222:
1219:
1186:
1183:
1155:
1154:
1151:
1150:
1145:
1135:
1134:
1128:
1127:
1124:
1123:
1121:
1119:
1112:
1111:
1104:
1097:
1093:
1092:
1089:
1088:
1062:
1061:
1035:
1032:
1031:
1013:
1012:
1006:
1002:
1001:
998:
997:
975:
974:
968:
965:
964:
942:
941:
935:
931:
930:
927:
926:
918:
917:
911:
908:
907:
899:
898:
892:
886:
885:
882:
881:
873:
872:
866:
863:
862:
854:
853:
847:
841:
840:
837:
836:
828:
827:
821:
818:
817:
809:
808:
802:
796:
795:
790:
785:
781:
780:
770:
769:
766:
765:
754:
753:
751:
750:
745:
740:
735:
730:
725:
720:
715:
709:
707:
703:
702:
700:
699:
694:
689:
684:
679:
674:
669:
664:
659:
654:
648:
646:
642:
641:
639:
638:
633:
628:
623:
618:
612:
610:
606:
605:
595:
594:
591:
590:
587:
586:
578:
577:
566:
560:
559:
556:
555:
547:
546:
543:
542:
540:
539:
535:
531:
527:
523:
519:
518:tail of embryo
515:
511:
507:
503:
502:primary oocyte
499:
496:
495:
484:
481:
480:
478:
477:
473:
469:
465:
461:
457:
453:
449:
445:
441:
437:
434:
433:
421:
420:
412:
401:
395:
394:
391:RNA expression
383:
382:
379:
378:
370:
366:
365:
357:
354:
349:
343:
342:
333:
326:
320:
316:
315:
310:
304:
303:
295:
294:
282:
281:
278:
277:
269:
265:
264:
256:
253:
248:
242:
241:
232:
225:
219:
215:
214:
207:
201:
200:
192:
191:
179:
178:
135:
131:
130:
122:
114:
113:
109:
108:
105:
104:
101:
100:
78:
77:
69:
68:
57:
51:
50:
42:
41:
33:
32:
15:
13:
10:
9:
6:
4:
3:
2:
2589:
2578:
2575:
2574:
2572:
2562:
2558:
2557:
2552:
2548:
2544:
2543:
2539:
2532:
2528:
2523:
2518:
2514:
2510:
2507:(3): 433–43.
2506:
2502:
2498:
2493:
2489:
2485:
2481:
2477:
2472:
2467:
2463:
2459:
2456:(2): 174–81.
2455:
2451:
2447:
2442:
2438:
2434:
2430:
2426:
2422:
2418:
2414:
2410:
2405:
2401:
2397:
2392:
2387:
2382:
2377:
2373:
2369:
2365:
2361:
2357:
2352:
2348:
2344:
2339:
2334:
2329:
2324:
2320:
2316:
2312:
2308:
2304:
2299:
2295:
2291:
2287:
2283:
2279:
2275:
2271:
2267:
2263:
2259:
2253:
2249:
2245:
2240:
2235:
2230:
2225:
2221:
2217:
2213:
2209:
2205:
2200:
2196:
2192:
2187:
2182:
2178:
2174:
2170:
2166:
2162:
2157:
2153:
2149:
2144:
2139:
2135:
2131:
2127:
2122:
2118:
2114:
2109:
2104:
2100:
2096:
2092:
2088:
2084:
2079:
2075:
2071:
2066:
2061:
2057:
2053:
2052:J. Biol. Chem
2049:
2044:
2040:
2036:
2031:
2026:
2022:
2018:
2014:
2010:
2006:
2001:
2000:
1995:
1987:
1983:
1978:
1973:
1969:
1965:
1961:
1957:
1953:
1949:
1945:
1938:
1935:
1930:
1926:
1921:
1916:
1912:
1908:
1904:
1900:
1896:
1889:
1886:
1881:
1877:
1873:
1869:
1865:
1861:
1857:
1853:
1849:
1845:
1837:
1834:
1829:
1825:
1820:
1815:
1811:
1807:
1804:(3): 433–43.
1803:
1799:
1795:
1788:
1785:
1780:
1776:
1772:
1768:
1763:
1758:
1754:
1750:
1746:
1742:
1735:
1733:
1729:
1724:
1720:
1715:
1710:
1707:(2): 118–20.
1706:
1702:
1698:
1691:
1689:
1687:
1685:
1683:
1679:
1674:
1670:
1665:
1660:
1656:
1652:
1648:
1644:
1640:
1633:
1630:
1625:
1621:
1616:
1611:
1607:
1603:
1599:
1595:
1591:
1589:
1580:
1577:
1572:
1568:
1563:
1558:
1554:
1550:
1547:(2): 377–90.
1546:
1542:
1538:
1531:
1529:
1527:
1525:
1523:
1519:
1514:
1510:
1505:
1500:
1497:(3): 420–32.
1496:
1492:
1488:
1481:
1478:
1473:
1469:
1465:
1461:
1457:
1453:
1446:
1443:
1438:
1432:
1430:
1426:
1421:
1417:
1412:
1407:
1404:(6): 877–85.
1403:
1399:
1395:
1388:
1386:
1382:
1377:
1373:
1367:
1364:
1359:
1355:
1349:
1346:
1342:
1338:
1333:
1331:
1329:
1325:
1321:
1317:
1312:
1310:
1308:
1304:
1298:
1293:
1290:
1286:
1282:
1279:
1276:
1272:
1269:
1266:
1262:
1258:
1255:
1254:
1250:
1248:
1244:
1237:
1235:
1233:
1228:
1220:
1218:
1216:
1212:
1208:
1202:
1200:
1196:
1192:
1184:
1182:
1180:
1176:
1173:
1169:
1165:
1161:
1149:
1144:
1140:
1136:
1133:
1129:
1122:
1120:
1117:
1113:
1109:
1105:
1102:
1098:
1094:
1087:
1085:
1081:
1077:
1073:
1069:
1063:
1059:
1055:
1051:
1047:
1043:
1039:
1036:
1030:
1028:
1024:
1020:
1014:
1010:
1007:
1003:
996:
994:
990:
986:
982:
976:
972:
969:
963:
961:
957:
953:
949:
943:
939:
936:
934:RefSeq (mRNA)
932:
925:
924:
919:
915:
912:
906:
905:
900:
896:
893:
891:
887:
880:
879:
874:
870:
867:
861:
860:
855:
851:
848:
846:
842:
835:
834:
829:
825:
822:
816:
815:
810:
806:
803:
801:
797:
794:
791:
789:
786:
782:
779:
775:
771:
764:
760:
755:
749:
746:
744:
741:
739:
736:
734:
731:
729:
726:
724:
723:cell division
721:
719:
716:
714:
711:
710:
708:
705:
704:
698:
695:
693:
690:
688:
685:
683:
680:
678:
675:
673:
670:
668:
665:
663:
660:
658:
655:
653:
650:
649:
647:
644:
643:
637:
634:
632:
629:
627:
624:
622:
619:
617:
614:
613:
611:
608:
607:
604:
603:Gene ontology
600:
596:
584:
579:
575:
570:
567:
565:
561:
553:
548:
537:
533:
529:
525:
521:
517:
513:
509:
505:
501:
500:
497:
493:
488:
485:
475:
471:
467:
463:
459:
455:
451:
447:
443:
439:
438:
435:
431:
426:
423:
422:
419:
417:
413:
411:
410:
406:
405:
402:
400:
396:
392:
388:
384:
376:
371:
367:
363:
358:
348:
344:
337:
330:
324:
317:
309:
305:
301:
296:
292:
287:
283:
275:
270:
266:
262:
257:
247:
243:
236:
229:
223:
216:
212:
206:
202:
198:
193:
189:
184:
180:
176:
172:
168:
164:
160:
156:
152:
148:
144:
140:
132:
127:
120:
115:
110:
99:
97:
93:
89:
85:
79:
74:
71:
70:
66:
63:
56:
52:
47:
43:
39:
34:
29:
24:
19:
2554:
2504:
2500:
2453:
2449:
2412:
2408:
2363:
2359:
2310:
2306:
2261:
2257:
2214:(2): 343–8.
2211:
2207:
2168:
2164:
2133:
2129:
2090:
2086:
2055:
2051:
2012:
2009:J. Cell Biol
2008:
1951:
1947:
1937:
1902:
1898:
1888:
1847:
1843:
1836:
1801:
1797:
1787:
1747:(5): 440–7.
1744:
1740:
1704:
1700:
1646:
1642:
1632:
1597:
1593:
1587:
1579:
1544:
1540:
1494:
1490:
1480:
1455:
1451:
1445:
1401:
1397:
1375:
1366:
1357:
1348:
1251:Interactions
1245:
1241:
1224:
1203:
1188:
1171:
1163:
1159:
1158:
1084:NP_001369319
1080:NP_001369318
1076:NP_001369317
1072:NP_001369316
1068:NP_001369315
1065:
1058:NP_001369314
1054:NP_001361553
1046:NP_001272927
1042:NP_001272926
1019:NP_001254509
1016:
993:NM_001374624
985:NM_001285998
981:NM_001285997
978:
948:NM_001267580
945:
921:
902:
876:
857:
831:
812:
792:
787:
682:cytoskeleton
662:spindle pole
510:spermatocyte
460:right testis
414:
407:
134:External IDs
81:
18:
1762:11336/20338
1179:cytokinesis
743:cytokinesis
667:microtubule
464:bone marrow
456:left testis
373:80,316,259
360:80,294,450
272:90,995,629
259:90,966,040
112:Identifiers
2450:Cancer Sci
1594:Curr. Biol
1588:Drosophila
1586:"Feo, the
1458:: 543–66.
1343:, May 2017
1322:, May 2017
1299:References
1289:C-terminus
1207:C. elegans
1195:interphase
718:cell cycle
418:(ortholog)
155:HomoloGene
1701:Dev. Cell
1398:Mol. Cell
1221:Structure
1209:, Feo in
1050:NP_660132
1027:NP_955445
1023:NP_003972
989:NM_145150
960:NM_199414
956:NM_199413
952:NM_003981
778:Orthologs
697:cytoplasm
163:GeneCards
2571:Category
2531:20691902
2480:17233835
2471:11159940
2437:14294292
2429:16964243
2400:16603632
2347:16565220
2286:16189514
2248:15625105
2195:15616196
2152:15531928
2130:Oncogene
2117:15297875
2074:14744859
2039:12082078
1986:26088160
1954:: 7290.
1929:22621898
1872:16189514
1828:20691902
1771:22552143
1723:23906062
1673:22278743
1624:15341744
1571:23870126
1513:20691901
1472:16756502
1339:–
1318:–
1261:interact
1227:spectrin
1217:(MAPs).
1185:Function
1132:Wikidata
757:Sources:
452:testicle
2561:PDBe-KB
2551:UniProt
2522:2966277
2488:1635584
2391:1458854
2368:Bibcode
2338:1459365
2315:Bibcode
2294:4427026
2266:Bibcode
2216:Bibcode
2030:2173564
1977:4557309
1956:Bibcode
1920:3395659
1880:4427026
1852:Bibcode
1819:2966277
1779:3355851
1664:3302730
1602:Bibcode
1562:3761943
1420:9885575
1341:Ensembl
1320:Ensembl
1193:during
1191:nucleus
1168:protein
1166:) is a
890:UniProt
845:Ensembl
784:Species
763:QuickGO
692:midbody
687:nucleus
657:cytosol
652:spindle
393:pattern
251:15q26.1
151:1858961
119:Aliases
2556:O43663
2529:
2519:
2486:
2478:
2468:
2435:
2427:
2398:
2388:
2345:
2335:
2292:
2284:
2258:Nature
2246:
2239:544298
2236:
2193:
2186:551472
2183:
2150:
2115:
2108:514520
2105:
2087:EMBO J
2072:
2037:
2027:
1984:
1974:
1927:
1917:
1878:
1870:
1844:Nature
1826:
1816:
1777:
1769:
1721:
1671:
1661:
1622:
1569:
1559:
1511:
1470:
1418:
1265:TRIM37
1118:search
1116:PubMed
923:Q99K43
904:O43663
833:233406
800:Entrez
564:BioGPS
534:morula
530:morula
514:zygote
143:603484
2484:S2CID
2433:S2CID
2290:S2CID
1876:S2CID
1775:S2CID
1263:with
1232:actin
793:Mouse
788:Human
759:Amigo
448:gonad
416:Mouse
409:Human
356:Start
291:Mouse
255:Start
188:Human
159:37868
2549:for
2527:PMID
2501:Cell
2476:PMID
2425:PMID
2396:PMID
2343:PMID
2282:PMID
2244:PMID
2191:PMID
2148:PMID
2113:PMID
2070:PMID
2035:PMID
1982:PMID
1925:PMID
1868:PMID
1824:PMID
1798:Cell
1767:PMID
1719:PMID
1669:PMID
1620:PMID
1567:PMID
1541:Cell
1509:PMID
1491:Cell
1468:PMID
1416:PMID
1281:PLK1
1275:CDK1
1175:gene
1172:PRC1
1164:PRC1
814:9055
399:Bgee
347:Band
308:Chr.
246:Band
205:Chr.
167:PRC1
139:OMIM
126:PRC1
96:4L6Y
92:4L3I
88:3NRY
84:3NRX
65:RCSB
62:PDBe
31:PRC1
2547:PDB
2517:PMC
2509:doi
2505:142
2466:PMC
2458:doi
2417:doi
2386:PMC
2376:doi
2364:103
2333:PMC
2323:doi
2311:103
2274:doi
2262:437
2234:PMC
2224:doi
2212:102
2181:PMC
2173:doi
2138:doi
2103:PMC
2095:doi
2060:doi
2056:279
2025:PMC
2017:doi
2013:157
1972:PMC
1964:doi
1915:PMC
1907:doi
1860:doi
1848:437
1814:PMC
1806:doi
1802:142
1757:hdl
1749:doi
1709:doi
1659:PMC
1651:doi
1610:doi
1557:PMC
1549:doi
1545:154
1499:doi
1495:142
1460:doi
1406:doi
369:End
268:End
171:OMA
147:MGI
55:PDB
2573::
2553::
2525:.
2515:.
2503:.
2499:.
2482:.
2474:.
2464:.
2454:98
2452:.
2448:.
2431:.
2423:.
2413:24
2411:.
2394:.
2384:.
2374:.
2362:.
2358:.
2341:.
2331:.
2321:.
2309:.
2305:.
2288:.
2280:.
2272:.
2260:.
2242:.
2232:.
2222:.
2210:.
2206:.
2189:.
2179:.
2169:16
2167:.
2163:.
2146:.
2134:23
2132:.
2128:.
2111:.
2101:.
2091:23
2089:.
2085:.
2068:.
2054:.
2050:.
2033:.
2023:.
2011:.
2007:.
1980:.
1970:.
1962:.
1950:.
1946:.
1923:.
1913:.
1903:23
1901:.
1897:.
1874:.
1866:.
1858:.
1846:.
1822:.
1812:.
1800:.
1796:.
1773:.
1765:.
1755:.
1745:14
1743:.
1731:^
1717:.
1705:26
1703:.
1699:.
1681:^
1667:.
1657:.
1647:23
1645:.
1641:.
1618:.
1608:.
1598:14
1596:.
1592:.
1565:.
1555:.
1543:.
1539:.
1521:^
1507:.
1493:.
1489:.
1466:.
1456:75
1454:.
1428:^
1414:.
1400:.
1396:.
1384:^
1374:.
1356:.
1327:^
1306:^
1234:.
1181:.
761:/
375:bp
362:bp
274:bp
261:bp
169:;
165::
161:;
157::
153:;
149::
145:;
141::
94:,
90:,
86:,
2563:.
2533:.
2511::
2490:.
2460::
2439:.
2419::
2402:.
2378::
2370::
2349:.
2325::
2317::
2296:.
2276::
2268::
2250:.
2226::
2218::
2197:.
2175::
2154:.
2140::
2119:.
2097::
2076:.
2062::
2041:.
2019::
1988:.
1966::
1958::
1952:6
1931:.
1909::
1882:.
1862::
1854::
1830:.
1808::
1781:.
1759::
1751::
1725:.
1711::
1675:.
1653::
1626:.
1612::
1604::
1573:.
1551::
1515:.
1501::
1474:.
1462::
1439:.
1422:.
1408::
1402:2
1378:.
1360:.
1277:.
1267:.
1162:(
293:)
190:)
173::
25:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.