1404:
WT, Chang YC, Gashi D, Beale E, Ozerov D, Nass K, Knopp G, Johnson PJ, Cirelli C, Milne C, Bacellar C, Sugahara M, Owada S, Joti Y, Yamashita A, Tanaka R, Tanaka T, Luo F, Tono K, Zarzycka W, Müller P, Alahmad MA, Bezold F, Fuchs V, Gnau P, Kiontke S, Korf L, Reithofer V, Rosner CJ, Seiler EM, Watad M, Werel L, Spadaccini R, Yamamoto J, Iwata S, Zhong D, Standfuss J, Royant A, Bessho Y, Essen LO, Tsai MD (December 2023). "Visualizing the DNA repair process by a photolyase at atomic resolution".
2587:
290:
42:
2003:
557:
bridge. Photolyases have a high affinity for these lesions and reversibly bind and convert them back to the original bases. The photolyase-catalyzed DNA repair process by which cyclobutane pyrimidine dimers are resolved has been studied by time-resolved crystallography and computational analysis to
1403:
Maestre-Reyna M, Wang PH, Nango E, Hosokawa Y, Saft M, Furrer A, Yang CH, Gusti Ngurah Putu EP, Wu WJ, Emmerich HJ, Caramello N, Franz-Badur S, Yang C, Engilberge S, Wranik M, Glover HL, Weinert T, Wu HY, Lee CC, Huang WC, Huang KF, Chang YK, Liao JH, Weng JH, Gad W, Chang CW, Pang AH, Yang KC, Lin
954:
The non-class 2 branch of CPDs tend to be grouped into class 1 in some systems such as PRINTS (PR00147). Although the members of the smaller groups are agreed upon, the phylogeny can vary greatly among authors due to differences in methodology, leading to some confusion with authors who try to fit
520:(from the violet/blue end of the spectrum) both for their own activation and for the actual DNA repair. The DNA repair mechanism involving photolyases is called photoreactivation. They mainly convert pyrimidine dimers into a normal pair of pyrimidine bases. Photo reactivation, the first
552:
linked. The bond length of this dimerization is shorter than the bond length of normal B-DNA structure which produces an incorrect template for replication and transcription. The more common covalent linkage involves the formation of a
1085:
Yamamoto J, Shimizu K, Kanda T, Hosokawa Y, Iwai S, Plaza P, Müller P (October 2017). "Loss of Fourth
Electron-Transferring Tryptophan in Animal (6-4) Photolyase Impairs DNA Repair Activity in Bacterial Cells".
586:. Photolyase is particularly important in repairing UV induced damage in plants. The photolyase mechanism is no longer working in humans and other placental mammals who instead rely on the less efficient
926:, four taxa initially thought to carry this family of lyases. The categorization has since changed. The "Cry" part of their name was due to initial assumptions that they were cryptochromes.
2035:
459:
159:
859:
Class 1 CPD photolyases are enzymes that process cyclobutane pyrimidine dimer (CPD) lesions from Gram-negative and Gram-positive bacteria, as well as the halophilic
884:
The plant and fungi cryptochromes are similar to Class 1 CPDs. They are blue light photoreceptors that mediate blue light-induced gene expression and modulation of
641:. Although only FAD is required for catalytic activity, the second cofactor significantly accelerates reaction rate in low-light conditions. The enzyme acts by
478:
1607:
Jaikumar NS, Dorn KM, Baas D, Wilke B, Kapp C, Snapp SS (December 2020). "Nucleic acid damage and DNA repair are affected by freezing stress in annual wheat (
1694:"The class III cyclobutane pyrimidine dimer photolyase structure reveals a new antenna chromophore binding site and alternative photoreduction pathways"
2130:
2154:
2028:
1901:
Eker AP, Fichtinger-Schepman AM (1975). "Studies on a DNA photoreactivating enzyme from
Streptomyces griseus II. Purification of the enzyme".
1379:
107:
2237:
2617:
2135:
2085:
298:
which is repaired by DNA photolyase. Note: The above diagram is incorrectly labelled as thymine as the structures lack 5-methyl groups.
2159:
2021:
1335:
Friedberg EC (September 2015). "A history of the DNA repair and mutagenesis field: I. The discovery of enzymatic photoreactivation".
2306:
2120:
1840:"Nuclear and cell membrane effects contribute independently to the induction of apoptosis in human cells exposed to UVB radiation"
2150:
2075:
618:
471:
524:
mechanism to be discovered, was described initially by Albert Kelner in 1949 and independently by Renato
Dulbecco also in 1949.
2125:
2182:
422:
398:
2462:
179:
645:
in which the reduced flavin FADH is activated by light energy and acts as an electron donor to break the pyrimidine dimer.
2204:
891:
Class 3 CPD lyases make up a sister group to the plant cryptochromes, which in turn are a sister group to class 1 CPDs.
2577:
2115:
2070:
933:
form a group with animal cryptochromes that control circadian rhythms. They are found in diverse species including
587:
2447:
2563:
2550:
2537:
2524:
2511:
2498:
2485:
2247:
2219:
2169:
2105:
2058:
2457:
416:
2411:
2354:
2271:
2049:
499:
309:
167:
1650:
Sancar A (June 2003). "Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors".
403:
2359:
2110:
2095:
2090:
959:
group including photolyases that have lost their DNA repair activity and instead control circadian rhythms.
614:
2622:
2140:
2080:
864:
602:
2007:
483:
2380:
2299:
391:
48:
2452:
163:
326:
2100:
1965:
Setlow JK, Bollum FJ (1968). "The minimum size of the substrate for yeast photoreactivating enzyme".
1851:
1512:
1413:
1252:
1197:
1134:
873:
120:
2612:
2416:
1932:"Action mechanism of Escherichia coli DNA photolyase. I. Formation of the enzyme-substrate complex"
1243:
Dulbecco R (June 1949). "Reactivation of ultra-violet-inactivated bacteriophage by visible light".
930:
419:
321:
1501:"A cryptochrome/photolyase class of enzymes with single-stranded DNA-specific photolyase activity"
343:
2607:
2349:
2209:
1123:"Kinetics of cyclobutane thymine dimer splitting by DNA photolyase directly monitored in the UV"
871:
Class 2 CPD photolyases also process CPD lesions. They are found in plants like the thale cress
625:; they are divided into two main classes based on the second cofactor, which may be either the
1982:
1953:
1918:
1879:
1820:
1781:
1725:
1667:
1632:
1589:
1540:
1478:
1429:
1385:
1375:
1352:
1317:
1268:
1225:
1162:
1103:
642:
410:
295:
186:
154:
1692:
Scheerer P, Zhang F, Kalms J, von
Stetten D, Krauß N, Oberpichler I, Lamparter T (May 2015).
2395:
2390:
2364:
2292:
2232:
1974:
1943:
1910:
1869:
1859:
1812:
1771:
1763:
1715:
1705:
1659:
1624:
1579:
1571:
1530:
1520:
1468:
1460:
1421:
1344:
1307:
1299:
1260:
1215:
1205:
1152:
1142:
1095:
968:
885:
537:
1288:"Experiments on photoreactivation of bacteriophages inactivated with ultraviolet radiation"
894:
The Cry-DASH group are CPD lyases highly specific for single-stranded DNA. Members include
379:
146:
2442:
2426:
2339:
1752:"Blue-light-receptive cryptochrome is expressed in a sponge eye lacking neurons and opsin"
1750:
Rivera AS, Ozturk N, Fahey B, Plachetzki DC, Degnan BM, Sancar A, Oakley TH (April 2012).
1560:"Evolution of mutation rates: phylogenomic analysis of the photolyase/cryptochrome family"
1002:
896:
648:
On the basis of sequence similarities DNA photolyases can be grouped into a few classes:
355:
1855:
1516:
1417:
1256:
1201:
1138:
950:), also known as the FeS-BCP group, form their own outgroup relative to all photolyases.
502:
314:
2591:
2480:
2421:
2062:
1776:
1751:
1720:
1693:
1584:
1559:
1535:
1500:
1473:
1448:
1220:
1181:
1157:
1122:
955:
everything (sparing FeS-BCP) into a two-class classification. The cryptochromes form a
902:
454:
1948:
1931:
1312:
1287:
434:
2601:
2385:
2344:
2256:
1978:
1914:
1874:
1839:
567:
549:
517:
429:
112:
76:
2334:
956:
610:
596:
591:
142:
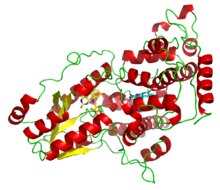
52:, illustrating the two light-harvesting cofactors: FADH (yellow) and 8-HDF (cyan).
1348:
1303:
946:
88:
17:
2558:
2493:
2329:
2013:
1099:
634:
554:
513:
438:
294:
A UV radiation induced thymine-thymine cyclobutane dimer (right) is the type of
289:
2586:
621:
that binds a second cofactor. All photolyases contain the two-electron-reduced
2197:
2192:
2187:
1838:
Kulms D, Pöppelmann B, Yarosh D, Luger TA, Krutmann J, Schwarz T (July 1999).
1449:"The native cyclobutane pyrimidine dimer photolyase of rice is phosphorylated"
521:
506:
41:
1389:
2532:
2506:
2266:
1864:
1710:
1575:
1525:
1425:
1147:
1883:
1824:
1785:
1729:
1671:
1636:
1593:
1544:
1482:
1433:
1356:
1321:
1272:
1229:
1166:
1107:
2002:
1986:
1957:
1464:
116:
2173:
1922:
1210:
941:
571:
545:
83:
367:
2223:
2145:
1816:
1767:
860:
541:
386:
100:
95:
1663:
1628:
2545:
2315:
1264:
626:
583:
579:
570:
old enzyme which is present and functional in many species, from the
509:
466:
362:
350:
338:
174:
606:
is accompanied by large increases in expression of DNA photolyases.
266:
259:
253:
247:
241:
235:
229:
223:
217:
211:
204:
198:
192:
1121:
Thiagarajan V, Byrdin M, Eker AP, Müller P, Brettel K (June 2011).
2519:
2045:
1066:
575:
1069:, specifically in the "catch-all" class of carbon-carbon lyases.
1447:
eranishi M, Nakamura K, Morioka H, Yamamoto K, Hidema J (2008).
991:
987:
975:
878:
622:
374:
136:
71:
2288:
2017:
1930:
Sancar GB, Smith FW, Reid R, Payne G, Levy M, Sancar A (1987).
937:
and humans. The cryptochromes have their own detailed grouping.
533:
1799:
McCready S, Marcello L (June 2003). "Repair of UV damage in
1611:) and by plant age and freezing in its perennial relative (
2284:
1063:
deoxyribonucleate pyrimidine dimer lyase (photosensitive)
978:
cells partially reduced DNA damage from UVB exposure.
637:
8-hydroxy-7,8-didemethyl-5-deazariboflavin (8-HDF) in
2575:
1051:
deoxyribonucleic cyclobutane dipyrimidine photolyase
2471:
2435:
2404:
2373:
2322:
2246:
2218:
2168:
2057:
477:
465:
453:
448:
428:
409:
397:
385:
373:
361:
349:
337:
332:
320:
308:
303:
282:
185:
173:
153:
135:
130:
106:
94:
82:
70:
62:
57:
34:
1494:
1492:
35:Cryptochrome/photolyase, C-terminal, FAD binding
1007:deoxyribocyclobutadipyrimidine pyrimidine-lyase
2300:
2029:
1745:
1743:
1741:
1739:
8:
1182:"Effect of Visible Light on the Recovery of
558:allow atomic visualization of the process.
2307:
2293:
2285:
2036:
2022:
2014:
445:
288:
127:
40:
1947:
1873:
1863:
1775:
1719:
1709:
1687:
1685:
1683:
1681:
1583:
1534:
1524:
1472:
1311:
1219:
1209:
1156:
1146:
512:that repair damage caused by exposure to
2131:Phosphoribosylaminoimidazole carboxylase
1059:dipyrimidine photolyase (photosensitive)
2582:
1077:
1065:. This enzyme belongs to the family of
1023:DNA cyclobutane dipyrimidine photolyase
548:bases on the same strand of DNA become
283:deoxyribodipyrimidine photo-lyase (CPD)
652:Cryptochrome/photolyase family (2015)
594:. Freezing stress in the annual wheat
279:
31:
1186:from Ultra-violet Irradiation Injury"
982:Human proteins containing this domain
7:
2238:3-hydroxy-3-methylglutaryl-CoA lyase
2155:Orotidine 5'-phosphate decarboxylase
1558:Lucas-Lledó JI, Lynch M (May 2009).
1499:Selby CP, Sancar A (November 2006).
1009:. Other names in common use include
588:nucleotide excision repair mechanism
2136:Pyrophosphomevalonate decarboxylase
2086:Aromatic L-amino acid decarboxylase
1756:The Journal of Experimental Biology
1698:The Journal of Biological Chemistry
795:
725:
705:
698:
678:
658:
629:methenyltetrahydrofolate (MTHF) in
536:strands and break certain types of
2160:Uroporphyrinogen III decarboxylase
25:
2121:Phosphoenolpyruvate carboxykinase
1374:. Brooks/Cole, Cengage Learning.
910:. DASH was initially named after
613:and contain two light-harvesting
2585:
2151:Uridine monophosphate synthetase
2076:Adenosylmethionine decarboxylase
2001:
1035:deoxyribodipyrimidine photolyase
2126:Phosphoenolpyruvate carboxylase
1564:Molecular Biology and Evolution
1370:Garrett RH, Grisham CM (2010).
759:
590:, although they do retain many
532:Photolyases bind complementary
2183:Fructose-bisphosphate aldolase
650:
600:and in its perennial relative
516:light. These enzymes require
46:A deazaflavin photolyase from
1:
1949:10.1016/S0021-9258(19)75952-3
131:Available protein structures:
2205:2-hydroxyphytanoyl-CoA lyase
1979:10.1016/0005-2787(68)90077-4
1915:10.1016/0005-2787(75)90136-7
1349:10.1016/j.dnarep.2015.06.007
1304:10.1128/jb.59.3.329-347.1950
1184:Streptomyces Griseus Conidia
1019:DNA-photoreactivating enzyme
617:. Many photolyases have an
1100:10.1021/acs.biochem.7b00366
1031:deoxyribonucleic photolyase
815:Eukaryotic 6-4; Animal Cry
2639:
2618:Enzymes of known structure
2116:Oxaloacetate decarboxylase
2071:Acetoacetate decarboxylase
1180:Kelner A (February 1949).
540:that arise when a pair of
2463:Michaelis–Menten kinetics
2106:Malonyl-CoA decarboxylase
1286:Dulbecco R (March 1950).
967:Adding photolyase from a
813:
800:
793:
756:
743:
730:
723:
710:
703:
696:
683:
676:
663:
444:
287:
126:
39:
2355:Diffusion-limited enzyme
2272:Spore photoproduct lyase
1844:Proc Natl Acad Sci U S A
1505:Proc Natl Acad Sci U S A
1190:Proc Natl Acad Sci U S A
1127:Proc Natl Acad Sci U S A
1011:photoreactivating enzyme
1005:of this enzyme class is
2111:Ornithine decarboxylase
2096:Histidine decarboxylase
2091:Glutamate decarboxylase
1865:10.1073/pnas.96.14.7974
1801:Halobacterium salinarum
1711:10.1074/jbc.M115.637868
1526:10.1073/pnas.0607993103
1426:10.1126/science.add7795
1148:10.1073/pnas.1101026108
639:deazaflavin photolyases
2141:Pyruvate decarboxylase
2081:Arginine decarboxylase
1967:Biochim. Biophys. Acta
1903:Biochim. Biophys. Acta
1613:Thinopyrum intermedium
940:Bacterial 6-4 lyases (
865:Halobacterium halobium
603:Thinopyrum intermedium
2448:Eadie–Hofstee diagram
2381:Allosteric regulation
1576:10.1093/molbev/msp029
1465:10.1104/pp.107.110189
2458:Lineweaver–Burk plot
2101:Lysine decarboxylase
2010:at Wikimedia Commons
1211:10.1073/pnas.35.2.73
931:(6-4)DNA photolyases
908:Arabidopsis thaliana
874:Arabidopsis thaliana
1856:1999PNAS...96.7974K
1517:2006PNAS..10317696S
1418:2023Sci...382d7795M
1257:1949Natur.163..949D
1202:1949PNAS...35...73K
1139:2011PNAS..108.9402T
653:
2417:Enzyme superfamily
2350:Enzyme promiscuity
2210:Threonine aldolase
1817:10.1042/bst0310694
1768:10.1242/jeb.067140
1412:(6674): eadd7795.
972:Anacystis nidulans
906:, and AtCry3 from
651:
631:folate photolyases
49:Anacystis nidulans
2573:
2572:
2282:
2281:
2006:Media related to
1805:Biochem Soc Trans
1762:(Pt 8): 1278–86.
1664:10.1021/cr0204348
1629:10.1002/ajb2.1584
1623:(12): 1693–1709.
1609:Triticum aestivum
1511:(47): 17696–700.
1381:978-0-495-10935-8
1337:DNA Repair (Amst)
1251:(4155): 949–950.
886:circadian rhythms
856:
855:
851:
850:
842:
841:
833:
832:
824:
823:
782:
781:
773:
772:
643:electron transfer
619:N-terminal domain
597:Triticum aestivum
538:pyrimidine dimers
493:
492:
489:
488:
392:metabolic pathway
278:
277:
274:
273:
180:structure summary
18:Photoreactivation
16:(Redirected from
2630:
2590:
2589:
2581:
2453:Hanes–Woolf plot
2396:Enzyme activator
2391:Enzyme inhibitor
2365:Enzyme catalysis
2309:
2302:
2295:
2286:
2233:Isocitrate lyase
2038:
2031:
2024:
2015:
2005:
1990:
1961:
1951:
1926:
1888:
1887:
1877:
1867:
1835:
1829:
1828:
1796:
1790:
1789:
1779:
1747:
1734:
1733:
1723:
1713:
1704:(18): 11504–14.
1689:
1676:
1675:
1652:Chemical Reviews
1647:
1641:
1640:
1604:
1598:
1597:
1587:
1555:
1549:
1548:
1538:
1528:
1496:
1487:
1486:
1476:
1453:Plant Physiology
1444:
1438:
1437:
1400:
1394:
1393:
1367:
1361:
1360:
1332:
1326:
1325:
1315:
1283:
1277:
1276:
1265:10.1038/163949b0
1240:
1234:
1233:
1223:
1213:
1177:
1171:
1170:
1160:
1150:
1118:
1112:
1111:
1082:
1055:phr A photolyase
969:blue-green algae
796:
763:
726:
706:
699:
679:
659:
654:
609:Photolyases are
568:phylogenetically
566:Photolyase is a
446:
292:
280:
269:
262:
256:
250:
244:
238:
232:
226:
220:
214:
207:
201:
195:
128:
44:
32:
27:Class of enzymes
21:
2638:
2637:
2633:
2632:
2631:
2629:
2628:
2627:
2598:
2597:
2596:
2584:
2576:
2574:
2569:
2481:Oxidoreductases
2467:
2443:Enzyme kinetics
2431:
2427:List of enzymes
2400:
2369:
2340:Catalytic triad
2318:
2313:
2283:
2278:
2242:
2214:
2164:
2053:
2042:
1998:
1993:
1964:
1929:
1900:
1896:
1894:Further reading
1891:
1837:
1836:
1832:
1811:(Pt 3): 694–8.
1798:
1797:
1793:
1749:
1748:
1737:
1691:
1690:
1679:
1649:
1648:
1644:
1606:
1605:
1601:
1557:
1556:
1552:
1498:
1497:
1490:
1446:
1445:
1441:
1402:
1401:
1397:
1382:
1369:
1368:
1364:
1334:
1333:
1329:
1285:
1284:
1280:
1242:
1241:
1237:
1179:
1178:
1174:
1120:
1119:
1115:
1094:(40): 5356–64.
1084:
1083:
1079:
1075:
1047:PhrB photolyase
1003:systematic name
999:
986:Cryptochromes:
984:
965:
897:Vibrio cholerae
852:
843:
834:
825:
783:
774:
758:P. tricornutum
564:
530:
299:
265:
258:
252:
246:
240:
234:
228:
222:
216:
210:
203:
197:
191:
53:
28:
23:
22:
15:
12:
11:
5:
2636:
2634:
2626:
2625:
2620:
2615:
2610:
2600:
2599:
2595:
2594:
2571:
2570:
2568:
2567:
2554:
2541:
2528:
2515:
2502:
2489:
2475:
2473:
2469:
2468:
2466:
2465:
2460:
2455:
2450:
2445:
2439:
2437:
2433:
2432:
2430:
2429:
2424:
2419:
2414:
2408:
2406:
2405:Classification
2402:
2401:
2399:
2398:
2393:
2388:
2383:
2377:
2375:
2371:
2370:
2368:
2367:
2362:
2357:
2352:
2347:
2342:
2337:
2332:
2326:
2324:
2320:
2319:
2314:
2312:
2311:
2304:
2297:
2289:
2280:
2279:
2277:
2276:
2275:
2274:
2269:
2259:
2253:
2251:
2244:
2243:
2241:
2240:
2235:
2229:
2227:
2216:
2215:
2213:
2212:
2207:
2202:
2201:
2200:
2195:
2190:
2179:
2177:
2166:
2165:
2163:
2162:
2157:
2148:
2143:
2138:
2133:
2128:
2123:
2118:
2113:
2108:
2103:
2098:
2093:
2088:
2083:
2078:
2073:
2067:
2065:
2063:Carboxy-lyases
2055:
2054:
2044:Carbon–carbon
2043:
2041:
2040:
2033:
2026:
2018:
2012:
2011:
1997:
1996:External links
1994:
1992:
1991:
1962:
1927:
1897:
1895:
1892:
1890:
1889:
1850:(14): 7974–9.
1830:
1791:
1735:
1677:
1658:(6): 2203–37.
1642:
1599:
1570:(5): 1143–53.
1550:
1488:
1459:(4): 1941–51.
1439:
1395:
1380:
1362:
1327:
1278:
1235:
1172:
1133:(23): 9402–7.
1113:
1076:
1074:
1071:
1027:DNA photolyase
1015:DNA photolyase
998:
995:
983:
980:
964:
961:
952:
951:
938:
927:
903:Xenopus laevis
892:
889:
882:
869:
854:
853:
849:
848:
845:
844:
840:
839:
836:
835:
831:
830:
827:
826:
822:
821:
818:
817:
812:
809:
808:
805:
804:
799:
794:
792:
789:
788:
785:
784:
780:
779:
776:
775:
771:
770:
767:
766:
755:
752:
751:
748:
747:
742:
739:
738:
735:
734:
729:
724:
722:
719:
718:
715:
714:
709:
704:
702:
697:
695:
692:
691:
688:
687:
682:
677:
675:
672:
671:
668:
667:
662:
657:
563:
560:
529:
526:
491:
490:
487:
486:
481:
475:
474:
469:
463:
462:
457:
451:
450:
442:
441:
432:
426:
425:
414:
407:
406:
401:
395:
394:
389:
383:
382:
377:
371:
370:
365:
359:
358:
353:
347:
346:
341:
335:
334:
330:
329:
324:
318:
317:
312:
306:
305:
301:
300:
293:
285:
284:
276:
275:
272:
271:
189:
183:
182:
177:
171:
170:
157:
151:
150:
140:
133:
132:
124:
123:
110:
104:
103:
98:
92:
91:
86:
80:
79:
74:
68:
67:
64:
60:
59:
55:
54:
45:
37:
36:
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
2635:
2624:
2623:Flavoproteins
2621:
2619:
2616:
2614:
2611:
2609:
2606:
2605:
2603:
2593:
2588:
2583:
2579:
2565:
2561:
2560:
2555:
2552:
2548:
2547:
2542:
2539:
2535:
2534:
2529:
2526:
2522:
2521:
2516:
2513:
2509:
2508:
2503:
2500:
2496:
2495:
2490:
2487:
2483:
2482:
2477:
2476:
2474:
2470:
2464:
2461:
2459:
2456:
2454:
2451:
2449:
2446:
2444:
2441:
2440:
2438:
2434:
2428:
2425:
2423:
2422:Enzyme family
2420:
2418:
2415:
2413:
2410:
2409:
2407:
2403:
2397:
2394:
2392:
2389:
2387:
2386:Cooperativity
2384:
2382:
2379:
2378:
2376:
2372:
2366:
2363:
2361:
2358:
2356:
2353:
2351:
2348:
2346:
2345:Oxyanion hole
2343:
2341:
2338:
2336:
2333:
2331:
2328:
2327:
2325:
2321:
2317:
2310:
2305:
2303:
2298:
2296:
2291:
2290:
2287:
2273:
2270:
2268:
2265:
2264:
2263:
2260:
2258:
2257:Tryptophanase
2255:
2254:
2252:
2249:
2245:
2239:
2236:
2234:
2231:
2230:
2228:
2225:
2221:
2217:
2211:
2208:
2206:
2203:
2199:
2196:
2194:
2191:
2189:
2186:
2185:
2184:
2181:
2180:
2178:
2175:
2171:
2167:
2161:
2158:
2156:
2152:
2149:
2147:
2144:
2142:
2139:
2137:
2134:
2132:
2129:
2127:
2124:
2122:
2119:
2117:
2114:
2112:
2109:
2107:
2104:
2102:
2099:
2097:
2094:
2092:
2089:
2087:
2084:
2082:
2079:
2077:
2074:
2072:
2069:
2068:
2066:
2064:
2060:
2056:
2051:
2047:
2039:
2034:
2032:
2027:
2025:
2020:
2019:
2016:
2009:
2004:
2000:
1999:
1995:
1988:
1984:
1980:
1976:
1972:
1968:
1963:
1959:
1955:
1950:
1945:
1942:(1): 478–85.
1941:
1937:
1936:J. Biol. Chem
1933:
1928:
1924:
1920:
1916:
1912:
1908:
1904:
1899:
1898:
1893:
1885:
1881:
1876:
1871:
1866:
1861:
1857:
1853:
1849:
1845:
1841:
1834:
1831:
1826:
1822:
1818:
1814:
1810:
1806:
1802:
1795:
1792:
1787:
1783:
1778:
1773:
1769:
1765:
1761:
1757:
1753:
1746:
1744:
1742:
1740:
1736:
1731:
1727:
1722:
1717:
1712:
1707:
1703:
1699:
1695:
1688:
1686:
1684:
1682:
1678:
1673:
1669:
1665:
1661:
1657:
1653:
1646:
1643:
1638:
1634:
1630:
1626:
1622:
1618:
1614:
1610:
1603:
1600:
1595:
1591:
1586:
1581:
1577:
1573:
1569:
1565:
1561:
1554:
1551:
1546:
1542:
1537:
1532:
1527:
1522:
1518:
1514:
1510:
1506:
1502:
1495:
1493:
1489:
1484:
1480:
1475:
1470:
1466:
1462:
1458:
1454:
1450:
1443:
1440:
1435:
1431:
1427:
1423:
1419:
1415:
1411:
1407:
1399:
1396:
1391:
1387:
1383:
1377:
1373:
1366:
1363:
1358:
1354:
1350:
1346:
1342:
1338:
1331:
1328:
1323:
1319:
1314:
1309:
1305:
1301:
1298:(3): 329–47.
1297:
1293:
1289:
1282:
1279:
1274:
1270:
1266:
1262:
1258:
1254:
1250:
1246:
1239:
1236:
1231:
1227:
1222:
1217:
1212:
1207:
1203:
1199:
1195:
1191:
1187:
1185:
1176:
1173:
1168:
1164:
1159:
1154:
1149:
1144:
1140:
1136:
1132:
1128:
1124:
1117:
1114:
1109:
1105:
1101:
1097:
1093:
1089:
1081:
1078:
1072:
1070:
1068:
1064:
1060:
1056:
1052:
1048:
1044:
1040:
1036:
1032:
1028:
1024:
1020:
1016:
1012:
1008:
1004:
996:
994:
993:
989:
981:
979:
977:
973:
970:
962:
960:
958:
949:
948:
943:
939:
936:
932:
928:
925:
921:
920:Synechocystis
917:
913:
909:
905:
904:
900:, X1Cry from
899:
898:
893:
890:
887:
883:
880:
876:
875:
870:
867:
866:
862:
858:
857:
847:
846:
838:
837:
829:
828:
820:
819:
816:
811:
810:
807:
806:
803:
798:
797:
791:
790:
787:
786:
778:
777:
769:
768:
765:
762:
754:
753:
750:
749:
746:
741:
740:
737:
736:
733:
728:
727:
721:
720:
717:
716:
713:
708:
707:
701:
700:
694:
693:
690:
689:
686:
681:
680:
674:
673:
670:
669:
666:
661:
660:
656:
655:
649:
646:
644:
640:
636:
632:
628:
624:
620:
616:
612:
611:flavoproteins
607:
605:
604:
599:
598:
593:
592:cryptochromes
589:
585:
581:
577:
573:
569:
561:
559:
556:
551:
547:
543:
539:
535:
527:
525:
523:
519:
518:visible light
515:
511:
508:
504:
501:
497:
485:
482:
480:
476:
473:
470:
468:
464:
461:
458:
456:
452:
447:
443:
440:
436:
433:
431:
430:Gene Ontology
427:
424:
421:
418:
415:
412:
408:
405:
402:
400:
396:
393:
390:
388:
384:
381:
378:
376:
372:
369:
368:NiceZyme view
366:
364:
360:
357:
354:
352:
348:
345:
342:
340:
336:
331:
328:
325:
323:
319:
316:
313:
311:
307:
302:
297:
291:
286:
281:
268:
264:
261:
255:
249:
243:
237:
231:
225:
219:
213:
206:
200:
194:
190:
188:
184:
181:
178:
176:
172:
169:
165:
161:
158:
156:
152:
148:
144:
141:
138:
134:
129:
125:
122:
118:
114:
111:
109:
105:
102:
99:
97:
93:
90:
87:
85:
81:
78:
75:
73:
69:
66:FAD_binding_7
65:
61:
56:
51:
50:
43:
38:
33:
30:
19:
2559:Translocases
2556:
2543:
2530:
2517:
2504:
2494:Transferases
2491:
2478:
2335:Binding site
2261:
1973:(2): 233–7.
1970:
1966:
1939:
1935:
1909:(1): 54–63.
1906:
1902:
1847:
1843:
1833:
1808:
1804:
1800:
1794:
1759:
1755:
1701:
1697:
1655:
1651:
1645:
1620:
1616:
1612:
1608:
1602:
1567:
1563:
1553:
1508:
1504:
1456:
1452:
1442:
1409:
1405:
1398:
1372:Biochemistry
1371:
1365:
1340:
1336:
1330:
1295:
1291:
1281:
1248:
1244:
1238:
1193:
1189:
1183:
1175:
1130:
1126:
1116:
1091:
1088:Biochemistry
1087:
1080:
1062:
1058:
1054:
1050:
1046:
1042:
1038:
1034:
1030:
1026:
1022:
1018:
1014:
1010:
1006:
1000:
997:Nomenclature
985:
971:
966:
957:polyphyletic
953:
945:
934:
923:
919:
915:
911:
907:
901:
895:
872:
863:
814:
801:
760:
757:
744:
731:
711:
684:
664:
647:
638:
630:
608:
601:
595:
565:
531:
495:
494:
356:BRENDA entry
257:A:207-472
251:A:207-472
233:A:207-472
221:B:202-469
215:A:176-418
209:
202:A:214-492
196:A:214-492
47:
29:
2330:Active site
1292:J Bacteriol
1196:(2): 73–9.
963:Application
929:Eukaryotic
916:Arabidopsis
635:deazaflavin
582:and to the
555:cyclobutane
514:ultraviolet
496:Photolyases
344:IntEnz view
327:37290-70-3
304:Identifiers
239::207-472
58:Identifiers
2613:DNA repair
2602:Categories
2533:Isomerases
2507:Hydrolases
2374:Regulation
2262:Photolyase
2198:Aldolase C
2193:Aldolase B
2188:Aldolase A
2008:Photolyase
1073:References
1039:photolyase
935:Drosophila
912:Drosophila
745:Plant Cry
550:covalently
522:DNA repair
507:DNA repair
413:structures
380:KEGG entry
296:DNA damage
263:A:207-472
245:A:207-472
227:D:207-472
208:A:176-418
143:structures
2608:EC 4.1.99
2412:EC number
2267:CPD lyase
1390:984382855
1343:: 35–42.
947:IPR007357
802:Cry-DASH
732:CPD-3gre
615:cofactors
562:Evolution
333:Databases
270:B:213-453
101:PDOC00331
89:IPR005101
2436:Kinetics
2360:Cofactor
2323:Activity
2224:Oxo-acid
2174:Aldehyde
1884:10393932
1825:12773185
1786:22442365
1730:25784552
1672:12797829
1637:33340368
1617:Am J Bot
1594:19228922
1545:17062752
1483:18235036
1434:38033054
1357:26151545
1322:15436402
1273:18229246
1230:16588862
1167:21606324
1108:28880077
942:InterPro
877:and the
665:FeS-BCP
572:bacteria
546:cytosine
528:Function
503:4.1.99.3
484:proteins
472:articles
460:articles
417:RCSB PDB
315:4.1.99.3
160:RCSB PDB
84:InterPro
2592:Biology
2546:Ligases
2316:Enzymes
2250:: Other
2226:-lyases
2176:-lyases
2146:RuBisCO
1987:5649902
1958:3539939
1852:Bibcode
1777:3309880
1721:4416854
1585:2668831
1536:1621107
1513:Bibcode
1474:2287361
1414:Bibcode
1406:Science
1253:Bibcode
1221:1062964
1198:Bibcode
1158:3111307
1135:Bibcode
944::
861:archaea
633:or the
584:animals
574:to the
542:thymine
510:enzymes
439:QuickGO
404:profile
387:MetaCyc
322:CAS no.
96:PROSITE
77:PF03441
2578:Portal
2520:Lyases
2248:4.1.99
2046:lyases
1985:
1956:
1923:804322
1921:
1882:
1872:
1823:
1784:
1774:
1728:
1718:
1670:
1635:
1592:
1582:
1543:
1533:
1481:
1471:
1432:
1388:
1378:
1355:
1320:
1313:385765
1310:
1271:
1245:Nature
1228:
1218:
1165:
1155:
1106:
1067:lyases
1061:, and
922:, and
712:CPD-1
685:CPD-2
627:pterin
580:plants
505:) are
467:PubMed
449:Search
435:AmiGO
423:PDBsum
363:ExPASy
351:BRENDA
339:IntEnz
310:EC no.
175:PDBsum
149:
139:
121:SUPFAM
63:Symbol
2472:Types
2220:4.1.3
2170:4.1.2
2059:4.1.1
1875:22172
974:, to
924:Human
576:fungi
399:PRIAM
117:SCOPe
108:SCOP2
2564:list
2557:EC7
2551:list
2544:EC6
2538:list
2531:EC5
2525:list
2518:EC4
2512:list
2505:EC3
2499:list
2492:EC2
2486:list
2479:EC1
2052:4.1)
1983:PMID
1954:PMID
1919:PMID
1880:PMID
1821:PMID
1782:PMID
1726:PMID
1668:PMID
1633:PMID
1615:)".
1590:PMID
1541:PMID
1479:PMID
1430:PMID
1386:OCLC
1376:ISBN
1353:PMID
1318:PMID
1269:PMID
1226:PMID
1163:PMID
1104:PMID
1001:The
992:CRY2
988:CRY1
976:HeLa
879:rice
761:CryP
623:FADH
479:NCBI
420:PDBe
375:KEGG
267:1np7
260:1own
254:1owl
248:1owo
242:1owp
236:1qnf
230:1owm
224:1tez
218:1dnp
212:1iqr
205:1iqu
199:1u3d
193:1u3c
168:PDBj
164:PDBe
147:ECOD
137:Pfam
113:1qnf
72:Pfam
1975:doi
1971:157
1944:doi
1940:262
1911:doi
1907:378
1870:PMC
1860:doi
1813:doi
1803:".
1772:PMC
1764:doi
1760:215
1716:PMC
1706:doi
1702:290
1660:doi
1656:103
1625:doi
1621:107
1580:PMC
1572:doi
1531:PMC
1521:doi
1509:103
1469:PMC
1461:doi
1457:146
1422:doi
1410:382
1345:doi
1308:PMC
1300:doi
1261:doi
1249:163
1216:PMC
1206:doi
1153:PMC
1143:doi
1131:108
1096:doi
1043:PRE
578:to
544:or
534:DNA
455:PMC
411:PDB
187:PDB
155:PDB
2604::
2222::
2172::
2061::
2050:EC
1981:.
1969:.
1952:.
1938:.
1934:.
1917:.
1905:.
1878:.
1868:.
1858:.
1848:96
1846:.
1842:.
1819:.
1809:31
1807:.
1780:.
1770:.
1758:.
1754:.
1738:^
1724:.
1714:.
1700:.
1696:.
1680:^
1666:.
1654:.
1631:.
1619:.
1588:.
1578:.
1568:26
1566:.
1562:.
1539:.
1529:.
1519:.
1507:.
1503:.
1491:^
1477:.
1467:.
1455:.
1451:.
1428:.
1420:.
1408:.
1384:.
1351:.
1341:33
1339:.
1316:.
1306:.
1296:59
1294:.
1290:.
1267:.
1259:.
1247:.
1224:.
1214:.
1204:.
1194:35
1192:.
1188:.
1161:.
1151:.
1141:.
1129:.
1125:.
1102:.
1092:56
1090:.
1057:,
1053:,
1049:,
1045:,
1041:,
1037:,
1033:,
1029:,
1025:,
1021:,
1017:,
1013:,
990:;
918:,
914:,
764:]
500:EC
437:/
166:;
162:;
145:/
119:/
115:/
2580::
2566:)
2562:(
2553:)
2549:(
2540:)
2536:(
2527:)
2523:(
2514:)
2510:(
2501:)
2497:(
2488:)
2484:(
2308:e
2301:t
2294:v
2153:/
2048:(
2037:e
2030:t
2023:v
1989:.
1977::
1960:.
1946::
1925:.
1913::
1886:.
1862::
1854::
1827:.
1815::
1788:.
1766::
1732:.
1708::
1674:.
1662::
1639:.
1627::
1596:.
1574::
1547:.
1523::
1515::
1485:.
1463::
1436:.
1424::
1416::
1392:.
1359:.
1347::
1324:.
1302::
1275:.
1263::
1255::
1232:.
1208::
1200::
1169:.
1145::
1137::
1110:.
1098::
888:.
881:.
868:.
498:(
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.