1940:
6108:
2002:
31:
1882:
Protein sequences contain sequence motifs that are recognized by modifying enzymes, and which can be documented or predicted in PTM databases. With the large number of different modifications being discovered, there is a need to document this sort of information in databases. PTM information can be
1875:
1331:
In 2011, statistics of each post-translational modification experimentally and putatively detected have been compiled using proteome-wide information from the Swiss-Prot database. The 10 most common experimentally found modifications were as follows:
2438:"Murine elongation factor 1 alpha (EF-1 alpha) is posttranslationally modified by novel amide-linked ethanolamine-phosphoglycerol moieties. Addition of ethanolamine-phosphoglycerol to specific glutamic acid residues on EF-1 alpha"
1883:
collected through experimental means or predicted from high-quality, manually curated data. Numerous databases have been created, often with a focus on certain taxonomic groups (e.g. human proteins) or other features.
4878:
5131:
4040:
503:
212:
is one example that targets the modified protein for degradation and can result in the formation of protein aggregates. Specific amino acid modifications can be used as
1100:(PEG) using a pegylation reagent, typically to the N-terminus or the side-chains of Lys residues. Pegylation is used to improve the efficacy of protein pharmaceuticals.
2019:
1083:
carbamylation: the addition of
Isocyanic acid to a protein's N-terminus or the side-chain of Lys or Cys residues, typically resulting from exposure to urea solutions.
3019:
Rabe von
Pappenheim, Fabian; Wensien, Marie; Ye, Jin; Uranga, Jon; Irisarri, Iker; de Vries, Jan; Funk, Lisa-Marie; Mata, Ricardo A.; Tittmann, Kai (April 2022).
2074:
1452:
Some common post-translational modifications to specific amino-acid residues are shown below. Modifications occur on the side-chain unless indicated otherwise.
4871:
2270:
5731:
6143:
3853:
2655:"On a potential global role for vitamin K-dependent gamma-carboxylation in animal systems. Evidence for a gamma-glutamyl carboxylase in Drosophila"
1931:– A database that shows different PTM's and information regarding their chemical components/structures and a frequency for amino acid modified site
4864:
4033:
5476:
1907:– A database for different modifications and understand different proteins, their class, and function/process related to disease causing proteins
5899:
2248:
3658:
Yang Y, Peng X, Ying P, Tian J, Li J, Ke J, Zhu Y, Gong Y, Zou D, Yang N, Wang X, Mei S, Zhong R, Gong J, Chang J, Miao X (January 2019).
2321:
1086:
oxidation: addition of one or more Oxygen atoms to a susceptible side-chain, principally of Met, Trp, His or Cys residues. Formation of
5783:
5763:
5694:
4026:
1150:
2610:
Eddé B, Rossier J, Le Caer JP, Desbruyères E, Gros F, Denoulet P (January 1990). "Posttranslational glutamylation of alpha-tubulin".
5871:
2883:
2420:
2142:
2041:
1904:
201:
is removed from the middle of the chain; the resulting protein consists of two polypeptide chains connected by disulfide bonds.
6025:
5964:
5087:
4053:
2984:
Brennan DF, Barford D (March 2009). "Eliminylation: a post-translational modification catalyzed by phosphothreonine lyases".
2023:
2759:"Carbamylation-derived products: bioactive compounds and potential biomarkers in chronic renal failure and atherosclerosis"
6138:
6052:
5983:
5724:
2101:
6098:
2871:
5788:
5300:
4703:
3846:
1916:
1829:
1080:: covalent attachment of a biotin moiety using a biotinylation reagent, typically for the purpose of labeling a protein.
5245:
3797:
3497:
Wulff-Fuentes E, Berendt RR, Massman L, Danner L, Malard F, Vora J, Kahsay R, Olivier-Van
Stichelen S (January 2021).
3119:
3064:"Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database"
2176:"Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database"
1895:– A database of comprehensive information and tools for the study of mammalian protein post-translational modification
1030:
Examples of non-enzymatic PTMs are glycation, glycoxidation, nitrosylation, oxidation, succination, and lipoxidation.
444:
219:
Sites that often undergo post-translational modification are those that have a functional group that can serve as a
6133:
6003:
2368:
Gianazza E, Crawford J, Miller I (July 2007). "Detecting oxidative post-translational modifications in proteins".
2012:
499:
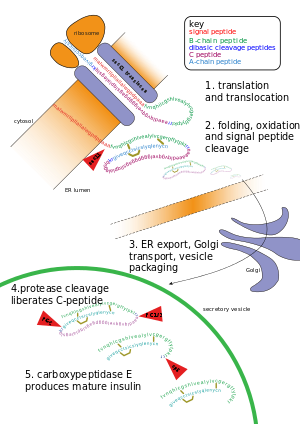
38:. At the top, the ribosome translates a mRNA sequence into a protein, insulin, and passes the protein through the
4743:
4154:
4049:
301:
Post-translational modification of proteins can be experimentally detected by a variety of techniques, including
4018:
46:, where it is packaged into a vesicle. In the vesicle, more parts are cut off, and it turns into mature insulin.
6128:
6047:
5835:
5748:
5717:
5185:
2070:
1278:
463:
135:
is highly effective for controlling the enzyme activity and is the most common change after translation. Many
124:
2896:
Malakhova OA, Yan M, Malakhov MP, Yuan Y, Ritchie KJ, Kim KI, Peterson LF, Shuai K, Zhang DE (February 2003).
2747:"The Advanced Lipoxidation End-Product Malondialdehyde-Lysine in Aging and Longevity" PMID 33203089 PMC7696601
1955:- A curated database for protein O-GlcNAcylation and referencing more than 14 000 protein entries and 10 000
684:
42:, where it is cut, folded, and held in shape by disulfide (-S-S-) bonds. Then the protein passes through the
6008:
5825:
5810:
5510:
4797:
3839:
3399:
Huang H, Arighi CN, Ross KE, Ren J, Li G, Chen SC, Wang Q, Cowart J, Vijay-Shanker K, Wu CH (January 2018).
2653:
Walker CS, Shetty RP, Clark K, Kazuko SG, Letsou A, Olivera BM, Bandyopadhyay PK, et al. (March 2001).
1250:
5682:
4681:
3768:
6013:
5938:
5830:
5551:
3301:
Sigrist CJ, Cerutti L, de Castro E, Langendijk-Genevaux PS, Bulliard V, Bairoch A, Hulo N (January 2010).
1804:
1780:
1517:
1409:
1369:
1061:
2798:
Kang HJ, Baker EN (April 2011). "Intramolecular isopeptide bonds: protein crosslinks built for stress?".
1253:, the covalent linkage of 1 lysine and 1 or 2 cystine residues via an oxygen atom (NOS and SONOS bridges)
6148:
5928:
5913:
5793:
4932:
3985:
2171:
910:
802:
440:
83:
39:
2701:"Cysteine oxidative posttranslational modifications: emerging regulation in the cardiovascular system"
6035:
5933:
5851:
4072:
3876:
3807:
3514:
3075:
2619:
2189:
1256:
1220:
1208:
71:
4087:
3812:
A Computational
Protocol for Identification of Post-Translational Modifications in Protein Sequences
5861:
4895:
4159:
3816:
3811:
3254:"Human Protein Reference Database and Human Proteinpedia as resources for phosphoproteome analysis"
2477:
Roy H, Zou SB, Bullwinkle TJ, Wolfe BS, Gilreath MS, Forsyth CJ, Navarre WW, Ibba M (August 2011).
1898:
1706:
1619:
1498:
1200:
1097:
929:
713:
527:
398:
94:
4508:
797:
708:
93:, which may then change to form the mature protein product. PTMs are important components in cell
5996:
5879:
5541:
4909:
4498:
4402:
4252:
3787:
2393:
2180:
2120:
1844:
1011:
190:
residues may also be referred to as a post-translational modification. For instance, the peptide
3826:
1925:– A database that integrates PTM information from several knowledgbases and text mining results.
3401:"iPTMnet: an integrated resource for protein post-translational modification network discovery"
1937:
has PTM information although that may be less comprehensive than in more specialized databases.
1934:
6080:
5211:
4356:
4242:
4201:
3917:
3891:
3827:
Overview and description of commonly used post-translational modification detection techniques
3730:
3689:
3640:
3589:
3540:
3479:
3430:
3381:
3332:
3283:
3231:
3182:
3101:
3001:
2966:
2927:
2879:
2815:
2780:
2730:
2676:
2635:
2592:
2557:
2508:
2459:
2416:
2385:
2350:
2297:
2244:
2215:
2148:
2138:
2096:
2080:
1615:
1583:
1429:
676:
302:
90:
51:
3560:"Automatization and self-maintenance of the O-GlcNAcome catalog: a smart scientific database"
2962:
1978:
5471:
5430:
5425:
5400:
5390:
5385:
5375:
5175:
4659:
4637:
4536:
4455:
4346:
4323:
4247:
4219:
3995:
3953:
3948:
3943:
3720:
3679:
3671:
3630:
3620:
3579:
3571:
3530:
3522:
3469:
3461:
3420:
3412:
3371:
3363:
3322:
3314:
3273:
3265:
3221:
3213:
3172:
3164:
3091:
3083:
3042:
3032:
2993:
2958:
2917:
2909:
2846:
2807:
2770:
2720:
2712:
2666:
2627:
2584:
2547:
2539:
2498:
2490:
2449:
2377:
2340:
2330:
2287:
2279:
2205:
2197:
2056:
1655:
1599:
1310:
1240:
1212:
1037:, the addition of a sugar molecule to a protein without the controlling action of an enzyme.
822:
818:
310:
306:
205:
128:
5446:
5380:
1928:
535:
517:
6057:
5894:
5740:
5677:
4900:
4770:
4470:
4361:
4237:
4196:
3901:
3886:
3775:
2875:
1800:
1776:
1718:
1591:
1567:
1540:
1521:
1349:
1057:
937:
845:
835:
827:
786:
690:
387:
295:
152:
132:
43:
3518:
3079:
2623:
2436:
Whiteheart SW, Shenbagamurthi P, Chen L, Cotter RJ, Hart GW, et al. (August 1989).
2193:
6112:
5884:
5856:
5224:
5219:
4632:
4526:
4503:
4445:
4295:
4277:
4164:
4130:
3922:
3684:
3659:
3635:
3608:
3584:
3559:
3535:
3498:
3474:
3449:
3425:
3400:
3327:
3302:
3278:
3253:
3226:
3201:
3177:
3152:
3096:
3063:
3047:
3020:
2725:
2700:
2552:
2527:
2503:
2478:
2345:
2316:
2292:
2283:
2265:
2210:
2175:
2132:
2124:
1698:
1635:
1559:
1494:
1292:
1162:
1044:
918:
371:
357:
339:
183:
3558:
Malard F, Wulff-Fuentes E, Berendt RR, Didier G, Olivier-Van
Stichelen S (July 2021).
3376:
3351:
2922:
2897:
2851:
2834:
2454:
2437:
6122:
6085:
5772:
4651:
4596:
4578:
4554:
4493:
4488:
4440:
4435:
4384:
4341:
4262:
4206:
4110:
3980:
3975:
2588:
2238:
1761:
1741:
1710:
1606:
1587:
1528:
1389:
1216:
1195:
1187:
1077:
1050:
1040:
998:
983:-sulfonylation, normally irreversible covalent addition of three oxygen atoms to the
923:
879:
812:
742:
722:
680:
640:
594:
393:
209:
164:
148:
3725:
3708:
3200:
Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E (January 2015).
2397:
1922:
5959:
5889:
5758:
5580:
5562:
4450:
4267:
4082:
4067:
3609:"PyTMs: a useful PyMOL plugin for modeling common post-translational modifications"
3303:"PROSITE, a protein domain database for functional characterization and annotation"
1984:
1702:
1268:
1262:
1124:
992:
969:-sulfinylation, normally irreversible covalent addition of two oxygen atoms to the
762:
666:
473:
436:
171:
144:
3817:
Functional analyses for site-specific phosphorylation of a target protein in cells
3660:"AWESOME: a database of SNPs that affect protein post-translational modifications"
2866:
2716:
17:
4856:
2775:
2758:
2528:"Lysine Acetylation Goes Global: From Epigenetics to Metabolism and Therapeutics"
2479:"The tRNA synthetase paralog PoxA modifies elongation factor-P with (R)-β-lysine"
2156:
1265:
formation, via the cyclisation of asparagine or aspartic acid amino-acid residues
811:: addition of an iodine atom to the aromatic ring of a tyrosine residue (e.g. in
6062:
5991:
5656:
5524:
4572:
4531:
4460:
4425:
4420:
4379:
4351:
4318:
4300:
4272:
4125:
4115:
4105:
4077:
4000:
3990:
3821:
2835:"Reactions of the Cyanate Present in Aqueous Urea with Amino Acids and Proteins"
2543:
2317:"Oxidative stress and covalent modification of protein with bioactive aldehydes"
2128:
2001:
1860:
1808:
1784:
1733:
1694:
1639:
1563:
1536:
1513:
1477:
1399:
1359:
1179:
1130:
1120:
624:
619:
600:
578:
523:
489:
476:
422:
375:
267:
220:
140:
6107:
3798:
Statistics of each post-translational modification from the Swiss-Prot database
3575:
3526:
3465:
3037:
2997:
2811:
2234:
1939:
1919:– A database consisting of a collection of annotations and structures for PTMs.
1874:
6020:
5486:
4914:
4812:
4718:
4695:
4691:
4647:
4611:
4588:
4371:
4175:
4141:
4097:
3927:
3625:
3021:"Widespread occurrence of covalent lysine–cysteine redox switches in proteins"
2413:
Posttranslational modification of proteins : expanding nature's inventory
2381:
2152:
1981:– Interactive tool to see the role of single nucleotide polymorphisms to PTM's
1825:
1816:
1725:
1662:
1505:
1302:
1298:
1288:
1191:
1174:
1146:
1093:
808:
758:
734:
660:
607:
586:
572:
470:, as in fatty acid, polyketide, non-ribosomal peptide and leucine biosynthesis
467:
409:
405:
379:
291:
283:
198:
179:
175:
160:
136:
120:
116:
112:
109:
98:
3153:"dbPTM: an information repository of protein post-translational modification"
155:
and improve stability as well as serving regulatory functions. Attachment of
5234:
5180:
4841:
4754:
4564:
4483:
4430:
4394:
4333:
4310:
4257:
4188:
4120:
4005:
3748:
2631:
2235:"17.6, Post-Translational Modifications and Quality Control in the Rough ER"
1901:– A database of proteins and post-translational modifications experimentally
1792:
1647:
1574:
1555:
1439:
1419:
1224:
1183:
1110:
1087:
1034:
895:
869:
857:
849:
782:-acetylglucosamine to serine or threonine residues in a β-glycosidic linkage
766:
750:
717:
651:
at C-terminus. Formed by oxidative dissociation of a C-terminal Gly residue.
548:
454:
361:
347:
343:
275:
271:
252:
232:
213:
79:
3734:
3693:
3644:
3593:
3544:
3483:
3385:
3336:
3287:
3235:
3186:
3127:
3105:
3005:
2970:
2931:
2819:
2784:
2734:
2680:
2671:
2654:
2561:
2512:
2389:
2354:
2335:
2301:
2219:
3434:
3416:
3217:
2639:
2596:
2463:
1913:– A database of Consensus patterns for many types of PTM's including sites
30:
6040:
6030:
5954:
5514:
5147:
5139:
4829:
4825:
4821:
4758:
4731:
4673:
4624:
4620:
4518:
4465:
4287:
4229:
4211:
3958:
3896:
3675:
3367:
3318:
3168:
2494:
2264:
Dalle-Donne I, Aldini G, Carini M, Colombo R, Rossi R, Milzani A (2006).
1836:
1736:(N-terminus), N-linked Ubiquitination, oxidation to sulfoxide or sulfone
1714:
1547:
1485:
1244:
1170:
1114:
1015:
1002:
988:
974:
960:
955:-sulphenylation), reversible covalent addition of one oxygen atom to the
887:
883:
861:
772:
754:
738:
730:
726:
636:
509:
458:
383:
263:
256:
248:
236:
224:
187:
63:
2913:
5776:
5265:
5152:
5112:
5043:
5038:
5033:
5028:
5023:
4978:
4789:
4781:
4762:
4735:
4546:
3968:
3963:
3862:
3782:
3269:
2066:
2060:
2026: in this section. Unsourced material may be challenged and removed.
1910:
1753:
1674:
1626:
1469:
1284:
698:
694:
426:
194:
191:
102:
75:
67:
35:
3822:
Detection of Post-Translational
Modifications after high-accuracy MSMS
3151:
Lee TY, Huang HD, Hung JH, Huang HY, Yang YS, Wang TH (January 2006).
3087:
2945:
Klareskog L, Rönnelid J, Lundberg K, Padyukov L, Alfredsson L (2008).
2266:"Protein carbonylation, cellular dysfunction, and disease progression"
2201:
805:: addition of an oxygen atom to the side-chain of a Pro or Lys residue
5481:
5456:
5451:
5441:
5405:
5395:
5370:
5355:
5350:
5345:
5340:
5335:
5330:
5325:
5320:
5285:
5270:
5117:
5082:
5067:
5062:
5057:
5018:
5013:
5008:
5003:
4998:
4993:
4988:
4983:
4973:
4968:
4963:
4958:
4953:
4833:
4785:
4727:
4669:
4412:
4184:
1852:
1768:
1686:
1595:
1314:
1274:
1228:
1204:
1053:
the addition of carbon monoxide to other organic/inorganic compounds.
1006:
899:
853:
746:
632:
628:
615:
590:
582:
564:
556:
450:
287:
244:
228:
683:
residues to the N-terminus of tubulin and some other proteins. (See
2946:
1558:-bond formation, oxidation to sulfenic, sulfinic or sulfonic acid,
170:
Other forms of post-translational modification consist of cleaving
5798:
5709:
5672:
5662:
5652:
5637:
5619:
5614:
5609:
5604:
5536:
5496:
5461:
5435:
5420:
5415:
5410:
5365:
5360:
5315:
5310:
5305:
5275:
5260:
5157:
5126:
5107:
5102:
5097:
5092:
5077:
5072:
5052:
4948:
4941:
4937:
4927:
4922:
4478:
4149:
1972:
1140:
1134:
984:
970:
956:
654:
611:
531:
513:
279:
259:
240:
204:
Some types of post-translational modification are consequences of
156:
29:
27:
Chemical changes in proteins following their translation from mRNA
5687:
5667:
5647:
5642:
5592:
5575:
5531:
5519:
5501:
5491:
5290:
5280:
5255:
5250:
5195:
5190:
3791:
3352:"The RESID Database of Protein Modifications: 2003 developments"
2086:
Cleavage of polypeptide chains as crucial for lectin specificity
1878:
Flowchart of the process and the data sources to predict PTMs.
790:
670:
493:
86:
5713:
4860:
4022:
3835:
1948:
1943:
Effect of PTMs on protein function and physiological processes.
286:
is a weak nucleophile, it can serve as an attachment point for
163:, often targets a protein or part of a protein attached to the
5768:
3607:
Warnecke A, Sandalova T, Achour A, Harris RA (November 2014).
1995:
1968:
List of software for visualization of proteins and their PTMs
1892:
3707:
Morris JH, Huang CC, Babbitt PC, Ferrin TE (September 2007).
2898:"Protein ISGylation modulates the JAK-STAT signaling pathway"
3769:
dbPTM - database of protein post-translational modifications
2947:"Immunity to citrullinated proteins in rheumatoid arthritis"
769:, which is regarded as a nonenzymatic attachment of sugars.
3831:
3202:"PhosphoSitePlus, 2014: mutations, PTMs and recalibrations"
2315:
Grimsrud PA, Xie H, Griffin TJ, Bernlohr DA (August 2008).
3120:"Proteome-Wide Post-Translational Modification Statistics"
2575:
Bradbury AF, Smyth DG (March 1991). "Peptide amidation".
2134:
4771:
4-(p-hydroxybenzylidene)-5-imidazolinone (HBI) formation
3252:
Goel R, Harsha HC, Pandey A, Prasad TS (February 2012).
909:
uridylylation, the addition of an uridylyl-group (i.e.
466:, the addition of a 4'-phosphopantetheinyl moiety from
278:; and the N- and C-termini. In addition, although the
6096:
1975:– introduce a set of common PTM's into protein models
197:
is cut twice after disulfide bonds are formed, and a
3062:
Khoury GA, Baliban RC, Floudas CA (September 2011).
2757:
Jaisson S, Pietrement C, Gillery P (November 2011).
123:
termini. They can expand the chemical set of the 22
6073:
5982:
5947:
5921:
5912:
5870:
5844:
5818:
5809:
5747:
5630:
5561:
5550:
5233:
5210:
5168:
4908:
4894:
4820:
4810:
4780:
4753:
4726:
4716:
4690:
4668:
4646:
4619:
4609:
4587:
4563:
4545:
4517:
4411:
4393:
4370:
4332:
4309:
4286:
4228:
4183:
4173:
4140:
4096:
4060:
3936:
3910:
3869:
3450:"In silico prediction tools and molecular modeling"
2526:Ali I, Conrad RJ, Verdin E, Ott M (February 2018).
2233:Lodish H, Berk A, Zipursky SL, et al. (2000).
1047:
to a protein's N-terminus or the side-chain of Lys.
4744:p-Hydroxybenzylidene-imidazolinone (HBI) formation
3709:"structureViz: linking Cytoscape and UCSF Chimera"
3454:Computational and Structural Biotechnology Journal
108:Post-translational modifications can occur on the
3783:List of posttranslational modifications in ExPASy
1060:formation, as found in many surface proteins of
78:or occur spontaneously. Proteins are created by
2868:Sumoylation: Molecular Biology and Biochemistry
147:molecules attached to them in a process called
412:formation via an amide bond to C-terminal tail
5725:
4872:
4034:
3847:
3247:
3245:
3146:
3144:
2075:RNA polymerase control by chromatin structure
1987:– Interactive Database to visualize molecules
8:
4704:Tryptophan tryptophylquinone (TTQ) formation
530:(EFP) in most bacteria. EFP is a homolog to
334:Hydrophobic groups for membrane localization
321:PTMs involving addition of functional groups
290:. Rarer modifications can occur at oxidized
1105:Conjugation with other proteins or peptides
178:to a mature form or removing the initiator
5918:
5880:Precursor mRNA (pre-mRNA / hnRNA)
5815:
5732:
5718:
5710:
5558:
4905:
4879:
4865:
4857:
4817:
4723:
4616:
4180:
4041:
4027:
4019:
3854:
3840:
3832:
2271:Journal of Cellular and Molecular Medicine
1454:
693:, covalent linkage of one to more than 40
3724:
3683:
3634:
3624:
3583:
3534:
3473:
3448:Audagnotto M, Dal Peraro M (2017-03-31).
3424:
3375:
3326:
3277:
3225:
3176:
3095:
3046:
3036:
2921:
2850:
2774:
2724:
2670:
2551:
2502:
2453:
2344:
2334:
2291:
2243:(4th ed.). New York: W. H. Freeman.
2209:
2042:Learn how and when to remove this message
1259:, cleavage of a protein at a peptide bond
991:residue, resulting in the formation of a
417:Cofactors for enhanced enzymatic activity
410:glycosylphosphatidylinositol (GPI) anchor
313:. Additional methods are provided in the
2963:10.1146/annurev.immunol.26.021607.090244
2694:
2692:
2690:
1938:
1873:
1334:
1139:ISGylation, the covalent linkage to the
6103:
4798:Methylidene-imidazolone (MIO) formation
2112:
1143:protein (Interferon-Stimulated Gene 15)
1014:, the addition of a sulfate group to a
425:(a type of acylation), attachment of a
131:or adding a new one such as phosphate.
3503:-GlcNAcome database and meta-analysis"
2699:Chung HS, et al. (January 2013).
526:addition on a conserved lysine of the
5900:Histone acetylation and deacetylation
4682:Lysine tyrosylquinone (LTQ) formation
1719:spontaneous isopeptide bond formation
1541:spontaneous isopeptide bond formation
1522:spontaneous isopeptide bond formation
360:(a type of acylation), attachment of
7:
5965:Ribosome-nascent chain complex (RNC)
2833:Stark GR, Stein WH, Moore X (1960).
2137:(2nd ed.). Hoboken, NJ: Wiley.
2024:adding citations to reliable sources
1824:mono- or di-oxidation, formation of
1157:Chemical modification of amino acids
484:Modifications of translation factors
4155:Glycosyl phosphatidylinositol (GPI)
2659:The Journal of Biological Chemistry
2442:The Journal of Biological Chemistry
2415:. Englewood: Roberts and Co. Publ.
2322:The Journal of Biological Chemistry
492:formation (on a histidine found in
314:
34:Post-translational modification of
5695:Prokaryotic ubiquitin-like protein
2284:10.1111/j.1582-4934.2006.tb00407.x
1151:prokaryotic ubiquitin-like protein
1127:(Small Ubiquitin-related MOdifier)
512:formation (on conserved lysine of
502:attachment (on glutamate found in
25:
4466:Oxidative deamination to aldehyde
1117:linkage to the protein ubiquitin.
6106:
3749:"1tp8 - Proteopedia, life in 3D"
2000:
1905:Human Protein Reference Database
639:residues. The reverse is called
593:residues. The reverse is called
6144:Post-translational modification
5970:Post-translational modification
4054:posttranslational modifications
3882:Post-translational modification
2011:needs additional citations for
56:post-translational modification
5225:Mitochondrial targeting signal
4888:Posttranslational modification
2986:Trends in Biochemical Sciences
2800:Trends in Biochemical Sciences
2577:Trends in Biochemical Sciences
2411:Walsh, Christopher T. (2006).
2083:as regulation of transcription
1379:
1243:, the covalent linkage of two
1149:, the covalent linkage to the
1133:, the covalent linkage to the
1123:, the covalent linkage to the
1:
3726:10.1093/bioinformatics/btm329
3350:Garavelli JS (January 2003).
2852:10.1016/S0021-9258(20)81332-5
2717:10.1161/CIRCRESAHA.112.268680
2455:10.1016/S0021-9258(18)71682-7
2102:Post-translational regulation
1586:(N-terminus), deamidation to
785:polysialylation, addition of
216:indicating oxidative damage.
5301:Ubiquitin-conjugating enzyme
2865:Van G. Wilson (Ed.) (2004).
2776:10.1373/clinchem.2011.163188
2589:10.1016/0968-0004(91)90044-v
1830:tryptophan tryptophylquinone
1317:analogous to mRNA processing
1313:, self-catalytic removal of
1305:, also a frog opioid peptide
1023:Non-enzymatic modifications
500:ethanolamine phosphoglycerol
447:) may be covalently attached
5589:E2 SUMO-conjugating enzyme
5246:Ubiquitin-activating enzyme
4273:Topaquinone (TPQ) formation
3212:(Database issue): D512-20.
2951:Annual Review of Immunology
2544:10.1021/acs.chemrev.7b00181
1430:Pyrrolidone carboxylic acid
1090:bonds between Cys residues.
913:, UMP), usually to tyrosine
6165:
5572:E1 SUMO-activating enzyme
3923:Protein structural domains
3788:Browse SCOP domains by PTM
3527:10.1038/s41597-021-00810-4
3466:10.1016/j.csbj.2017.03.004
3313:(Database issue): D161-6.
3163:(Database issue): D622-7.
3038:10.1038/s41589-021-00966-5
2998:10.1016/j.tibs.2008.11.005
2812:10.1016/j.tibs.2010.09.007
2055:Cleavage and formation of
182:residue. The formation of
4717:Crosslinks between three
4050:Protein primary structure
3626:10.1186/s12859-014-0370-6
2382:10.1007/s00726-006-0410-2
2059:during the production of
1096:: covalent attachment of
6031:sequestration (P-bodies)
5186:Survival of motor neuron
4811:Crosslinks between four
3576:10.1093/database/baab039
1327:Common PTMs by frequency
1279:protein-serine epimerase
1069:Non-enzymatic additions
464:phosphopantetheinylation
127:by changing an existing
6009:Gene regulatory network
5552:Ubiquitin-like proteins
5511:Deubiquitinating enzyme
4610:Crosslinks between two
3803:(Wayback Machine copy)
3025:Nature Chemical Biology
2902:Genes & Development
2632:10.1126/science.1967194
2483:Nature Chemical Biology
2170:Khoury GA, Baliban RC,
1251:lysine-cysteine bridges
1203:, the conversion to an
685:tubulin polyglutamylase
543:Smaller chemical groups
538:(archaeal) (see above).
6014:cis-regulatory element
4258:Porphyrin ring linkage
3664:Nucleic Acids Research
3405:Nucleic Acids Research
3356:Nucleic Acids Research
3307:Nucleic Acids Research
3206:Nucleic Acids Research
3157:Nucleic Acids Research
2878:. Horizon Bioscience.
2672:10.1074/jbc.M009576200
2336:10.1074/jbc.R700019200
2240:Molecular Cell Biology
1944:
1879:
1805:O-linked glycosylation
1781:O-linked glycosylation
1518:N-linked glycosylation
1448:Common PTMs by residue
1410:O-linked glycosylation
1370:N-linked glycosylation
1062:Gram-positive bacteria
679:, covalent linkage of
326:Addition by an enzyme
97:, as for example when
47:
4319:Succinimide formation
3986:Photoreceptor protein
1942:
1877:
1539:to isoaspartic acid,
911:uridine monophosphate
882:, the addition of an
610:, the addition of an
589:of the protein or at
585:group, either at the
581:, the addition of an
378:, the addition of an
223:in the reaction: the
174:, as in processing a
40:endoplasmic reticulum
33:
6139:Protein biosynthesis
6036:alternative splicing
6026:Post-transcriptional
5852:Transcription factor
4073:Protein biosynthesis
3877:Protein biosynthesis
3258:Molecular BioSystems
3124:selene.princeton.edu
2705:Circulation Research
2495:10.1038/nchembio.632
2020:improve this article
1516:to Asp or iso(Asp),
1257:proteolytic cleavage
1182:, the conversion of
1169:, the conversion of
848:, the addition of a
725:, the addition of a
159:molecules, known as
151:, which can promote
115:or at the protein's
72:protein biosynthesis
66:process of changing
5960:Transfer RNA (tRNA)
4910:Heat shock proteins
3753:www.proteopedia.org
3519:2021NatSD...8...25W
3417:10.1093/nar/gkx1104
3218:10.1093/nar/gku1267
3080:2011NatSR...1E..90K
2914:10.1101/gad.1056303
2624:1990Sci...247...83E
2194:2011NatSR...1E..90K
2121:Pratt, Charlotte W.
1870:Databases and tools
1620:gamma-carboxylation
1098:polyethylene glycol
886:moiety, usually to
842:-linked) formation
819:nucleotide addition
714:gamma-carboxylation
673:-mediation addition
528:elongation factor P
399:geranylgeranylation
143:proteins also have
74:. PTMs may involve
6074:Influential people
6053:Post-translational
5872:Post-transcription
4403:Transglutamination
3676:10.1093/nar/gky821
3613:BMC Bioinformatics
3368:10.1093/nar/gkg038
3319:10.1093/nar/gkp885
3270:10.1039/c1mb05340j
3169:10.1093/nar/gkj083
3068:Scientific Reports
2874:2005-02-09 at the
2763:Clinical Chemistry
2181:Scientific Reports
2174:(September 2011).
1945:
1880:
1847:, phosphorylation
1235:Structural changes
933:-glutathionylation
852:group, usually to
631:group, usually at
627:the addition of a
433:) functional group
91:polypeptide chains
48:
18:Post-translational
6134:Protein structure
6094:
6093:
5978:
5977:
5908:
5907:
5784:Special transfers
5707:
5706:
5703:
5702:
5212:Protein targeting
5206:
5205:
4854:
4853:
4850:
4849:
4806:
4805:
4712:
4711:
4605:
4604:
4357:Polyglutamylation
4243:Dephosphorylation
4202:Dephosphorylation
4016:
4015:
3918:Protein structure
3892:Protein targeting
3670:(D1): D874–D880.
3564:Database (Oxford)
3088:10.1038/srep00090
2845:(11): 3177–3181.
2250:978-0-7167-3136-8
2202:10.1038/srep00090
2097:Protein targeting
2081:RNA polymerase II
2069:as regulation of
2057:disulfide bridges
2052:
2051:
2044:
1887:List of resources
1867:
1866:
1616:Pyroglutamic acid
1584:pyroglutamic acid
1445:
1444:
1241:disulfide bridges
677:polyglutamylation
534:(eukaryotic) and
516:(eukaryotic) and
346:), attachment of
303:mass spectrometry
101:are converted to
52:molecular biology
16:(Redirected from
6156:
6111:
6110:
6102:
5919:
5816:
5734:
5727:
5720:
5711:
5559:
5472:Ubiquitin ligase
5238:(ubiquitylation)
5176:Alpha crystallin
4906:
4881:
4874:
4867:
4858:
4818:
4724:
4660:Sulfilimine bond
4638:ADP-ribosylation
4617:
4537:ADP-ribosylation
4456:ADP-ribosylation
4347:ADP-ribosylation
4324:ADP-ribosylation
4248:ADP-ribosylation
4220:ADP-ribosylation
4181:
4174:Single specific
4043:
4036:
4029:
4020:
3996:Phycobiliprotein
3954:Globular protein
3949:Membrane protein
3944:List of proteins
3856:
3849:
3842:
3833:
3808:AutoMotif Server
3757:
3756:
3745:
3739:
3738:
3728:
3704:
3698:
3697:
3687:
3655:
3649:
3648:
3638:
3628:
3604:
3598:
3597:
3587:
3555:
3549:
3548:
3538:
3494:
3488:
3487:
3477:
3445:
3439:
3438:
3428:
3411:(1): D542–D550.
3396:
3390:
3389:
3379:
3347:
3341:
3340:
3330:
3298:
3292:
3291:
3281:
3249:
3240:
3239:
3229:
3197:
3191:
3190:
3180:
3148:
3139:
3138:
3136:
3135:
3126:. Archived from
3116:
3110:
3109:
3099:
3059:
3053:
3052:
3050:
3040:
3016:
3010:
3009:
2981:
2975:
2974:
2942:
2936:
2935:
2925:
2893:
2887:
2863:
2857:
2856:
2854:
2830:
2824:
2823:
2795:
2789:
2788:
2778:
2769:(11): 1499–505.
2754:
2748:
2745:
2739:
2738:
2728:
2696:
2685:
2684:
2674:
2650:
2644:
2643:
2607:
2601:
2600:
2572:
2566:
2565:
2555:
2538:(3): 1216–1252.
2523:
2517:
2516:
2506:
2474:
2468:
2467:
2457:
2448:(24): 14334–41.
2433:
2427:
2426:
2408:
2402:
2401:
2365:
2359:
2358:
2348:
2338:
2329:(32): 21837–41.
2312:
2306:
2305:
2295:
2261:
2255:
2254:
2230:
2224:
2223:
2213:
2167:
2161:
2160:
2159:on 13 July 2012.
2155:. Archived from
2117:
2047:
2040:
2036:
2033:
2027:
2004:
1996:
1953:-GlcNAc Database
1600:transglutaminase
1455:
1335:
1311:protein splicing
1213:phosphothreonine
1209:beta-elimination
1043:the addition of
948:-sulfenylation (
823:ADP-ribosylation
765:. Distinct from
729:group to either
697:residues to the
650:
311:Western blotting
307:Eastern blotting
298:in side chains.
296:methylene groups
206:oxidative stress
129:functional group
21:
6164:
6163:
6159:
6158:
6157:
6155:
6154:
6153:
6129:Gene expression
6119:
6118:
6117:
6105:
6097:
6095:
6090:
6069:
6004:Transcriptional
5974:
5943:
5904:
5895:Polyadenylation
5866:
5840:
5805:
5799:Protein→Protein
5750:
5743:
5741:Gene expression
5738:
5708:
5699:
5626:
5601:E3 SUMO ligase
5565:
5554:
5546:
5237:
5229:
5202:
5164:
5143:
5135:
4913:
4901:protein folding
4899:
4890:
4885:
4855:
4846:
4802:
4776:
4749:
4708:
4686:
4664:
4642:
4601:
4597:C-mannosylation
4583:
4559:
4541:
4513:
4479:Imine formation
4407:
4389:
4366:
4362:Polyglycylation
4328:
4305:
4282:
4238:Phosphorylation
4224:
4197:Phosphorylation
4169:
4136:
4092:
4056:
4047:
4017:
4012:
3976:Fibrous protein
3932:
3906:
3902:Protein methods
3887:Protein folding
3865:
3860:
3776:Wayback Machine
3765:
3760:
3747:
3746:
3742:
3706:
3705:
3701:
3657:
3656:
3652:
3606:
3605:
3601:
3557:
3556:
3552:
3507:Scientific Data
3496:
3495:
3491:
3447:
3446:
3442:
3398:
3397:
3393:
3349:
3348:
3344:
3300:
3299:
3295:
3251:
3250:
3243:
3199:
3198:
3194:
3150:
3149:
3142:
3133:
3131:
3118:
3117:
3113:
3061:
3060:
3056:
3018:
3017:
3013:
2983:
2982:
2978:
2944:
2943:
2939:
2895:
2894:
2890:
2876:Wayback Machine
2864:
2860:
2832:
2831:
2827:
2797:
2796:
2792:
2756:
2755:
2751:
2746:
2742:
2698:
2697:
2688:
2665:(11): 7769–74.
2652:
2651:
2647:
2609:
2608:
2604:
2574:
2573:
2569:
2525:
2524:
2520:
2476:
2475:
2471:
2435:
2434:
2430:
2423:
2410:
2409:
2405:
2367:
2366:
2362:
2314:
2313:
2309:
2263:
2262:
2258:
2251:
2232:
2231:
2227:
2169:
2168:
2164:
2145:
2125:Voet, Judith G.
2119:
2118:
2114:
2110:
2093:
2048:
2037:
2031:
2028:
2017:
2005:
1994:
1966:
1893:PhosphoSitePlus
1889:
1872:
1801:Phosphorylation
1777:Phosphorylation
1656:Phosphorylation
1614:cyclization to
1594:formation to a
1592:isopeptide bond
1582:cyclization to
1568:S-nitrosylation
1493:deimination to
1450:
1350:Phosphorylation
1329:
1324:
1237:
1159:
1107:
1074:
1058:isopeptide bond
1028:
846:phosphorylation
836:phosphoramidate
828:phosphate ester
787:polysialic acid
761:resulting in a
701:C-terminal tail
691:polyglycylation
657:bond formation
648:
545:
486:
453:attachment via
432:
419:
388:geranylgeraniol
367:
353:
336:
331:
323:
315:#External links
184:disulfide bonds
153:protein folding
133:Phosphorylation
44:golgi apparatus
28:
23:
22:
15:
12:
11:
5:
6162:
6160:
6152:
6151:
6146:
6141:
6136:
6131:
6121:
6120:
6116:
6115:
6092:
6091:
6089:
6088:
6083:
6081:François Jacob
6077:
6075:
6071:
6070:
6068:
6067:
6066:
6065:
6060:
6050:
6045:
6044:
6043:
6038:
6033:
6023:
6018:
6017:
6016:
6011:
6001:
6000:
5999:
5988:
5986:
5980:
5979:
5976:
5975:
5973:
5972:
5967:
5962:
5957:
5951:
5949:
5945:
5944:
5942:
5941:
5936:
5931:
5925:
5923:
5916:
5910:
5909:
5906:
5905:
5903:
5902:
5897:
5892:
5887:
5882:
5876:
5874:
5868:
5867:
5865:
5864:
5859:
5857:RNA polymerase
5854:
5848:
5846:
5842:
5841:
5839:
5838:
5833:
5828:
5822:
5820:
5813:
5807:
5806:
5804:
5803:
5802:
5801:
5796:
5791:
5781:
5780:
5779:
5761:
5755:
5753:
5745:
5744:
5739:
5737:
5736:
5729:
5722:
5714:
5705:
5704:
5701:
5700:
5698:
5697:
5691:
5690:
5685:
5680:
5675:
5670:
5665:
5660:
5650:
5645:
5640:
5634:
5632:
5628:
5627:
5625:
5624:
5623:
5622:
5617:
5612:
5607:
5598:
5597:
5596:
5595:
5586:
5585:
5584:
5583:
5578:
5569:
5567:
5556:
5548:
5547:
5545:
5544:
5539:
5534:
5528:
5527:
5522:
5517:
5507:
5506:
5505:
5504:
5499:
5494:
5489:
5484:
5479:
5467:
5466:
5465:
5464:
5459:
5454:
5449:
5444:
5439:
5433:
5428:
5423:
5418:
5413:
5408:
5403:
5398:
5393:
5388:
5383:
5378:
5373:
5368:
5363:
5358:
5353:
5348:
5343:
5338:
5333:
5328:
5323:
5318:
5313:
5308:
5296:
5295:
5294:
5293:
5288:
5283:
5278:
5273:
5268:
5263:
5258:
5253:
5241:
5239:
5231:
5230:
5228:
5227:
5222:
5220:Signal peptide
5216:
5214:
5208:
5207:
5204:
5203:
5201:
5200:
5199:
5198:
5193:
5183:
5178:
5172:
5170:
5166:
5165:
5163:
5162:
5161:
5160:
5155:
5150:
5145:
5141:
5137:
5133:
5123:
5122:
5121:
5120:
5115:
5110:
5105:
5100:
5095:
5090:
5085:
5080:
5075:
5070:
5065:
5060:
5049:
5048:
5047:
5046:
5041:
5036:
5031:
5026:
5021:
5016:
5011:
5006:
5001:
4996:
4991:
4986:
4981:
4976:
4971:
4966:
4961:
4956:
4945:
4944:
4935:
4930:
4925:
4919:
4917:
4903:
4892:
4891:
4886:
4884:
4883:
4876:
4869:
4861:
4852:
4851:
4848:
4847:
4845:
4844:
4838:
4836:
4815:
4808:
4807:
4804:
4803:
4801:
4800:
4794:
4792:
4778:
4777:
4775:
4774:
4767:
4765:
4751:
4750:
4748:
4747:
4740:
4738:
4721:
4714:
4713:
4710:
4709:
4707:
4706:
4700:
4698:
4688:
4687:
4685:
4684:
4678:
4676:
4666:
4665:
4663:
4662:
4656:
4654:
4644:
4643:
4641:
4640:
4635:
4633:Disulfide bond
4629:
4627:
4614:
4607:
4606:
4603:
4602:
4600:
4599:
4593:
4591:
4585:
4584:
4582:
4581:
4576:
4569:
4567:
4561:
4560:
4558:
4557:
4551:
4549:
4543:
4542:
4540:
4539:
4534:
4529:
4527:Citrullination
4523:
4521:
4515:
4514:
4512:
4511:
4506:
4504:Propionylation
4501:
4496:
4491:
4486:
4481:
4476:
4474:-glycosylation
4468:
4463:
4458:
4453:
4448:
4446:Ubiquitination
4443:
4438:
4433:
4428:
4423:
4417:
4415:
4409:
4408:
4406:
4405:
4399:
4397:
4391:
4390:
4388:
4387:
4382:
4376:
4374:
4368:
4367:
4365:
4364:
4359:
4354:
4349:
4344:
4338:
4336:
4330:
4329:
4327:
4326:
4321:
4315:
4313:
4307:
4306:
4304:
4303:
4298:
4296:Palmitoylation
4292:
4290:
4284:
4283:
4281:
4280:
4278:Detyrosination
4275:
4270:
4268:Flavin linkage
4265:
4260:
4255:
4250:
4245:
4240:
4234:
4232:
4226:
4225:
4223:
4222:
4217:
4209:
4204:
4199:
4193:
4191:
4178:
4171:
4170:
4168:
4167:
4165:Detyrosination
4162:
4157:
4152:
4146:
4144:
4138:
4137:
4135:
4134:
4131:Myristoylation
4128:
4123:
4118:
4113:
4108:
4102:
4100:
4094:
4093:
4091:
4090:
4088:N–O acyl shift
4085:
4080:
4075:
4070:
4064:
4062:
4058:
4057:
4048:
4046:
4045:
4038:
4031:
4023:
4014:
4013:
4011:
4010:
4009:
4008:
4003:
3998:
3988:
3983:
3978:
3973:
3972:
3971:
3966:
3961:
3951:
3946:
3940:
3938:
3934:
3933:
3931:
3930:
3925:
3920:
3914:
3912:
3908:
3907:
3905:
3904:
3899:
3894:
3889:
3884:
3879:
3873:
3871:
3867:
3866:
3861:
3859:
3858:
3851:
3844:
3836:
3830:
3829:
3824:
3819:
3814:
3801:
3800:
3795:
3785:
3772:
3771:
3764:
3763:External links
3761:
3759:
3758:
3740:
3719:(17): 2345–7.
3713:Bioinformatics
3699:
3650:
3599:
3550:
3489:
3440:
3391:
3362:(1): 499–501.
3342:
3293:
3241:
3192:
3140:
3111:
3054:
3031:(4): 368–375.
3011:
2976:
2937:
2888:
2858:
2825:
2790:
2749:
2740:
2686:
2645:
2618:(4938): 83–5.
2602:
2567:
2518:
2469:
2428:
2421:
2403:
2360:
2307:
2278:(2): 389–406.
2256:
2249:
2225:
2162:
2143:
2111:
2109:
2106:
2105:
2104:
2099:
2092:
2089:
2088:
2087:
2084:
2077:
2063:
2050:
2049:
2008:
2006:
1999:
1993:
1990:
1989:
1988:
1982:
1976:
1965:
1962:
1961:
1960:
1959:-GlcNAc sites.
1946:
1932:
1926:
1920:
1914:
1908:
1902:
1896:
1888:
1885:
1871:
1868:
1865:
1864:
1858:
1855:
1849:
1848:
1842:
1839:
1833:
1832:
1822:
1819:
1813:
1812:
1798:
1795:
1789:
1788:
1774:
1771:
1765:
1764:
1759:
1756:
1750:
1749:
1747:
1744:
1738:
1737:
1731:
1728:
1722:
1721:
1699:ubiquitylation
1692:
1689:
1683:
1682:
1680:
1677:
1671:
1670:
1668:
1665:
1659:
1658:
1653:
1650:
1644:
1643:
1638:(N-terminus),
1636:Myristoylation
1632:
1629:
1623:
1622:
1618:(N-terminus),
1612:
1609:
1603:
1602:
1580:
1577:
1571:
1570:
1566:(N-terminus),
1560:palmitoylation
1553:
1550:
1544:
1543:
1534:
1531:
1525:
1524:
1511:
1508:
1502:
1501:
1491:
1488:
1482:
1481:
1475:
1472:
1466:
1465:
1462:
1459:
1449:
1446:
1443:
1442:
1437:
1433:
1432:
1427:
1423:
1422:
1420:Ubiquitylation
1417:
1413:
1412:
1407:
1403:
1402:
1397:
1393:
1392:
1387:
1383:
1382:
1377:
1373:
1372:
1367:
1363:
1362:
1357:
1353:
1352:
1347:
1343:
1342:
1339:
1328:
1325:
1323:
1320:
1319:
1318:
1308:
1307:
1306:
1295:
1293:opioid peptide
1281:
1266:
1260:
1254:
1248:
1236:
1233:
1232:
1231:
1198:
1177:
1163:citrullination
1158:
1155:
1154:
1153:
1144:
1137:
1128:
1118:
1111:ubiquitination
1106:
1103:
1102:
1101:
1091:
1084:
1081:
1073:
1067:
1066:
1065:
1054:
1048:
1045:Isocyanic acid
1038:
1027:
1021:
1020:
1019:
1009:
1001:addition of a
996:
978:
964:
943:
941:-nitrosylation
935:
927:
921:
919:propionylation
916:
915:
914:
907:
877:
825:
816:
806:
800:
795:
794:
793:
783:
778:, addition of
720:
711:
706:
705:
704:
703:
702:
688:
674:
652:
646:
645:
644:
605:
604:
603:
598:
544:
541:
540:
539:
521:
507:
497:
485:
482:
481:
480:
471:
461:
448:
434:
430:
418:
415:
414:
413:
403:
402:
401:
396:
372:isoprenylation
369:
368:saturated acid
365:
358:palmitoylation
355:
354:saturated acid
351:
340:myristoylation
335:
332:
330:
324:
322:
319:
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
6161:
6150:
6147:
6145:
6142:
6140:
6137:
6135:
6132:
6130:
6127:
6126:
6124:
6114:
6109:
6104:
6100:
6087:
6086:Jacques Monod
6084:
6082:
6079:
6078:
6076:
6072:
6064:
6061:
6059:
6056:
6055:
6054:
6051:
6049:
6048:Translational
6046:
6042:
6039:
6037:
6034:
6032:
6029:
6028:
6027:
6024:
6022:
6019:
6015:
6012:
6010:
6007:
6006:
6005:
6002:
5998:
5995:
5994:
5993:
5990:
5989:
5987:
5985:
5981:
5971:
5968:
5966:
5963:
5961:
5958:
5956:
5953:
5952:
5950:
5946:
5940:
5937:
5935:
5932:
5930:
5927:
5926:
5924:
5920:
5917:
5915:
5911:
5901:
5898:
5896:
5893:
5891:
5888:
5886:
5883:
5881:
5878:
5877:
5875:
5873:
5869:
5863:
5860:
5858:
5855:
5853:
5850:
5849:
5847:
5843:
5837:
5834:
5832:
5829:
5827:
5824:
5823:
5821:
5817:
5814:
5812:
5811:Transcription
5808:
5800:
5797:
5795:
5792:
5790:
5787:
5786:
5785:
5782:
5778:
5774:
5770:
5767:
5766:
5765:
5764:Central dogma
5762:
5760:
5757:
5756:
5754:
5752:
5746:
5742:
5735:
5730:
5728:
5723:
5721:
5716:
5715:
5712:
5696:
5693:
5692:
5689:
5686:
5684:
5681:
5679:
5676:
5674:
5671:
5669:
5666:
5664:
5661:
5658:
5654:
5651:
5649:
5646:
5644:
5641:
5639:
5636:
5635:
5633:
5629:
5621:
5618:
5616:
5613:
5611:
5608:
5606:
5603:
5602:
5600:
5599:
5594:
5591:
5590:
5588:
5587:
5582:
5579:
5577:
5574:
5573:
5571:
5570:
5568:
5566:(SUMOylation)
5564:
5560:
5557:
5553:
5549:
5543:
5540:
5538:
5535:
5533:
5530:
5529:
5526:
5523:
5521:
5518:
5516:
5512:
5509:
5508:
5503:
5500:
5498:
5495:
5493:
5490:
5488:
5485:
5483:
5480:
5478:
5475:
5474:
5473:
5469:
5468:
5463:
5460:
5458:
5455:
5453:
5450:
5448:
5445:
5443:
5440:
5437:
5434:
5432:
5429:
5427:
5424:
5422:
5419:
5417:
5414:
5412:
5409:
5407:
5404:
5402:
5399:
5397:
5394:
5392:
5389:
5387:
5384:
5382:
5379:
5377:
5374:
5372:
5369:
5367:
5364:
5362:
5359:
5357:
5354:
5352:
5349:
5347:
5344:
5342:
5339:
5337:
5334:
5332:
5329:
5327:
5324:
5322:
5319:
5317:
5314:
5312:
5309:
5307:
5304:
5303:
5302:
5298:
5297:
5292:
5289:
5287:
5284:
5282:
5279:
5277:
5274:
5272:
5269:
5267:
5264:
5262:
5259:
5257:
5254:
5252:
5249:
5248:
5247:
5243:
5242:
5240:
5236:
5232:
5226:
5223:
5221:
5218:
5217:
5215:
5213:
5209:
5197:
5194:
5192:
5189:
5188:
5187:
5184:
5182:
5179:
5177:
5174:
5173:
5171:
5167:
5159:
5156:
5154:
5151:
5149:
5146:
5144:
5138:
5136:
5130:
5129:
5128:
5125:
5124:
5119:
5116:
5114:
5111:
5109:
5106:
5104:
5101:
5099:
5096:
5094:
5091:
5089:
5086:
5084:
5081:
5079:
5076:
5074:
5071:
5069:
5066:
5064:
5061:
5059:
5056:
5055:
5054:
5051:
5050:
5045:
5042:
5040:
5037:
5035:
5032:
5030:
5027:
5025:
5022:
5020:
5017:
5015:
5012:
5010:
5007:
5005:
5002:
5000:
4997:
4995:
4992:
4990:
4987:
4985:
4982:
4980:
4977:
4975:
4972:
4970:
4967:
4965:
4962:
4960:
4957:
4955:
4952:
4951:
4950:
4947:
4946:
4943:
4939:
4936:
4934:
4931:
4929:
4926:
4924:
4921:
4920:
4918:
4916:
4911:
4907:
4904:
4902:
4897:
4893:
4889:
4882:
4877:
4875:
4870:
4868:
4863:
4862:
4859:
4843:
4840:
4839:
4837:
4835:
4831:
4827:
4823:
4819:
4816:
4814:
4809:
4799:
4796:
4795:
4793:
4791:
4787:
4783:
4779:
4773:(chromophore)
4772:
4769:
4768:
4766:
4764:
4760:
4756:
4752:
4746:(chromophore)
4745:
4742:
4741:
4739:
4737:
4733:
4729:
4725:
4722:
4720:
4715:
4705:
4702:
4701:
4699:
4697:
4693:
4689:
4683:
4680:
4679:
4677:
4675:
4671:
4667:
4661:
4658:
4657:
4655:
4653:
4652:Hydroxylysine
4649:
4645:
4639:
4636:
4634:
4631:
4630:
4628:
4626:
4622:
4618:
4615:
4613:
4608:
4598:
4595:
4594:
4592:
4590:
4586:
4580:
4579:Adenylylation
4577:
4574:
4571:
4570:
4568:
4566:
4562:
4556:
4555:Hydroxylation
4553:
4552:
4550:
4548:
4544:
4538:
4535:
4533:
4530:
4528:
4525:
4524:
4522:
4520:
4516:
4510:
4507:
4505:
4502:
4500:
4497:
4495:
4494:Succinylation
4492:
4490:
4489:Carbamylation
4487:
4485:
4482:
4480:
4477:
4475:
4473:
4469:
4467:
4464:
4462:
4459:
4457:
4454:
4452:
4449:
4447:
4444:
4442:
4441:Hydroxylation
4439:
4437:
4436:Adenylylation
4434:
4432:
4429:
4427:
4424:
4422:
4419:
4418:
4416:
4414:
4410:
4404:
4401:
4400:
4398:
4396:
4392:
4386:
4385:Glycosylation
4383:
4381:
4378:
4377:
4375:
4373:
4369:
4363:
4360:
4358:
4355:
4353:
4350:
4348:
4345:
4343:
4342:Carboxylation
4340:
4339:
4337:
4335:
4331:
4325:
4322:
4320:
4317:
4316:
4314:
4312:
4308:
4302:
4299:
4297:
4294:
4293:
4291:
4289:
4285:
4279:
4276:
4274:
4271:
4269:
4266:
4264:
4263:Adenylylation
4261:
4259:
4256:
4254:
4251:
4249:
4246:
4244:
4241:
4239:
4236:
4235:
4233:
4231:
4227:
4221:
4218:
4216:
4214:
4210:
4208:
4207:Glycosylation
4205:
4203:
4200:
4198:
4195:
4194:
4192:
4190:
4186:
4182:
4179:
4177:
4172:
4166:
4163:
4161:
4160:O-methylation
4158:
4156:
4153:
4151:
4148:
4147:
4145:
4143:
4139:
4132:
4129:
4127:
4124:
4122:
4119:
4117:
4114:
4112:
4111:Carbamylation
4109:
4107:
4104:
4103:
4101:
4099:
4095:
4089:
4086:
4084:
4081:
4079:
4076:
4074:
4071:
4069:
4066:
4065:
4063:
4059:
4055:
4051:
4044:
4039:
4037:
4032:
4030:
4025:
4024:
4021:
4007:
4004:
4002:
3999:
3997:
3994:
3993:
3992:
3989:
3987:
3984:
3982:
3981:Chromoprotein
3979:
3977:
3974:
3970:
3967:
3965:
3962:
3960:
3957:
3956:
3955:
3952:
3950:
3947:
3945:
3942:
3941:
3939:
3935:
3929:
3926:
3924:
3921:
3919:
3916:
3915:
3913:
3909:
3903:
3900:
3898:
3895:
3893:
3890:
3888:
3885:
3883:
3880:
3878:
3875:
3874:
3872:
3868:
3864:
3857:
3852:
3850:
3845:
3843:
3838:
3837:
3834:
3828:
3825:
3823:
3820:
3818:
3815:
3813:
3809:
3806:
3805:
3804:
3799:
3796:
3793:
3789:
3786:
3784:
3781:
3780:
3779:
3777:
3770:
3767:
3766:
3762:
3754:
3750:
3744:
3741:
3736:
3732:
3727:
3722:
3718:
3714:
3710:
3703:
3700:
3695:
3691:
3686:
3681:
3677:
3673:
3669:
3665:
3661:
3654:
3651:
3646:
3642:
3637:
3632:
3627:
3622:
3618:
3614:
3610:
3603:
3600:
3595:
3591:
3586:
3581:
3577:
3573:
3569:
3565:
3561:
3554:
3551:
3546:
3542:
3537:
3532:
3528:
3524:
3520:
3516:
3512:
3508:
3504:
3502:
3493:
3490:
3485:
3481:
3476:
3471:
3467:
3463:
3459:
3455:
3451:
3444:
3441:
3436:
3432:
3427:
3422:
3418:
3414:
3410:
3406:
3402:
3395:
3392:
3387:
3383:
3378:
3373:
3369:
3365:
3361:
3357:
3353:
3346:
3343:
3338:
3334:
3329:
3324:
3320:
3316:
3312:
3308:
3304:
3297:
3294:
3289:
3285:
3280:
3275:
3271:
3267:
3264:(2): 453–63.
3263:
3259:
3255:
3248:
3246:
3242:
3237:
3233:
3228:
3223:
3219:
3215:
3211:
3207:
3203:
3196:
3193:
3188:
3184:
3179:
3174:
3170:
3166:
3162:
3158:
3154:
3147:
3145:
3141:
3130:on 2012-08-30
3129:
3125:
3121:
3115:
3112:
3107:
3103:
3098:
3093:
3089:
3085:
3081:
3077:
3073:
3069:
3065:
3058:
3055:
3049:
3044:
3039:
3034:
3030:
3026:
3022:
3015:
3012:
3007:
3003:
2999:
2995:
2992:(3): 108–14.
2991:
2987:
2980:
2977:
2972:
2968:
2964:
2960:
2956:
2952:
2948:
2941:
2938:
2933:
2929:
2924:
2919:
2915:
2911:
2908:(4): 455–60.
2907:
2903:
2899:
2892:
2889:
2885:
2884:0-9545232-8-8
2881:
2877:
2873:
2870:
2869:
2862:
2859:
2853:
2848:
2844:
2840:
2836:
2829:
2826:
2821:
2817:
2813:
2809:
2806:(4): 229–37.
2805:
2801:
2794:
2791:
2786:
2782:
2777:
2772:
2768:
2764:
2760:
2753:
2750:
2744:
2741:
2736:
2732:
2727:
2722:
2718:
2714:
2711:(2): 382–92.
2710:
2706:
2702:
2695:
2693:
2691:
2687:
2682:
2678:
2673:
2668:
2664:
2660:
2656:
2649:
2646:
2641:
2637:
2633:
2629:
2625:
2621:
2617:
2613:
2606:
2603:
2598:
2594:
2590:
2586:
2582:
2578:
2571:
2568:
2563:
2559:
2554:
2549:
2545:
2541:
2537:
2533:
2529:
2522:
2519:
2514:
2510:
2505:
2500:
2496:
2492:
2489:(10): 667–9.
2488:
2484:
2480:
2473:
2470:
2465:
2461:
2456:
2451:
2447:
2443:
2439:
2432:
2429:
2424:
2422:9780974707730
2418:
2414:
2407:
2404:
2399:
2395:
2391:
2387:
2383:
2379:
2375:
2371:
2364:
2361:
2356:
2352:
2347:
2342:
2337:
2332:
2328:
2324:
2323:
2318:
2311:
2308:
2303:
2299:
2294:
2289:
2285:
2281:
2277:
2273:
2272:
2267:
2260:
2257:
2252:
2246:
2242:
2241:
2236:
2229:
2226:
2221:
2217:
2212:
2207:
2203:
2199:
2195:
2191:
2187:
2183:
2182:
2177:
2173:
2166:
2163:
2158:
2154:
2150:
2146:
2144:9780471214953
2140:
2136:
2135:
2130:
2126:
2122:
2116:
2113:
2107:
2103:
2100:
2098:
2095:
2094:
2090:
2085:
2082:
2078:
2076:
2072:
2071:transcription
2068:
2064:
2062:
2058:
2054:
2053:
2046:
2043:
2035:
2025:
2021:
2015:
2014:
2009:This section
2007:
2003:
1998:
1997:
1992:Case examples
1991:
1986:
1983:
1980:
1977:
1974:
1971:
1970:
1969:
1963:
1958:
1954:
1952:
1947:
1941:
1936:
1933:
1930:
1927:
1924:
1921:
1918:
1915:
1912:
1909:
1906:
1903:
1900:
1899:ProteomeScout
1897:
1894:
1891:
1890:
1886:
1884:
1876:
1869:
1863:(N-terminus)
1862:
1861:N-acetylation
1859:
1856:
1854:
1851:
1850:
1846:
1843:
1840:
1838:
1835:
1834:
1831:
1827:
1823:
1820:
1818:
1815:
1814:
1811:(N-terminus)
1810:
1809:N-acetylation
1806:
1802:
1799:
1796:
1794:
1791:
1790:
1787:(N-terminus)
1786:
1785:N-acetylation
1782:
1778:
1775:
1772:
1770:
1767:
1766:
1763:
1762:hydroxylation
1760:
1757:
1755:
1752:
1751:
1748:
1745:
1743:
1742:Phenylalanine
1740:
1739:
1735:
1734:N-acetylation
1732:
1729:
1727:
1724:
1723:
1720:
1716:
1712:
1711:hydroxylation
1708:
1704:
1700:
1696:
1693:
1690:
1688:
1685:
1684:
1681:
1678:
1676:
1673:
1672:
1669:
1666:
1664:
1661:
1660:
1657:
1654:
1651:
1649:
1646:
1645:
1642:(N-terminus)
1641:
1640:N-acetylation
1637:
1633:
1630:
1628:
1625:
1624:
1621:
1617:
1613:
1610:
1608:
1607:Glutamic acid
1605:
1604:
1601:
1597:
1593:
1589:
1588:Glutamic acid
1585:
1581:
1578:
1576:
1573:
1572:
1569:
1565:
1564:N-acetylation
1561:
1557:
1554:
1551:
1549:
1546:
1545:
1542:
1538:
1537:isomerization
1535:
1532:
1530:
1529:Aspartic acid
1527:
1526:
1523:
1519:
1515:
1512:
1509:
1507:
1504:
1503:
1500:
1496:
1492:
1489:
1487:
1484:
1483:
1480:(N-terminus)
1479:
1478:N-acetylation
1476:
1473:
1471:
1468:
1467:
1464:Modification
1463:
1460:
1457:
1456:
1453:
1447:
1441:
1438:
1435:
1434:
1431:
1428:
1425:
1424:
1421:
1418:
1415:
1414:
1411:
1408:
1405:
1404:
1401:
1398:
1395:
1394:
1391:
1390:Hydroxylation
1388:
1385:
1384:
1381:
1378:
1375:
1374:
1371:
1368:
1365:
1364:
1361:
1358:
1355:
1354:
1351:
1348:
1345:
1344:
1341:Modification
1340:
1337:
1336:
1333:
1326:
1321:
1316:
1312:
1309:
1304:
1300:
1296:
1294:
1290:
1286:
1282:
1280:
1276:
1272:
1271:
1270:
1267:
1264:
1261:
1258:
1255:
1252:
1249:
1246:
1242:
1239:
1238:
1234:
1230:
1226:
1222:
1218:
1217:phosphoserine
1214:
1210:
1206:
1202:
1201:eliminylation
1199:
1197:
1196:aspartic acid
1193:
1189:
1188:glutamic acid
1185:
1181:
1178:
1176:
1172:
1168:
1164:
1161:
1160:
1156:
1152:
1148:
1145:
1142:
1138:
1136:
1132:
1129:
1126:
1122:
1119:
1116:
1112:
1109:
1108:
1104:
1099:
1095:
1092:
1089:
1085:
1082:
1079:
1078:biotinylation
1076:
1075:
1072:
1068:
1063:
1059:
1055:
1052:
1051:carbonylation
1049:
1046:
1042:
1041:carbamylation
1039:
1036:
1033:
1032:
1031:
1026:
1022:
1017:
1013:
1010:
1008:
1004:
1000:
999:succinylation
997:
994:
990:
986:
982:
979:
976:
972:
968:
965:
962:
958:
954:
951:
947:
944:
942:
940:
936:
934:
932:
928:
925:
924:pyroglutamate
922:
920:
917:
912:
908:
905:
901:
897:
894:-linked), or
893:
889:
885:
881:
880:adenylylation
878:
875:
871:
868:-linked), or
867:
863:
859:
855:
851:
847:
844:
843:
841:
837:
833:
829:
826:
824:
820:
817:
814:
813:thyroglobulin
810:
807:
804:
803:hydroxylation
801:
799:
796:
792:
788:
784:
781:
777:
775:
771:
770:
768:
764:
760:
756:
752:
748:
744:
743:hydroxylysine
740:
736:
732:
728:
724:
723:glycosylation
721:
719:
716:dependent on
715:
712:
710:
707:
700:
696:
692:
689:
686:
682:
681:glutamic acid
678:
675:
672:
668:
665:
664:
662:
659:
658:
656:
653:
647:
642:
641:demethylation
638:
634:
630:
626:
623:
622:
621:
617:
613:
609:
606:
602:
599:
596:
595:deacetylation
592:
588:
584:
580:
577:
576:
574:
570:
566:
562:
558:
554:
550:
547:
546:
542:
537:
533:
529:
525:
522:
519:
515:
511:
508:
505:
501:
498:
495:
491:
488:
487:
483:
478:
475:
472:
469:
465:
462:
460:
456:
452:
449:
446:
442:
438:
435:
428:
424:
421:
420:
416:
411:
407:
404:
400:
397:
395:
394:farnesylation
392:
391:
389:
385:
381:
377:
373:
370:
363:
359:
356:
349:
345:
341:
338:
337:
333:
329:
325:
320:
318:
316:
312:
308:
304:
299:
297:
293:
289:
285:
281:
277:
273:
269:
265:
261:
258:
254:
250:
246:
242:
238:
234:
230:
226:
222:
217:
215:
211:
210:Carbonylation
207:
202:
200:
196:
193:
189:
185:
181:
177:
173:
172:peptide bonds
168:
166:
165:cell membrane
162:
158:
154:
150:
149:glycosylation
146:
142:
138:
134:
130:
126:
122:
118:
114:
111:
106:
104:
100:
96:
92:
88:
85:
81:
77:
73:
69:
65:
61:
57:
53:
45:
41:
37:
32:
19:
6149:Cell biology
6063:irreversible
5969:
5948:Key elements
5845:Key elements
5759:Genetic code
5749:Introduction
5563:SUMO protein
4887:
4509:Butyrylation
4471:
4212:
4083:Racemization
4068:Peptide bond
3881:
3802:
3773:
3752:
3743:
3716:
3712:
3702:
3667:
3663:
3653:
3616:
3612:
3602:
3567:
3563:
3553:
3510:
3506:
3500:
3492:
3457:
3453:
3443:
3408:
3404:
3394:
3359:
3355:
3345:
3310:
3306:
3296:
3261:
3257:
3209:
3205:
3195:
3160:
3156:
3132:. Retrieved
3128:the original
3123:
3114:
3071:
3067:
3057:
3028:
3024:
3014:
2989:
2985:
2979:
2954:
2950:
2940:
2905:
2901:
2891:
2867:
2861:
2842:
2838:
2828:
2803:
2799:
2793:
2766:
2762:
2752:
2743:
2708:
2704:
2662:
2658:
2648:
2615:
2611:
2605:
2583:(3): 112–5.
2580:
2576:
2570:
2535:
2531:
2521:
2486:
2482:
2472:
2445:
2441:
2431:
2412:
2406:
2373:
2369:
2363:
2326:
2320:
2310:
2275:
2269:
2259:
2239:
2228:
2185:
2179:
2165:
2157:the original
2133:
2129:Voet, Donald
2115:
2038:
2032:January 2016
2029:
2018:Please help
2013:verification
2010:
1967:
1956:
1950:
1881:
1451:
1330:
1269:racemization
1263:isoaspartate
1166:
1135:Nedd protein
1125:SUMO protein
1070:
1056:spontaneous
1029:
1024:
993:cysteic acid
980:
966:
952:
949:
945:
938:
930:
903:
891:
873:
865:
839:
834:-linked) or
831:
798:malonylation
779:
773:
763:glycoprotein
709:butyrylation
667:arginylation
614:group, e.g.
571:-acylation (
568:
563:-acylation (
560:
555:-acylation (
552:
474:retinylidene
382:group (e.g.
327:
300:
294:and at some
268:carboxylates
218:
203:
169:
145:carbohydrate
107:
59:
55:
49:
5914:Translation
5751:to genetics
5657:neddylation
4923:Hsp10/GroES
4915:Chaperonins
4573:Diphthamide
4532:Methylation
4499:Lactylation
4461:Deamination
4451:Sumoylation
4426:Acetylation
4421:Methylation
4380:Deamidation
4352:Methylation
4301:Prenylation
4126:Methylation
4116:Formylation
4106:Acetylation
4078:Proteolysis
4001:Phytochrome
3991:Biliprotein
3790:— from the
3499:"The human
3460:: 307–319.
2839:J Biol Chem
2376:(1): 51–6.
2370:Amino Acids
1713:leading to
1707:methylation
1703:SUMOylation
1695:acetylation
1514:deamidation
1499:methylation
1400:Methylation
1360:Acetylation
1247:amino acids
1221:dehydration
1180:deamidation
1167:deimination
1131:neddylation
1121:SUMOylation
987:group of a
973:group of a
959:group of a
625:methylation
601:formylation
579:acetylation
524:beta-Lysine
520:(archaeal))
490:diphthamide
477:Schiff base
457:bonds with
423:lipoylation
376:prenylation
342:(a type of
292:methionines
221:nucleophile
141:prokaryotic
125:amino acids
113:side chains
99:prohormones
6123:Categories
6058:reversible
6021:lac operon
5997:imprinting
5992:Epigenetic
5984:Regulation
5939:Eukaryotic
5885:5' capping
5836:Eukaryotic
4949:Hsp40/DnaJ
4896:Chaperones
4696:Tryptophan
4692:Tryptophan
4648:Methionine
4589:Tryptophan
4372:Asparagine
4142:C terminus
4098:N terminus
3928:Proteasome
3911:Structures
3619:(1): 370.
3134:2011-07-22
3074:(90): 90.
2957:: 651–75.
2172:Floudas CA
2153:1280801548
2108:References
1826:kynurenine
1817:Tryptophan
1726:Methionine
1663:Isoleucine
1506:Asparagine
1495:citrulline
1458:Amino Acid
1338:Frequency
1322:Statistics
1303:deltorphin
1299:methionine
1289:dermorphin
1192:asparagine
1175:citrulline
1147:pupylation
1094:pegylation
809:iodination
789:, PSA, to
759:tryptophan
735:asparagine
661:amino acid
608:alkylation
587:N-terminus
573:thioesters
468:coenzyme A
406:glypiation
380:isoprenoid
284:asparagine
227:groups of
214:biomarkers
199:propeptide
180:methionine
176:propeptide
161:lipidation
137:eukaryotic
110:amino acid
95:signalling
70:following
5929:Bacterial
5826:Bacterial
5235:Ubiquitin
5181:Clusterin
4842:Desmosine
4755:Histidine
4575:formation
4565:Histidine
4484:Glycation
4431:Acylation
4395:Glutamine
4334:Glutamate
4311:Aspartate
4253:Sulfation
4189:Threonine
4150:Amidation
4121:Glycation
4006:Lipocalin
3870:Processes
3513:(1): 25.
1857:Val or V
1845:sulfation
1841:Tyr or Y
1821:Trp or W
1797:Thr or T
1793:Threonine
1773:Ser or S
1758:Pro or P
1746:Phe or F
1730:Met or M
1691:Lys or K
1679:Leu or L
1667:Ile or I
1652:His or H
1648:Histidine
1631:Gly or G
1611:Glu or E
1579:Gln or Q
1575:Glutamine
1556:disulfide
1552:Cys or C
1533:Asp or D
1510:Asn or N
1490:Arg or R
1474:Ala or A
1440:Sulfation
1380:Amidation
1291:, a frog
1225:threonine
1184:glutamine
1088:disulfide
1035:glycation
1012:sulfation
1005:group to
926:formation
896:histidine
870:histidine
858:threonine
850:phosphate
767:glycation
751:threonine
718:Vitamin K
663:addition
649:amidation
549:acylation
479:formation
459:cysteines
455:thioether
362:palmitate
348:myristate
344:acylation
317:section.
276:glutamate
272:aspartate
253:histidine
243:forms of
233:threonine
84:translate
80:ribosomes
62:) is the
6041:microRNA
5955:Ribosome
5934:Archaeal
5890:Splicing
5862:Promoter
5831:Archaeal
5775: →
5771: →
5515:Ataxin 3
4830:Allysine
4826:Allysine
4822:Allysine
4759:Tyrosine
4732:Tyrosine
4674:Tyrosine
4625:Cysteine
4621:Cysteine
4519:Arginine
4288:Cysteine
4230:Tyrosine
3959:Globulin
3897:Proteome
3863:Proteins
3794:database
3735:17623706
3694:30215764
3645:25431162
3594:34279596
3545:33479245
3484:28458782
3386:12520062
3337:19858104
3288:22159132
3236:25514926
3187:16381945
3106:22034591
3006:19233656
2971:18173373
2932:12600939
2872:Archived
2820:21055949
2785:21768218
2735:23329793
2681:11110799
2562:29405707
2532:Chem Rev
2513:21841797
2398:23819101
2390:17021655
2355:18445586
2302:16796807
2220:22034591
2131:(2006).
2091:See also
2067:histones
1837:Tyrosine
1715:allysine
1548:Cysteine
1486:Arginine
1245:cysteine
1171:arginine
1115:covalent
1071:in vitro
1016:tyrosine
1003:succinyl
989:cysteine
975:cysteine
961:cysteine
906:-linked)
888:tyrosine
884:adenylyl
876:-linked)
862:tyrosine
821:such as
755:tyrosine
739:cysteine
731:arginine
727:glycosyl
637:arginine
510:hypusine
439:moiety (
384:farnesol
264:cysteine
257:thiolate
249:arginine
237:tyrosine
225:hydroxyl
188:cysteine
103:hormones
82:, which
68:proteins
64:covalent
6113:Biology
5794:RNA→DNA
5789:RNA→RNA
5777:Protein
5438:(CDC34)
4790:Glycine
4782:Alanine
4763:Glycine
4736:Glycine
4547:Proline
4215:-GlcNAc
4061:General
3969:Albumin
3964:Edestin
3685:6324025
3636:4256751
3585:8288053
3536:7820439
3515:Bibcode
3475:5397102
3435:2914561
3426:5753337
3328:2808866
3279:3804167
3227:4383998
3178:1347446
3097:3201773
3076:Bibcode
3048:8964421
2726:4340704
2640:1967194
2620:Bibcode
2612:Science
2597:2057999
2553:6609103
2504:3177975
2464:2569467
2346:2494933
2293:3933129
2211:3201773
2190:Bibcode
2079:PTM of
2065:PTM of
2061:insulin
1985:Chimera
1979:AWESOME
1935:Uniprot
1923:iPTMnet
1911:PROSITE
1754:Proline
1675:Leucine
1627:Glycine
1470:Alanine
1461:Abbrev.
1315:inteins
1285:alanine
1025:in vivo
995:residue
977:residue
963:residue
776:-GlcNAc
699:tubulin
695:glycine
551:, e.g.
427:lipoate
328:in vivo
288:glycans
195:insulin
192:hormone
76:enzymes
36:insulin
6099:Portal
5482:Cullin
4834:Lysine
4786:Serine
4728:Serine
4670:Lysine
4413:Lysine
4185:Serine
3778:copy)
3733:
3692:
3682:
3643:
3633:
3592:
3582:
3543:
3533:
3482:
3472:
3433:
3423:
3384:
3377:165485
3374:
3335:
3325:
3286:
3276:
3234:
3224:
3185:
3175:
3104:
3094:
3045:
3004:
2969:
2930:
2923:195994
2920:
2882:
2818:
2783:
2733:
2723:
2679:
2638:
2595:
2560:
2550:
2511:
2501:
2462:
2419:
2396:
2388:
2353:
2343:
2300:
2290:
2247:
2218:
2208:
2188:: 90.
2151:
2141:
1853:Valine
1769:Serine
1687:Lysine
1596:lysine
1346:58383
1275:serine
1229:serine
1205:alkene
1113:, the
1007:lysine
900:lysine
860:, and
854:serine
747:serine
633:lysine
629:methyl
616:methyl
591:lysine
583:acetyl
565:amides
557:esters
451:heme C
437:flavin
309:, and
266:; the
255:; the
251:, and
245:lysine
239:; the
235:, and
229:serine
5922:Types
5819:Types
5673:ATG12
5663:FAT10
5653:NEDD8
5638:ISG15
5631:Other
5620:PIAS4
5615:PIAS3
5610:PIAS2
5605:PIAS1
5555:(UBL)
5537:BIRC6
5497:FANCL
5169:Other
5158:TRAP1
5127:Hsp90
5053:Hsp70
4942:GroEL
4938:HSP60
4933:Hsp47
4928:Hsp27
4133:(Gly)
3937:Types
3570:: 1.
2394:S2CID
1973:PyMOL
1964:Tools
1929:dbPTM
1917:RESID
1598:by a
1406:1133
1396:1523
1386:1619
1376:2844
1366:5526
1356:6751
1219:, or
1165:, or
1141:ISG15
985:thiol
971:thiol
957:thiol
757:, or
655:amide
620:ethyl
612:alkyl
536:aIF5A
532:eIF5A
518:aIF5A
514:eIF5A
504:eEF1α
364:, a C
350:, a C
280:amide
260:anion
241:amine
186:from
157:lipid
89:into
5688:UBL5
5678:FUB1
5668:ATG8
5648:UFM1
5643:URM1
5593:UBC9
5581:SAE2
5576:SAE1
5542:UFC1
5532:ATG3
5525:CYLD
5520:USP6
5502:UBR1
5492:MDM2
5291:SAE1
5286:NAE1
5281:ATG7
5276:UBA7
5271:UBA6
5266:UBA5
5261:UBA3
5256:UBA2
5251:UBA1
5196:SMN2
5191:SMN1
4052:and
3792:dcGO
3731:PMID
3690:PMID
3641:PMID
3590:PMID
3568:2021
3541:PMID
3480:PMID
3431:PMID
3382:PMID
3333:PMID
3284:PMID
3232:PMID
3183:PMID
3102:PMID
3002:PMID
2967:PMID
2928:PMID
2880:ISBN
2816:PMID
2781:PMID
2731:PMID
2677:PMID
2636:PMID
2593:PMID
2558:PMID
2509:PMID
2460:PMID
2417:ISBN
2386:PMID
2351:PMID
2298:PMID
2245:ISBN
2216:PMID
2149:OCLC
2139:ISBN
1949:The
1436:504
1426:826
1416:878
1227:and
1215:and
898:and
791:NCAM
671:tRNA
669:, a
494:eEF2
386:and
274:and
139:and
87:mRNA
5773:RNA
5769:DNA
5683:MUB
5487:CBL
5477:VHL
5470:E3
5299:E2
5244:E1
5113:12A
5044:C19
5039:C14
5034:C13
5029:C11
5024:C10
4979:B11
4813:AAs
4719:AAs
4612:AAs
4176:AAs
3721:doi
3680:PMC
3672:doi
3631:PMC
3621:doi
3580:PMC
3572:doi
3531:PMC
3523:doi
3470:PMC
3462:doi
3421:PMC
3413:doi
3372:PMC
3364:doi
3323:PMC
3315:doi
3274:PMC
3266:doi
3222:PMC
3214:doi
3173:PMC
3165:doi
3092:PMC
3084:doi
3043:PMC
3033:doi
2994:doi
2959:doi
2918:PMC
2910:doi
2847:doi
2843:235
2808:doi
2771:doi
2721:PMC
2713:doi
2709:112
2667:doi
2663:276
2628:doi
2616:247
2585:doi
2548:PMC
2540:doi
2536:118
2499:PMC
2491:doi
2450:doi
2446:264
2378:doi
2341:PMC
2331:doi
2327:283
2288:PMC
2280:doi
2206:PMC
2198:doi
2022:by
1590:or
1301:in
1297:of
1287:in
1283:of
1277:by
1273:of
1223:of
1211:of
1207:by
1194:to
1190:or
1186:to
1173:to
950:aka
635:or
567:),
559:),
445:FAD
443:or
441:FMN
374:or
282:of
270:of
262:of
119:or
60:PTM
50:In
6125::
5513::
5457:V2
5452:V1
5442:R2
5436:R1
5431:Q2
5426:Q1
5406:L6
5401:L4
5396:L3
5391:L2
5386:L1
5376:J2
5371:J1
5356:G2
5351:G1
5346:E3
5341:E2
5336:E1
5331:D3
5326:D2
5321:D1
5153:ER
5118:14
5083:4L
5068:1L
5063:1B
5058:1A
5019:C7
5014:C6
5009:C5
5004:C3
4999:C1
4994:B9
4989:B6
4984:B4
4974:B2
4969:B1
4964:A3
4959:A2
4954:A1
3810:-
3751:.
3729:.
3717:23
3715:.
3711:.
3688:.
3678:.
3668:47
3666:.
3662:.
3639:.
3629:.
3617:15
3615:.
3611:.
3588:.
3578:.
3566:.
3562:.
3539:.
3529:.
3521:.
3509:.
3505:.
3478:.
3468:.
3458:15
3456:.
3452:.
3429:.
3419:.
3409:46
3407:.
3403:.
3380:.
3370:.
3360:31
3358:.
3354:.
3331:.
3321:.
3311:38
3309:.
3305:.
3282:.
3272:.
3260:.
3256:.
3244:^
3230:.
3220:.
3210:43
3208:.
3204:.
3181:.
3171:.
3161:34
3159:.
3155:.
3143:^
3122:.
3100:.
3090:.
3082:.
3070:.
3066:.
3041:.
3029:18
3027:.
3023:.
3000:.
2990:34
2988:.
2965:.
2955:26
2953:.
2949:.
2926:.
2916:.
2906:17
2904:.
2900:.
2841:.
2837:.
2814:.
2804:36
2802:.
2779:.
2767:57
2765:.
2761:.
2729:.
2719:.
2707:.
2703:.
2689:^
2675:.
2661:.
2657:.
2634:.
2626:.
2614:.
2591:.
2581:16
2579:.
2556:.
2546:.
2534:.
2530:.
2507:.
2497:.
2485:.
2481:.
2458:.
2444:.
2440:.
2392:.
2384:.
2374:33
2372:.
2349:.
2339:.
2325:.
2319:.
2296:.
2286:.
2276:10
2274:.
2268:.
2237:.
2214:.
2204:.
2196:.
2184:.
2178:.
2147:.
2127:;
2123:;
2073::
1828:,
1807:,
1803:,
1783:,
1779:,
1717:,
1709:,
1705:,
1701:,
1697:,
1634:N-
1562:,
1520:,
1497:,
856:,
753:,
749:,
745:,
741:,
737:,
733:,
618:,
575:)
429:(C
408:,
390:)
366:16
352:14
305:,
247:,
231:,
208:.
167:.
121:N-
117:C-
105:.
54:,
6101::
5733:e
5726:t
5719:v
5659:)
5655:(
5462:Z
5447:S
5421:O
5416:N
5411:M
5381:K
5366:I
5361:H
5316:C
5311:B
5306:A
5148:β
5142:2
5140:α
5134:1
5132:α
5108:9
5103:8
5098:7
5093:6
5088:5
5078:4
5073:2
4940:/
4912:/
4898:/
4880:e
4873:t
4866:v
4832:–
4828:–
4824:–
4788:–
4784:–
4761:–
4757:–
4734:–
4730:–
4694:–
4672:–
4650:–
4623:–
4472:O
4213:O
4187:/
4042:e
4035:t
4028:v
3855:e
3848:t
3841:v
3774:(
3755:.
3737:.
3723::
3696:.
3674::
3647:.
3623::
3596:.
3574::
3547:.
3525::
3517::
3511:8
3501:O
3486:.
3464::
3437:.
3415::
3388:.
3366::
3339:.
3317::
3290:.
3268::
3262:8
3238:.
3216::
3189:.
3167::
3137:.
3108:.
3086::
3078::
3072:1
3051:.
3035::
3008:.
2996::
2973:.
2961::
2934:.
2912::
2886:.
2855:.
2849::
2822:.
2810::
2787:.
2773::
2737:.
2715::
2683:.
2669::
2642:.
2630::
2622::
2599:.
2587::
2564:.
2542::
2515:.
2493::
2487:7
2466:.
2452::
2425:.
2400:.
2380::
2357:.
2333::
2304:.
2282::
2253:.
2222:.
2200::
2192::
2186:1
2045:)
2039:(
2034:)
2030:(
2016:.
1957:O
1951:O
1064:.
1018:.
981:S
967:S
953:S
946:S
939:S
931:S
904:N
902:(
892:O
890:(
874:N
872:(
866:O
864:(
840:N
838:(
832:O
830:(
815:)
780:N
774:O
687:)
643:.
597:.
569:S
561:N
553:O
506:)
496:)
431:8
58:(
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.