325:
fungi. TRFLP works by PCR amplification of DNA using primer pairs that have been labeled with fluorescent tags. The PCR products are then digested using RFLP enzymes and the resulting patterns visualized using a DNA sequencer. The results are analyzed either by simply counting and comparing bands or peaks in the TRFLP profile, or by matching bands from one or more TRFLP runs to a database of known species. A number of different software tools have been developed to automate the process of band matching, comparison and data basing of TRFLP profiles.
611:
247:
Analysis of RFLP variation in genomes was formerly a vital tool in genome mapping and genetic disease analysis. If researchers were trying to initially determine the chromosomal location of a particular disease gene, they would analyze the DNA of members of a family afflicted by the disease, and look
324:
RFLP is still used in marker-assisted selection. Terminal restriction fragment length polymorphism (TRFLP or sometimes T-RFLP) is a technique initially developed for characterizing bacterial communities in mixed-species samples. The technique has also been applied to other groups including soil
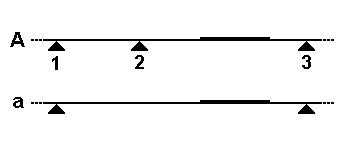
204:, so the probe now detects the larger fused fragment running from sites 1 to 3. The second diagram shows how this fragment size variation would look on a Southern blot, and how each allele (two per individual) might be inherited in members of a family.
168:
339:
The sequence changes directly involved with an RFLP can also be analyzed more quickly by PCR. Amplification can be directed across the altered restriction site, and the products digested with the restriction enzyme. This method has been called
284:
The technique for RFLP analysis is, however, slow and cumbersome. It requires a large amount of sample DNA, and the combined process of probe labeling, DNA fragmentation, electrophoresis, blotting, hybridization, washing, and
256:
of the mutant genes. RFLP test is used in identification and differentiation of organisms by analyzing unique patterns in genome. It is also used in identification of recombination rate in the loci between restriction sites.
147:
to the probe. A restriction fragment length polymorphism is said to occur when the length of a detected fragment varies between individuals, indicating non-identical sequence homologies. Each fragment length is considered an
391:
Saiki, R.; Scharf, S; Faloona, F; Mullis, K.; Horn, G.; Erlich, H.; Arnheim, N (1985). "Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia".
507:
301:(SNPs) in that project (as well as the direct identification of many disease genes and mutations) has replaced the need for RFLP disease linkage analysis (see
196:, the genome is cleaved by a restriction enzyme at three nearby sites (triangles), but only the rightmost fragment will be detected by the probe. In allele
219:, there are only two repeats in the VNTR, so the probe detects a shorter fragment between the same two restriction sites. Other genetic processes, such as
500:
329:
637:
367:
333:
188:
There are two common mechanisms by which the size of a particular restriction fragment can vary. In the first schematic, a small segment of the
361:
341:
252:). Once a disease gene was localized, RFLP analysis of other families could reveal who was at risk for the disease, or who was likely to be a
493:
144:
176:
160:
345:
298:
215:, there are five repeats in the VNTR, and the probe detects a longer fragment between the two restriction sites. In allele
208:
642:
615:
128:
207:
In the third schematic, the probe and restriction enzyme are chosen to detect a region of the genome that includes a
647:
136:
558:
306:
228:
57:
within a sequence. The term may refer to a polymorphism itself, as detected through the differing locations of
112:
The basic technique for the detection of RFLPs involves fragmenting a sample of DNA with the application of a
581:
261:
88:
technique inexpensive enough to see widespread application. RFLP analysis was an important early tool in
480:
310:
269:
232:
120:
101:
444:
401:
294:
290:
117:
443:
Heras, J.; Dominguez, C.; Mata, E.; Pascual, V.; Lozano, C.; Torres, C.; Zarazaga, M. (2015-03-29).
533:
236:
140:
127:. The DNA fragments produced by the digest are then separated by length through a process known as
74:
553:
516:
314:
124:
113:
66:
43:
538:
460:
425:
417:
273:
220:
50:
31:
548:
452:
409:
224:
93:
58:
485:
286:
253:
249:
97:
405:
248:
for RFLP alleles that show a similar pattern of inheritance as that of the disease (see
235:, can also lead to polymorphisms. RFLP tests require much larger samples of DNA than do
61:, or to a related laboratory technique by which such differences can be illustrated. In
573:
302:
89:
81:
631:
563:
543:
132:
85:
596:
372:
289:
can take up to a month to complete. A limited version of the RFLP method that used
297:
have largely replaced the need for RFLP mapping, and the identification of many
586:
525:
167:
153:
464:
421:
413:
305:). The analysis of VNTR alleles continues, but is now usually performed by
429:
80:
RFLP analysis is now largely obsolete due to the emergence of inexpensive
456:
349:
265:
201:
175:
159:
481:
https://www.ncbi.nlm.nih.gov/projects/genome/probe/doc/TechRFLP.shtml
189:
149:
17:
318:
174:
166:
158:
344:(CAPS). Alternatively, the amplified segment can be analyzed by
116:, which can selectively cleave a DNA molecule wherever a short,
54:
489:
46:
348:(ASO) probes, a process that can often be done by a simple
192:
is being detected by a DNA probe (thicker line). In allele
53:, populations, or species or to pinpoint the locations of
264:, useful in the identification of samples retrieved from
65:, a DNA sample is digested into fragments by one or more
171:
Analysis and inheritance of allelic RFLP fragments (NIH)
156:
or not, and can be used in subsequent genetic analysis.
211:(VNTR) segment (boxes in schematic diagram). In allele
260:
RFLP analysis was also the basis for early methods of
143:
then determines the length of the fragments which are
572:
524:
445:"A survey of tools for analysing DNA fingerprints"
501:
42:) is a technique that exploits variations in
8:
328:The technique is similar in some aspects to
276:or breeding patterns in animal populations.
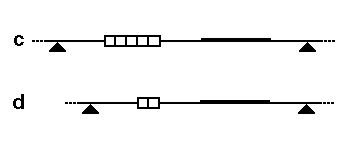
179:Schematic for RFLP by VNTR length variation
508:
494:
486:
309:(PCR) methods. For example, the standard
293:was reported in 1985. The results of the
592:Restriction fragment length polymorphism
200:, restriction site 2 has been lost by a
163:Schematic for RFLP by cleavage site loss
36:restriction fragment length polymorphism
383:
334:denaturing gradient gel electrophoresis
362:Amplified fragment length polymorphism
342:Cleaved Amplified Polymorphic Sequence
131:and transferred to a membrane via the
123:is recognized in a process known as a
7:
84:technologies, but it was the first
25:
272:, and in the characterization of
152:, whether it actually contains a
610:
609:
268:scenes, in the determination of
346:allele-specific oligonucleotide
299:single-nucleotide polymorphisms
638:Biochemistry detection methods
1:
209:variable number tandem repeat
139:of the membrane to a labeled
321:of more than a dozen VNTRs.
92:, localization of genes for
449:Briefings in Bioinformatics
129:agarose gel electrophoresis
27:Molecular biology technique
664:
605:
559:Site-directed mutagenesis
307:polymerase chain reaction
77:according to their size.
317:involve PCR analysis of
59:restriction enzyme sites
414:10.1126/science.2999980
519:: key methods of study
291:oligonucleotide probes
262:genetic fingerprinting
180:
172:
164:
73:are then separated by
178:
170:
162:
71:restriction fragments
330:temperature gradient
295:Human Genome Project
69:, and the resulting
49:sequences, known as
643:Genomics techniques
534:Gel electrophoresis
406:1985Sci...230.1350S
400:(4732): 1350–1354.
237:short tandem repeat
96:, determination of
75:gel electrophoresis
67:restriction enzymes
554:Restriction digest
517:Molecular genetics
457:10.1093/bib/bbv016
315:DNA fingerprinting
181:
173:
165:
125:restriction digest
114:restriction enzyme
648:Molecular biology
625:
624:
539:Molecular cloning
336:(TGGE and DGGE).
274:genetic diversity
102:paternity testing
100:for disease, and
94:genetic disorders
32:molecular biology
16:(Redirected from
655:
613:
612:
549:Promoter bashing
510:
503:
496:
487:
469:
468:
440:
434:
433:
388:
21:
663:
662:
658:
657:
656:
654:
653:
652:
628:
627:
626:
621:
601:
582:Gene sequencing
568:
520:
514:
477:
472:
442:
441:
437:
390:
389:
385:
381:
358:
287:autoradiography
282:
250:genetic linkage
245:
186:
110:
28:
23:
22:
15:
12:
11:
5:
661:
659:
651:
650:
645:
640:
630:
629:
623:
622:
620:
619:
606:
603:
602:
600:
599:
594:
589:
584:
578:
576:
574:Bioinformatics
570:
569:
567:
566:
561:
556:
551:
546:
541:
536:
530:
528:
522:
521:
515:
513:
512:
505:
498:
490:
484:
483:
476:
475:External links
473:
471:
470:
435:
382:
380:
377:
376:
375:
370:
365:
357:
354:
303:SNP genotyping
281:
278:
244:
241:
229:translocations
185:
182:
109:
106:
90:genome mapping
82:DNA sequencing
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
660:
649:
646:
644:
641:
639:
636:
635:
633:
618:
617:
608:
607:
604:
598:
595:
593:
590:
588:
585:
583:
580:
579:
577:
575:
571:
565:
564:Southern blot
562:
560:
557:
555:
552:
550:
547:
545:
544:Northern blot
542:
540:
537:
535:
532:
531:
529:
527:
523:
518:
511:
506:
504:
499:
497:
492:
491:
488:
482:
479:
478:
474:
466:
462:
458:
454:
450:
446:
439:
436:
431:
427:
423:
419:
415:
411:
407:
403:
399:
395:
387:
384:
378:
374:
371:
369:
366:
363:
360:
359:
355:
353:
351:
347:
343:
337:
335:
331:
326:
322:
320:
316:
312:
308:
304:
300:
296:
292:
288:
279:
277:
275:
271:
267:
263:
258:
255:
251:
242:
240:
239:(STR) tests.
238:
234:
230:
226:
222:
218:
214:
210:
205:
203:
199:
195:
191:
183:
177:
169:
161:
157:
155:
154:coding region
151:
146:
145:complementary
142:
138:
137:Hybridization
134:
133:Southern blot
130:
126:
122:
119:
115:
108:RFLP analysis
107:
105:
103:
99:
95:
91:
87:
86:DNA profiling
83:
78:
76:
72:
68:
64:
63:RFLP analysis
60:
56:
52:
51:polymorphisms
48:
45:
41:
37:
33:
19:
614:
597:STR analysis
591:
526:Experimental
448:
438:
397:
393:
386:
373:STR analysis
338:
327:
323:
283:
280:Alternatives
259:
246:
243:Applications
216:
212:
206:
197:
193:
187:
111:
79:
70:
62:
39:
35:
29:
135:procedure.
632:Categories
587:Microarray
451:: bbv016.
379:References
233:inversions
221:insertions
44:homologous
465:1467-5463
422:0036-8075
311:protocols
270:paternity
225:deletions
141:DNA probe
616:Category
356:See also
350:dot blot
202:mutation
184:Examples
121:sequence
118:specific
430:2999980
402:Bibcode
394:Science
254:carrier
463:
428:
420:
364:(AFLP)
319:panels
231:, and
190:genome
150:allele
266:crime
55:genes
461:ISSN
426:PMID
418:ISSN
368:RAPD
313:for
98:risk
40:RFLP
18:RFLP
453:doi
410:doi
398:230
332:or
47:DNA
30:In
634::
459:.
447:.
424:.
416:.
408:.
396:.
352:.
227:,
223:,
104:.
34:,
509:e
502:t
495:v
467:.
455::
432:.
412::
404::
217:d
213:c
198:a
194:A
38:(
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.