1138:. Recombination appears to occur by a copy choice mechanism in which the RdRp switches (+)ssRNA templates during negative strand synthesis. Recombination frequency is determined in part by the fidelity of RdRp replication. RdRp variants with high replication fidelity show reduced recombination, and low fidelity RdRps exhibit increased recombination. Recombination by RdRp strand switching occurs frequently during replication in the (+)ssRNA plant
357:
49:
346:
492:
949:
1161:) has a linear, single-stranded, negative-sense, nonsegmented RNA genome. The viral RdRp consists of two virus-encoded subunits, a smaller one P and a larger one L. Testing different inactive RdRp mutants with defects throughout the length of the L subunit in pairwise combinations, restoration of viral RNA synthesis was observed in some combinations. This positive L–L interaction is referred to as
4785:
388:. From there, the RNA acts as a template for complementary RNA synthesis. The complementary strand acts as a template for the production of new viral genomes that are packaged and released from the cell ready to infect more host cells. The advantage of this method of replication is that no DNA stage complicates replication. The disadvantage is that no 'back-up' DNA copy is available.
453:
formation – two Mg ions are present in the catalytically active state and arrange themselves around the newly synthesized RNA chain such that the substrate NTP undergoes a phosphatidyl transfer and forms a phosphodiester bond with the new chain. Without the use of these Mg ions, the active site is no
527:
triphosphates over dNTPs and, thus, determines whether RNA rather than DNA is synthesized. The domain organization and the 3D structure of the catalytic centre of a wide range of RdRps, even those with a low overall sequence homology, are conserved. The catalytic center is formed by several motifs
478:
initiation consists in the addition of a NTP to the 3'-OH of the first initiating NTP. During the following elongation phase, this nucleotidyl transfer reaction is repeated with subsequent NTPs to generate the complementary RNA product. Termination of the nascent RNA chain produced by RdRp is not
1098:
found in positive-strand ssRNA viruses are related to each other, forming three large superfamilies. Birnaviral RNA replicase is unique in that it lacks motif C (GDD) in the palm. Mononegaviral RdRp (PDB 5A22) has been automatically classified as similar to (+)−ssRNA RdRps, specifically one from
482:
One major drawback of RNA-dependent RNA polymerase replication is the transcription error rate. RdRps lack fidelity on the order of 10 nucleotides, which is thought to be a direct result of inadequate proofreading. This variation rate is favored in viral genomes as it allows for the pathogen to
1217:
that inhibits RdRp function by covalently binding to and interrupting termination of the nascent RNA through early or delayed termination or preventing further elongation of the RNA polynucleotide. This early termination leads to nonfunctional RNA that gets degraded through normal cellular
1212:
triphosphate is a substrate for RdRp, but not mammalian polymerases. It results in premature chain termination and inhibition of viral replication. GS-441524 triphosphate is the biologically active form of remdesivir. Remdesivir is classified as a
1178:
RdRps can be used as drug targets for viral pathogens as their function is not necessary for eukaryotic survival. By inhibiting RdRp function, new RNAs cannot be replicated from an RNA template strand, however, DNA-dependent RNA polymerase remains
499:
Viral/prokaryotic RdRp, along with many single-subunit DdRp, employ a fold whose organization has been linked to the shape of a right hand with three subdomains termed fingers, palm, and thumb. Only the palm subdomain, composed of a four-stranded
444:(NTP) binding – initially, the RdRp presents with a vacant active site in which an NTP binds, complementary to the corresponding nucleotide on the template strand. Correct NTP binding causes the RdRp to undergo a conformational change.
535:
requires a cellular RdRp (c RdRp). Unlike the "hand" polymerases, they resemble simplified multi-subunit DdRPs, specifically in the catalytic β/β' subunits, in that they use two sets of double-psi β-barrels in the active site. QDE1
3925:
457:
Translocation – once the active site is open, the RNA template strand moves by one position through the RdRp protein complex and continues chain elongation by binding a new NTP, unless otherwise specified by the
1732:
Zong J, Yao X, Yin J, Zhang D, Ma H (November 2009). "Evolution of the RNA-dependent RNA polymerase (RdRP) genes: duplications and possible losses before and after the divergence of major eukaryotic groups".
1235:) binding to mRNA rendering them inactive. Eukaryotic RdRp becomes active in the presence of dsRNA, and is less widely distributed than other RNAi components as it lost in some animals, though still found in
1323:
Koonin EV, Gorbalenya AE, Chumakov KM (July 1989). "Tentative identification of RNA-dependent RNA polymerases of dsRNA viruses and their relationship to positive strand RNA viral polymerases".
3935:
3930:
447:
Active site closure – the conformational change, initiated by the correct NTP binding, results in the restriction of active site access and produces a catalytically competent state.
425:
as primers. These RdRps are used in the defense mechanisms and can be appropriated by RNA viruses. Their evolutionary history predates the divergence of major eukaryotic groups.
332:, a drug that inhibits cellular DNA-directed RNA synthesis. This lack of sensitivity suggested the action of a virus-specific enzyme that could copy RNA from an RNA template.
4050:
4028:
1634:"Evolutionary connection between the catalytic subunits of DNA-dependent RNA polymerases and eukaryotic RNA-dependent RNA polymerases and the origin of RNA polymerases"
3846:
907:
786:
675:
218:
1252:. This presence of dsRNA triggers the activation of RdRp and RNAi processes by priming the initiation of RNA transcription through the introduction of siRNAs. In
237:
3956:
2120:"Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure"
377:, though the reason for this was an ongoing question as of 2009. The similarity led to speculation that viral RdRps are ancestral to human telomerase.
4408:
3851:
2967:"Distinct RNA-dependent RNA polymerases are required for RNAi triggered by double-stranded RNA versus truncated transgenes in Paramecium tetraurelia"
4018:
1127:
2421:
1946:"Mechanistic analysis of RNA synthesis by RNA-dependent RNA polymerase from two promoters reveals similarities to DNA-dependent RNA polymerase"
1260:, which works alongside mRNAs targeted for interference to recruit more RdRps to synthesize more secondary siRNAs and repress gene expression.
4576:
3657:
2786:
Waheed Y, Bhatti A, Ashraf M (March 2013). "RNA dependent RNA polymerase of HCV: a potential target for the development of antiviral drugs".
1564:
623:
2698:"Mechanism of RNA recombination in carmo- and tombusviruses: evidence for template switching by the RNA-dependent RNA polymerase in vitro"
2079:"Poliovirus RNA-dependent RNA polymerase (3Dpol): structural, biochemical, and biological analysis of conserved structural motifs A and B"
278:
4460:
4440:
3791:
437:
as it catalyzes RNA synthesis of strands complementary to a given RNA template. The RNA replication process is a four-step mechanism:
4548:
4117:
3361:
3155:
1092:
is cleaved to a number of products, one of which is NS5, an RdRp. It possesses short regions and motifs homologous to other RdRps.
3502:
230:
4702:
4646:
4641:
181:
157:
4273:
927:
806:
695:
4729:
4660:
4401:
4075:
4070:
3630:
3448:
2927:
Simaan JA, Aviado DM (November 1975). "Hemodynamic effects of aerosol propellants. II. Pulmonary circulation in the dog".
2302:"Recombinant dengue type 1 virus NS5 protein expressed in Escherichia coli exhibits RNA-dependent RNA polymerase activity"
1257:
3940:
3828:
3796:
1203:
434:
282:
33:
391:
Many RdRps associate tightly with membranes making them difficult to study. The best-known RdRps are polioviral 3Dpol,
3463:
1118:
Since it is a protein universal to RNA-containing viruses, RdRp is a useful marker for understanding their evolution.
501:
4775:
4680:
1162:
463:
392:
4258:
4374:
4361:
4348:
4335:
4322:
4309:
4296:
4061:
4039:
4014:
3989:
3909:
3517:
3477:
3422:
3399:
3371:
3339:
3205:
4268:
3292:
2200:
Werner F, Grohmann D (February 2011). "Evolution of multisubunit RNA polymerases in the three domains of life".
175:
4819:
4724:
4512:
4425:
4394:
4222:
4165:
3526:
3507:
3438:
3172:
3125:
3119:
286:
68:
915:
794:
683:
162:
4685:
4502:
4487:
4170:
3384:
1269:
441:
4690:
4615:
4507:
3838:
3611:
3521:
1237:
242:
4605:
4590:
4470:
4191:
4110:
3692:
3647:
3349:
3334:
3270:
3250:
3148:
2418:
1232:
1135:
422:
150:
4263:
2380:"Birnavirus VP1 proteins form a distinct subgroup of RNA-dependent RNA polymerases lacking a GDD motif"
911:
790:
679:
3133:
4712:
4610:
4528:
3606:
3601:
3481:
3344:
3324:
3233:
2834:
2549:
2256:
1840:
1465:
636:
85:
2747:"Intragenic complementation and oligomerization of the L subunit of the sendai virus RNA polymerase"
4538:
4227:
3869:
3861:
3642:
3309:
3245:
3209:
2823:"Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir"
2649:"Biochemical and genetic analysis of the role of the viral polymerase in enterovirus recombination"
1214:
1154:
450:
418:
178:
80:
102:
4673:
4556:
4160:
3732:
3670:
3621:
3356:
3183:
2487:
Wolf YI, Kazlauskas D, Iranzo J, Lucía-Sanz A, Kuhn JH, Krupovic M, et al. (November 2018).
2225:
1985:
Fitzsimmons WJ, Woods RJ, McCrone JT, Woodman A, Arnold JJ, Yennawar M, et al. (June 2018).
1532:
1489:
1348:
414:
370:
491:
1507:
Weiner AM (January 1988). "Eukaryotic nuclear telomeres: molecular fossils of the RNP world?".
4829:
4757:
3747:
3737:
3727:
3652:
3636:
3595:
3581:
3576:
3314:
3277:
3094:
3045:
2996:
2944:
2909:
2860:
2803:
2768:
2727:
2678:
2626:
2577:
2518:
2469:
2401:
2360:
2323:
2282:
2217:
2182:
2141:
2100:
2059:
2018:
1967:
1926:
1868:
1806:
1750:
1714:
1665:
1611:
1560:
1524:
1481:
1438:
1397:
1340:
902:
813:
781:
670:
544:
396:
169:
1551:
4206:
4201:
4175:
4103:
4080:
3742:
3722:
3664:
3560:
3458:
3443:
3389:
3287:
3282:
3141:
3084:
3076:
3035:
3027:
2986:
2978:
2936:
2899:
2891:
2850:
2842:
2795:
2758:
2717:
2709:
2668:
2660:
2616:
2608:
2567:
2557:
2508:
2500:
2459:
2451:
2391:
2350:
2313:
2272:
2264:
2209:
2172:
2131:
2090:
2049:
2008:
1998:
1957:
1916:
1906:
1858:
1848:
1796:
1786:
1742:
1704:
1696:
1655:
1645:
1601:
1593:
1516:
1473:
1428:
1387:
1379:
1332:
1228:
1056:
532:
406:
309:
270:
48:
894:
773:
662:
138:
4734:
4571:
4417:
4253:
4237:
4150:
4003:
3993:
3815:
3786:
3412:
3319:
3304:
2425:
1028:
3115:
2440:"Structural Insights into Bunyavirus Replication and Its Regulation by the vRNA Promoter"
193:
114:
2838:
2553:
2260:
2245:"Single-peptide DNA-dependent RNA polymerase homologous to multi-subunit RNA polymerase"
1844:
1469:
356:
73:
4789:
4561:
4533:
4291:
4232:
3971:
3810:
3717:
3709:
3586:
3549:
3089:
3064:
3040:
3015:
2991:
2966:
2904:
2879:
2855:
2822:
2713:
2673:
2648:
2621:
2596:
2572:
2537:
2513:
2488:
2464:
2439:
2277:
2244:
2013:
1987:"A speed-fidelity trade-off determines the mutation rate and virulence of an RNA virus"
1986:
1962:
1945:
1921:
1894:
1863:
1828:
1801:
1774:
1709:
1684:
1606:
1581:
1274:
1112:
1019:
524:
512:
384:. The viral genome is composed of RNA, which enters the cell through receptor-mediated
345:
213:
2722:
2697:
2161:"The Extended "Two-Barrel" Polymerases Superfamily: Structure, Function and Evolution"
2054:
2037:
1660:
1633:
1433:
1416:
1392:
1367:
4813:
4762:
4449:
4196:
4155:
3240:
2940:
2612:
1520:
1336:
1231:
in eukaryotes, a process used to silence gene expression via small interfering RNAs (
1095:
981:
974:
628:
549:
516:
329:
188:
2229:
1829:"Structural basis of viral RNA-dependent RNA polymerase catalysis and translocation"
1536:
1383:
1352:
856:
735:
591:
515:(A, B, and C). Motif A (D-x(4,5)-D) and motif C (GDD) are spatially juxtaposed; the
483:
overcome host defenses trying to avoid infection, allowing for evolutionary growth.
4824:
4636:
4566:
4435:
4145:
3329:
3299:
3228:
1493:
1417:"A New Ribonucleic Acid Polymerase Appearing after Mengovirus Infection of L-Cells"
1067:
1060:
986:
966:
948:
890:
769:
658:
548:, which has both barrels in the same chain, is an example of such a c RdRp enzyme.
197:
956:
Four superfamilies of viruses cover all RNA-containing viruses with no DNA stage:
604:
4805:
2799:
2562:
2003:
1775:"Visualizing the Nucleotide Addition Cycle of Viral RNA-Dependent RNA Polymerase"
1077:
1051:
1038:
868:
747:
616:
454:
longer catalytically stable and the RdRp complex changes to an open conformation.
4739:
4668:
4369:
4304:
4140:
3895:
3885:
3486:
3223:
3164:
2355:
2342:
1895:"RNA Dependent RNA Polymerases: Insights from Structure, Function and Evolution"
1183:
1143:
1107:
1089:
1013:
1007:
511:, is well conserved. In RdRp, the palm subdomain comprises three well-conserved
508:
385:
325:
4784:
2455:
2343:"The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses"
1833:
Proceedings of the
National Academy of Sciences of the United States of America
1746:
1582:"Robust kinetics of an RNA virus: Transcription rates are set by genome levels"
495:
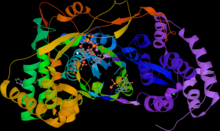
Overview of the flavivirus RdRp structure based on West Nile Virus (WNV) NS5Pol
17:
4697:
3697:
3540:
3453:
3218:
3168:
2177:
2160:
2077:
Gohara DW, Crotty S, Arnold JJ, Yoder JD, Andino R, Cameron CE (August 2000).
1243:
1199:
1191:
1139:
1131:
1101:
1085:
1001:
992:
961:
520:
504:
381:
374:
321:
305:
301:
54:
277:
from an RNA template. Specifically, it catalyzes synthesis of the RNA strand
4343:
4317:
3976:
3889:
3426:
3192:
2846:
1853:
1209:
1195:
3098:
3049:
3000:
2913:
2878:
Malin JJ, Suárez I, Priesner V, Fätkenheuer G, Rybniker J (December 2020).
2864:
2807:
2772:
2763:
2746:
2731:
2682:
2581:
2522:
2473:
2405:
2396:
2379:
2318:
2301:
2286:
2221:
2186:
2136:
2119:
2104:
2095:
2078:
2022:
1930:
1872:
1810:
1754:
1718:
1669:
1615:
1485:
1442:
1368:"A reevaluation of the higher taxonomy of viruses based on RNA polymerases"
296:
RdRp is an essential protein encoded in the genomes of most RNA-containing
2630:
2504:
2364:
2327:
2145:
2063:
1971:
1700:
1650:
1528:
1401:
1344:
632:
4801:
4717:
4707:
4631:
3823:
3403:
2982:
2895:
2664:
1187:
1166:
1072:
1046:
1033:
863:
742:
611:
554:
538:
519:
residues of these motifs are implied in the binding of Mg and/or Mn. The
410:
3128:
2268:
2213:
1477:
126:
4453:
3774:
3759:
3375:
2948:
145:
3080:
2821:
Yin W, Mao C, Luan X, Shen DD, Shen Q, Su H, et al. (June 2020).
1911:
1791:
1597:
972:
All positive-strand RNA eukaryotic viruses with no DNA stage, such as
952:
Structure and evolution of RdRp in RNA viruses and their superfamilies
4356:
4126:
3752:
3187:
3031:
2536:Černý J, Černá Bolfíková B, Valdés JJ, Grubhoffer L, Růžek D (2014).
1249:
922:
801:
690:
479:
completely known, however, RdRp termination is sequence-independent.
266:
225:
121:
109:
97:
2538:"Evolution of tertiary structure of viral RNA dependent polymerases"
960:
Viruses containing positive-strand RNA or double-strand RNA, except
552:
homologs of c RdRp, including the similarly single-chain DdRp yonO (
4475:
4386:
4330:
3769:
3764:
3616:
2300:
Tan BH, Fu J, Sugrue RJ, Yap EH, Chan YC, Tan YH (February 1996).
1944:
Adkins S, Stawicki SS, Faurote G, Siegel RW, Kao CC (April 1998).
947:
490:
297:
3926:
CDP-diacylglycerol—glycerol-3-phosphate 3-phosphatidyltransferase
2378:
Shwed PS, Dobos P, Cameron LA, Vakharia VN, Duncan R (May 2002).
4797:
3680:
3675:
3571:
3566:
3265:
3260:
3255:
1256:, siRNAs are integrated into the RNA-induced silencing complex,
884:
851:
763:
730:
652:
598:
586:
399:
133:
4390:
4099:
3137:
3016:"The Functions of RNA-Dependent RNA Polymerases in Arabidopsis"
320:
Viral RdRps were discovered in the early 1960s from studies on
4445:
471:
328:
when it was observed that these viruses were not sensitive to
290:
274:
2038:"Structure of the RNA-dependent RNA polymerase of poliovirus"
1559:(3d ed.). London: W.W. Norton&Company. p. 129.
1366:
Zanotto PM, Gibbs MJ, Gould EA, Holmes EC (September 1996).
4095:
2647:
Woodman A, Arnold JJ, Cameron CE, Evans DJ (August 2016).
1685:"RNAi, a new therapeutic strategy against viral infection"
3014:
Willmann MR, Endres MW, Cook RT, Gregory BD (July 2011).
1893:
Venkataraman S, Prasad BV, Selvarajan R (February 2018).
1032:(negative-strand RNA viruses with non-segmented genomes;
984:; the two families of RNA-containing bacteriophages are
362:
Structure and replication elongation mechanism of a RdRp
281:
to a given RNA template. This is in contrast to typical
2243:
Forrest D, James K, Yuzenkova Y, Zenkin N (June 2017).
4796:
This article incorporates text from the public domain
2880:"Remdesivir against COVID-19 and Other Viral Diseases"
1115:
4773:
3936:
CDP-diacylglycerol—inositol 3-phosphatidyltransferase
2438:
Gerlach P, Malet H, Cusack S, Reguera J (June 2015).
3941:
CDP-diacylglycerol—choline O-phosphatidyltransferase
2965:
Marker S, Le Mouël A, Meyer E, Simon M (July 2010).
2419:
1045:
Negative-strand RNA viruses with segmented genomes (
4750:
4659:
4624:
4598:
4589:
4547:
4521:
4495:
4486:
4424:
4282:
4246:
4215:
4184:
4133:
4060:
4038:
4013:
3988:
3969:
3949:
3931:
CDP-diacylglycerol—serine O-phosphatidyltransferase
3918:
3908:
3878:
3860:
3837:
3809:
3708:
3548:
3539:
3516:
3476:
3421:
3398:
3370:
3204:
3180:
1111:. Bunyaviral RdRp monomer (PDB 5AMQ) resembles the
921:
901:
883:
878:
862:
850:
842:
837:
832:
812:
800:
780:
762:
757:
741:
729:
721:
716:
711:
689:
669:
651:
646:
622:
610:
597:
585:
577:
572:
567:
236:
224:
212:
207:
187:
168:
156:
144:
132:
120:
108:
96:
91:
79:
67:
62:
41:
2597:"The mechanism of RNA recombination in poliovirus"
1456:Suttle CA (September 2005). "Viruses in the sea".
558:), appear to be closer to c RdRps than DdRPs are.
470:) or a primer-dependent mechanism that utilizes a
53:Stalled HCV RNA replicase (NS5B), in complex with
1088:produce a polyprotein from the ssRNA genome. The
3065:"New insights into siRNA amplification and RNAi"
2745:Smallwood S, Cevik B, Moyer SA (December 2002).
2489:"Origins and Evolution of the Global RNA Virome"
1293:See Pfam clan for other (+)ssRNA/dsRNA families.
405:Many eukaryotes have RdRps that are involved in
27:Enzyme that synthesizes RNA from an RNA template
523:residue of motif B is involved in selection of
2642:
2640:
2036:Hansen JL, Long AM, Schultz SC (August 1997).
1632:Iyer LM, Koonin EV, Aravind L (January 2003).
1627:
1625:
4402:
4111:
3149:
712:RNA-dependent RNA polymerase, eukaryotic-type
8:
3957:N-acetylglucosamine-1-phosphate transferase
3847:UTP—glucose-1-phosphate uridylyltransferase
2595:Kirkegaard K, Baltimore D (November 1986).
4595:
4557:Precursor mRNA (pre-mRNA / hnRNA)
4492:
4409:
4395:
4387:
4118:
4104:
4096:
3985:
3915:
3545:
3536:
3201:
3156:
3142:
3134:
875:
754:
643:
528:containing conserved amino acid residues.
380:The most famous example of RdRp is in the
312:and differ structurally from viral RdRps.
308:also contain RdRps, which are involved in
285:, which all organisms use to catalyze the
204:
47:
4051:serine/threonine-specific protein kinases
4029:serine/threonine-specific protein kinases
3852:Galactose-1-phosphate uridylyltransferase
3118:at the U.S. National Library of Medicine
3088:
3039:
2990:
2903:
2854:
2762:
2721:
2672:
2620:
2571:
2561:
2512:
2463:
2395:
2354:
2317:
2276:
2176:
2135:
2094:
2053:
2012:
2002:
1961:
1920:
1910:
1862:
1852:
1800:
1790:
1708:
1659:
1649:
1605:
1432:
1415:Baltimore D, Franklin RM (October 1963).
1391:
1190:specifically target RdRp. These include
4780:
1315:
1286:
1165:and indicates that the L protein is an
1580:Timm C, Gupta A, Yin J (August 2015).
1227:The use of RdRp plays a major role in
829:
708:
564:
38:
4577:Histone acetylation and deacetylation
2960:
2958:
2118:O'Reilly EK, Kao CC (December 1998).
1888:
1886:
1884:
1882:
1822:
1820:
1768:
1766:
1764:
1169:in the viral RNA polymerase complex.
7:
4642:Ribosome-nascent chain complex (RNC)
462:RNA synthesis can be performed by a
2696:Cheng CP, Nagy PD (November 2003).
2083:The Journal of Biological Chemistry
1421:The Journal of Biological Chemistry
2714:10.1128/jvi.77.22.12033-12047.2003
25:
3362:Glucose-1,6-bisphosphate synthase
3063:Zhang C, Ruvkun G (August 2012).
2788:Infection, Genetics and Evolution
300:that lack a DNA stage, including
4783:
3503:Ribose-phosphate diphosphokinase
1683:Tan FL, Yin JQ (December 2004).
1586:Biotechnology and Bioengineering
355:
344:
4647:Post-translational modification
2347:The Journal of General Virology
1384:10.1128/JVI.70.9.6083-6096.1996
373:in viruses and are related to
1:
4076:Protein-histidine tele-kinase
4071:Protein-histidine pros-kinase
3950:Glycosyl-1-phosphotransferase
2884:Clinical Microbiology Reviews
2349:. 72 ( Pt 9) (9): 2197–2206.
2055:10.1016/S0969-2126(97)00261-X
1773:Wu J, Gong P (January 2018).
1434:10.1016/S0021-9258(18)48679-6
1182:Some antiviral drugs against
879:Available protein structures:
758:Available protein structures:
647:Available protein structures:
283:DNA-dependent RNA polymerases
3782:RNA-dependent RNA polymerase
2941:10.1016/0300-483x(75)90110-9
2800:10.1016/j.meegid.2012.12.004
2613:10.1016/0092-8674(86)90600-8
2563:10.1371/journal.pone.0096070
2341:Koonin EV (September 1991).
2202:Nature Reviews. Microbiology
2165:Journal of Molecular Biology
2159:Sauguet L (September 2019).
2004:10.1371/journal.pbio.2006459
1521:10.1016/0092-8674(88)90501-6
1337:10.1016/0014-5793(89)80886-5
1206:approved drug against COVID-
990:(positive ssRNA phages) and
568:RNA dependent RNA polymerase
435:DNA dependent RNA polymerase
255:RNA-dependent RNA polymerase
42:RNA-dependent RNA polymerase
34:DNA-dependent RNA polymerase
3689:RNA-directed DNA polymerase
3557:DNA-directed DNA polymerase
2356:10.1099/0022-1317-72-9-2197
1827:Shu B, Gong P (July 2016).
472:viral protein genome-linked
4846:
4795:
4042:: protein-dual-specificity
2456:10.1016/j.cell.2015.05.006
1747:10.1016/j.gene.2009.07.004
1163:intragenic complementation
1150:Intragenic complementation
1134:RdRp is able to carry out
562:
393:vesicular stomatitis virus
31:
4274:Michaelis–Menten kinetics
2178:10.1016/j.jmb.2019.05.017
874:
753:
642:
203:
46:
4708:sequestration (P-bodies)
4166:Diffusion-limited enzyme
4019:protein-serine/threonine
3919:Phosphatidyltransferases
3508:Thiamine diphosphokinase
3120:Medical Subject Headings
1198:against Hepatitis C and
833:Bunyavirus RNA replicase
32:Not to be confused with
4686:Gene regulatory network
2847:10.1126/science.abc1560
1854:10.1073/pnas.1602591113
442:Nucleoside triphosphate
4691:cis-regulatory element
3839:Nucleotidyltransferase
3522:nucleotidyltransferase
3449:Nucleoside-diphosphate
2971:Nucleic Acids Research
2764:10.1006/viro.2002.1720
2653:Nucleic Acids Research
2397:10.1006/viro.2001.1334
2319:10.1006/viro.1996.0067
2137:10.1006/viro.1998.9463
2096:10.1074/jbc.M002671200
1638:BMC Structural Biology
1302:A (−)ssRNA polymerase.
953:
496:
423:small interfering RNAs
4259:Eadie–Hofstee diagram
4192:Allosteric regulation
3693:Reverse transcriptase
2505:10.1128/mBio.02329-18
2249:Nature Communications
1701:10.1038/sj.cr.7290248
1651:10.1186/1472-6807-3-1
1126:When replicating its
951:
494:
4713:alternative splicing
4703:Post-transcriptional
4529:Transcription factor
4269:Lineweaver–Burk plot
3482:diphosphotransferase
3464:Thiamine-diphosphate
3171:-containing groups (
3020:The Arabidopsis Book
2896:10.1128/CMR.00162-20
1553:The Blind Watchmaker
1270:Spiegelman's Monster
4637:Transfer RNA (tRNA)
4064:: protein-histidine
3982:; protein acceptor)
3870:mRNA capping enzyme
3862:Guanylyltransferase
2839:2020Sci...368.1499Y
2833:(6498): 1499–1504.
2708:(22): 12033–12047.
2702:Journal of Virology
2554:2014PLoSO...996070C
2269:10.1038/ncomms15774
2261:2017NatCo...815774F
2214:10.1038/nrmicro2507
2089:(33): 25523–25532.
1845:2016PNAS..113E4005S
1839:(28): E4005–E4014.
1478:10.1038/nature04160
1470:2005Natur.437..356S
1372:Journal of Virology
1065:dsRNA virus family
999:dsRNA virus family
980:All RNA-containing
451:Phosphodiester bond
419:double-stranded RNA
415:small temporal RNAs
269:that catalyzes the
4751:Influential people
4730:Post-translational
4549:Post-transcription
4228:Enzyme superfamily
4161:Enzyme promiscuity
3340:Phosphoinositide 3
3184:phosphotransferase
2983:10.1093/nar/gkq131
2665:10.1093/nar/gkw567
2424:2019-04-03 at the
1550:Dawkins R (1996).
954:
497:
474:(VPg) primer. The
433:RdRp differs from
4771:
4770:
4655:
4654:
4585:
4584:
4461:Special transfers
4384:
4383:
4093:
4092:
4089:
4088:
3965:
3964:
3904:
3903:
3805:
3804:
3718:Template-directed
3472:
3471:
3439:Phosphomevalonate
3081:10.4161/rna.21246
2977:(12): 4092–4107.
2659:(14): 6883–6895.
2171:(20): 4167–4183.
1912:10.3390/v10020076
1792:10.3390/v10010024
1598:10.1002/bit.25578
1566:978-0-393-35309-9
1464:(7057): 356–361.
1427:(10): 3395–3400.
1215:nucleotide analog
941:
940:
937:
936:
933:
932:
928:structure summary
826:
825:
822:
821:
807:structure summary
705:
704:
701:
700:
696:structure summary
545:Neurospora crassa
397:hepatitis C virus
369:RdRps are highly
252:
251:
248:
247:
151:metabolic pathway
16:(Redirected from
4837:
4788:
4787:
4779:
4596:
4493:
4411:
4404:
4397:
4388:
4264:Hanes–Woolf plot
4207:Enzyme activator
4202:Enzyme inhibitor
4176:Enzyme catalysis
4120:
4113:
4106:
4097:
4081:Histidine kinase
4004:tyrosine kinases
3994:protein-tyrosine
3986:
3916:
3723:RNA polymerase I
3546:
3537:
3390:Aspartate kinase
3385:Phosphoglycerate
3202:
3158:
3151:
3144:
3135:
3103:
3102:
3092:
3075:(8): 1045–1049.
3060:
3054:
3053:
3043:
3032:10.1199/tab.0146
3011:
3005:
3004:
2994:
2962:
2953:
2952:
2924:
2918:
2917:
2907:
2875:
2869:
2868:
2858:
2818:
2812:
2811:
2783:
2777:
2776:
2766:
2742:
2736:
2735:
2725:
2693:
2687:
2686:
2676:
2644:
2635:
2634:
2624:
2592:
2586:
2585:
2575:
2565:
2533:
2527:
2526:
2516:
2484:
2478:
2477:
2467:
2450:(6): 1267–1279.
2435:
2429:
2416:
2410:
2409:
2399:
2375:
2369:
2368:
2358:
2338:
2332:
2331:
2321:
2297:
2291:
2290:
2280:
2240:
2234:
2233:
2197:
2191:
2190:
2180:
2156:
2150:
2149:
2139:
2115:
2109:
2108:
2098:
2074:
2068:
2067:
2057:
2048:(8): 1109–1122.
2033:
2027:
2026:
2016:
2006:
1982:
1976:
1975:
1965:
1941:
1935:
1934:
1924:
1914:
1890:
1877:
1876:
1866:
1856:
1824:
1815:
1814:
1804:
1794:
1770:
1759:
1758:
1729:
1723:
1722:
1712:
1680:
1674:
1673:
1663:
1653:
1629:
1620:
1619:
1609:
1592:(8): 1655–1662.
1577:
1571:
1570:
1558:
1547:
1541:
1540:
1504:
1498:
1497:
1453:
1447:
1446:
1436:
1412:
1406:
1405:
1395:
1378:(9): 6083–6096.
1363:
1357:
1356:
1320:
1303:
1300:
1294:
1291:
1229:RNA interference
1223:RNA interference
1057:orthomyxoviruses
876:
830:
755:
709:
644:
565:
561:
560:
557:
541:
533:RNA interference
409:: these amplify
407:RNA interference
359:
348:
310:RNA interference
205:
51:
39:
21:
4845:
4844:
4840:
4839:
4838:
4836:
4835:
4834:
4820:Gene expression
4810:
4809:
4808:
4794:
4782:
4774:
4772:
4767:
4746:
4681:Transcriptional
4651:
4620:
4581:
4572:Polyadenylation
4543:
4517:
4482:
4476:Protein→Protein
4427:
4420:
4418:Gene expression
4415:
4385:
4380:
4292:Oxidoreductases
4278:
4254:Enzyme kinetics
4242:
4238:List of enzymes
4211:
4180:
4151:Catalytic triad
4129:
4124:
4094:
4085:
4056:
4034:
4009:
3980:
3974:
3970:2.7.10-2.7.13:
3961:
3945:
3912:: miscellaneous
3900:
3874:
3856:
3833:
3816:exoribonuclease
3813:
3801:
3787:Polyadenylation
3704:
3530:
3524:
3512:
3494:
3490:
3484:
3468:
3430:
3417:
3394:
3366:
3196:
3190:
3182:
3176:
3162:
3112:
3107:
3106:
3062:
3061:
3057:
3013:
3012:
3008:
2964:
2963:
2956:
2926:
2925:
2921:
2877:
2876:
2872:
2820:
2819:
2815:
2785:
2784:
2780:
2744:
2743:
2739:
2695:
2694:
2690:
2646:
2645:
2638:
2594:
2593:
2589:
2535:
2534:
2530:
2486:
2485:
2481:
2437:
2436:
2432:
2426:Wayback Machine
2417:
2413:
2377:
2376:
2372:
2340:
2339:
2335:
2299:
2298:
2294:
2242:
2241:
2237:
2199:
2198:
2194:
2158:
2157:
2153:
2117:
2116:
2112:
2076:
2075:
2071:
2035:
2034:
2030:
1997:(6): e2006459.
1984:
1983:
1979:
1943:
1942:
1938:
1892:
1891:
1880:
1826:
1825:
1818:
1772:
1771:
1762:
1731:
1730:
1726:
1682:
1681:
1677:
1631:
1630:
1623:
1579:
1578:
1574:
1567:
1556:
1549:
1548:
1544:
1506:
1505:
1501:
1455:
1454:
1450:
1414:
1413:
1409:
1365:
1364:
1360:
1322:
1321:
1317:
1312:
1307:
1306:
1301:
1297:
1292:
1288:
1283:
1266:
1225:
1175:
1159:Paramyxoviridae
1152:
1128:(+)ssRNA genome
1124:
1029:Mononegavirales
946:
553:
537:
489:
431:
367:
366:
365:
364:
363:
360:
351:
350:
349:
338:
318:
58:
37:
28:
23:
22:
18:RNA replication
15:
12:
11:
5:
4843:
4841:
4833:
4832:
4827:
4822:
4812:
4811:
4793:
4792:
4769:
4768:
4766:
4765:
4760:
4758:François Jacob
4754:
4752:
4748:
4747:
4745:
4744:
4743:
4742:
4737:
4727:
4722:
4721:
4720:
4715:
4710:
4700:
4695:
4694:
4693:
4688:
4678:
4677:
4676:
4665:
4663:
4657:
4656:
4653:
4652:
4650:
4649:
4644:
4639:
4634:
4628:
4626:
4622:
4621:
4619:
4618:
4613:
4608:
4602:
4600:
4593:
4587:
4586:
4583:
4582:
4580:
4579:
4574:
4569:
4564:
4559:
4553:
4551:
4545:
4544:
4542:
4541:
4536:
4534:RNA polymerase
4531:
4525:
4523:
4519:
4518:
4516:
4515:
4510:
4505:
4499:
4497:
4490:
4484:
4483:
4481:
4480:
4479:
4478:
4473:
4468:
4458:
4457:
4456:
4438:
4432:
4430:
4422:
4421:
4416:
4414:
4413:
4406:
4399:
4391:
4382:
4381:
4379:
4378:
4365:
4352:
4339:
4326:
4313:
4300:
4286:
4284:
4280:
4279:
4277:
4276:
4271:
4266:
4261:
4256:
4250:
4248:
4244:
4243:
4241:
4240:
4235:
4230:
4225:
4219:
4217:
4216:Classification
4213:
4212:
4210:
4209:
4204:
4199:
4194:
4188:
4186:
4182:
4181:
4179:
4178:
4173:
4168:
4163:
4158:
4153:
4148:
4143:
4137:
4135:
4131:
4130:
4125:
4123:
4122:
4115:
4108:
4100:
4091:
4090:
4087:
4086:
4084:
4083:
4078:
4073:
4067:
4065:
4058:
4057:
4055:
4054:
4045:
4043:
4036:
4035:
4033:
4032:
4023:
4021:
4011:
4010:
4008:
4007:
3998:
3996:
3983:
3978:
3972:protein kinase
3967:
3966:
3963:
3962:
3960:
3959:
3953:
3951:
3947:
3946:
3944:
3943:
3938:
3933:
3928:
3922:
3920:
3913:
3906:
3905:
3902:
3901:
3899:
3898:
3893:
3882:
3880:
3876:
3875:
3873:
3872:
3866:
3864:
3858:
3857:
3855:
3854:
3849:
3843:
3841:
3835:
3834:
3832:
3831:
3826:
3820:
3818:
3811:Phosphorolytic
3807:
3806:
3803:
3802:
3800:
3799:
3794:
3789:
3784:
3779:
3778:
3777:
3772:
3767:
3757:
3756:
3755:
3745:
3740:
3735:
3730:
3725:
3720:
3714:
3712:
3710:RNA polymerase
3706:
3705:
3703:
3702:
3701:
3700:
3690:
3686:
3685:
3684:
3683:
3678:
3673:
3662:
3661:
3660:
3655:
3650:
3645:
3634:
3628:
3627:
3626:
3619:
3614:
3609:
3604:
3593:
3592:
3591:
3584:
3579:
3574:
3569:
3558:
3554:
3552:
3550:DNA polymerase
3543:
3534:
3528:
3514:
3513:
3511:
3510:
3505:
3499:
3497:
3492:
3488:
3474:
3473:
3470:
3469:
3467:
3466:
3461:
3456:
3451:
3446:
3441:
3435:
3433:
3428:
3419:
3418:
3416:
3415:
3409:
3407:
3396:
3395:
3393:
3392:
3387:
3381:
3379:
3368:
3367:
3365:
3364:
3359:
3354:
3353:
3352:
3347:
3337:
3335:Diacylglycerol
3332:
3327:
3322:
3317:
3312:
3307:
3302:
3297:
3296:
3295:
3285:
3280:
3275:
3274:
3273:
3268:
3263:
3258:
3253:
3246:Phosphofructo-
3243:
3238:
3237:
3236:
3226:
3221:
3215:
3213:
3199:
3194:
3178:
3177:
3163:
3161:
3160:
3153:
3146:
3138:
3132:
3131:
3123:
3111:
3110:External links
3108:
3105:
3104:
3055:
3006:
2954:
2935:(2): 139–146.
2919:
2870:
2813:
2778:
2757:(2): 235–245.
2737:
2688:
2636:
2607:(3): 433–443.
2587:
2528:
2479:
2430:
2411:
2390:(2): 241–250.
2370:
2333:
2312:(2): 317–325.
2292:
2235:
2192:
2151:
2130:(2): 287–303.
2110:
2069:
2028:
1977:
1956:(4): 455–470.
1936:
1878:
1816:
1760:
1724:
1695:(6): 460–466.
1675:
1621:
1572:
1565:
1542:
1515:(2): 155–158.
1499:
1448:
1407:
1358:
1331:(1–2): 42–46.
1314:
1313:
1311:
1308:
1305:
1304:
1295:
1285:
1284:
1282:
1279:
1278:
1277:
1275:NS5B inhibitor
1272:
1265:
1262:
1244:P. tetraurelia
1224:
1221:
1220:
1219:
1207:
1180:
1174:
1173:Drug therapies
1171:
1151:
1148:
1123:
1120:
1113:heterotrimeric
1083:
1082:
1063:
1043:
1025:
1024:
1023:
1020:Partitiviridae
997:
996:(dsRNA phages)
982:bacteriophages
978:
945:
942:
939:
938:
935:
934:
931:
930:
925:
919:
918:
905:
899:
898:
888:
881:
880:
872:
871:
866:
860:
859:
854:
848:
847:
844:
840:
839:
835:
834:
827:
824:
823:
820:
819:
816:
810:
809:
804:
798:
797:
784:
778:
777:
767:
760:
759:
751:
750:
745:
739:
738:
733:
727:
726:
723:
719:
718:
714:
713:
706:
703:
702:
699:
698:
693:
687:
686:
673:
667:
666:
656:
649:
648:
640:
639:
626:
620:
619:
614:
608:
607:
602:
595:
594:
589:
583:
582:
579:
575:
574:
570:
569:
525:ribonucleoside
488:
485:
466:-independent (
460:
459:
455:
448:
445:
430:
427:
361:
354:
353:
352:
343:
342:
341:
340:
339:
337:
334:
317:
314:
289:of RNA from a
250:
249:
246:
245:
240:
234:
233:
228:
222:
221:
216:
210:
209:
201:
200:
191:
185:
184:
173:
166:
165:
160:
154:
153:
148:
142:
141:
136:
130:
129:
124:
118:
117:
112:
106:
105:
100:
94:
93:
89:
88:
83:
77:
76:
71:
65:
64:
60:
59:
52:
44:
43:
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
4842:
4831:
4828:
4826:
4823:
4821:
4818:
4817:
4815:
4807:
4803:
4799:
4791:
4786:
4781:
4777:
4764:
4763:Jacques Monod
4761:
4759:
4756:
4755:
4753:
4749:
4741:
4738:
4736:
4733:
4732:
4731:
4728:
4726:
4725:Translational
4723:
4719:
4716:
4714:
4711:
4709:
4706:
4705:
4704:
4701:
4699:
4696:
4692:
4689:
4687:
4684:
4683:
4682:
4679:
4675:
4672:
4671:
4670:
4667:
4666:
4664:
4662:
4658:
4648:
4645:
4643:
4640:
4638:
4635:
4633:
4630:
4629:
4627:
4623:
4617:
4614:
4612:
4609:
4607:
4604:
4603:
4601:
4597:
4594:
4592:
4588:
4578:
4575:
4573:
4570:
4568:
4565:
4563:
4560:
4558:
4555:
4554:
4552:
4550:
4546:
4540:
4537:
4535:
4532:
4530:
4527:
4526:
4524:
4520:
4514:
4511:
4509:
4506:
4504:
4501:
4500:
4498:
4494:
4491:
4489:
4488:Transcription
4485:
4477:
4474:
4472:
4469:
4467:
4464:
4463:
4462:
4459:
4455:
4451:
4447:
4444:
4443:
4442:
4441:Central dogma
4439:
4437:
4434:
4433:
4431:
4429:
4423:
4419:
4412:
4407:
4405:
4400:
4398:
4393:
4392:
4389:
4376:
4372:
4371:
4366:
4363:
4359:
4358:
4353:
4350:
4346:
4345:
4340:
4337:
4333:
4332:
4327:
4324:
4320:
4319:
4314:
4311:
4307:
4306:
4301:
4298:
4294:
4293:
4288:
4287:
4285:
4281:
4275:
4272:
4270:
4267:
4265:
4262:
4260:
4257:
4255:
4252:
4251:
4249:
4245:
4239:
4236:
4234:
4233:Enzyme family
4231:
4229:
4226:
4224:
4221:
4220:
4218:
4214:
4208:
4205:
4203:
4200:
4198:
4197:Cooperativity
4195:
4193:
4190:
4189:
4187:
4183:
4177:
4174:
4172:
4169:
4167:
4164:
4162:
4159:
4157:
4156:Oxyanion hole
4154:
4152:
4149:
4147:
4144:
4142:
4139:
4138:
4136:
4132:
4128:
4121:
4116:
4114:
4109:
4107:
4102:
4101:
4098:
4082:
4079:
4077:
4074:
4072:
4069:
4068:
4066:
4063:
4059:
4053:
4052:
4047:
4046:
4044:
4041:
4037:
4031:
4030:
4025:
4024:
4022:
4020:
4016:
4012:
4006:
4005:
4000:
3999:
3997:
3995:
3991:
3987:
3984:
3981:
3973:
3968:
3958:
3955:
3954:
3952:
3948:
3942:
3939:
3937:
3934:
3932:
3929:
3927:
3924:
3923:
3921:
3917:
3914:
3911:
3907:
3897:
3894:
3891:
3887:
3884:
3883:
3881:
3877:
3871:
3868:
3867:
3865:
3863:
3859:
3853:
3850:
3848:
3845:
3844:
3842:
3840:
3836:
3830:
3827:
3825:
3822:
3821:
3819:
3817:
3812:
3808:
3798:
3795:
3793:
3790:
3788:
3785:
3783:
3780:
3776:
3773:
3771:
3768:
3766:
3763:
3762:
3761:
3758:
3754:
3751:
3750:
3749:
3746:
3744:
3741:
3739:
3736:
3734:
3731:
3729:
3726:
3724:
3721:
3719:
3716:
3715:
3713:
3711:
3707:
3699:
3696:
3695:
3694:
3691:
3688:
3687:
3682:
3679:
3677:
3674:
3672:
3669:
3668:
3666:
3663:
3659:
3656:
3654:
3651:
3649:
3646:
3644:
3641:
3640:
3638:
3635:
3632:
3629:
3625:
3624:
3620:
3618:
3615:
3613:
3610:
3608:
3605:
3603:
3600:
3599:
3597:
3594:
3590:
3589:
3585:
3583:
3580:
3578:
3575:
3573:
3570:
3568:
3565:
3564:
3562:
3559:
3556:
3555:
3553:
3551:
3547:
3544:
3542:
3538:
3535:
3532:
3523:
3519:
3515:
3509:
3506:
3504:
3501:
3500:
3498:
3495:
3483:
3479:
3475:
3465:
3462:
3460:
3457:
3455:
3452:
3450:
3447:
3445:
3442:
3440:
3437:
3436:
3434:
3431:
3424:
3420:
3414:
3411:
3410:
3408:
3405:
3401:
3397:
3391:
3388:
3386:
3383:
3382:
3380:
3377:
3373:
3369:
3363:
3360:
3358:
3355:
3351:
3350:Class II PI 3
3348:
3346:
3343:
3342:
3341:
3338:
3336:
3333:
3331:
3328:
3326:
3325:Deoxycytidine
3323:
3321:
3318:
3316:
3313:
3311:
3308:
3306:
3303:
3301:
3298:
3294:
3293:ADP-thymidine
3291:
3290:
3289:
3286:
3284:
3281:
3279:
3276:
3272:
3269:
3267:
3264:
3262:
3259:
3257:
3254:
3252:
3249:
3248:
3247:
3244:
3242:
3239:
3235:
3232:
3231:
3230:
3227:
3225:
3222:
3220:
3217:
3216:
3214:
3211:
3207:
3203:
3200:
3197:
3189:
3185:
3179:
3174:
3170:
3166:
3159:
3154:
3152:
3147:
3145:
3140:
3139:
3136:
3130:
3127:
3124:
3121:
3117:
3116:RNA+Replicase
3114:
3113:
3109:
3100:
3096:
3091:
3086:
3082:
3078:
3074:
3070:
3066:
3059:
3056:
3051:
3047:
3042:
3037:
3033:
3029:
3025:
3021:
3017:
3010:
3007:
3002:
2998:
2993:
2988:
2984:
2980:
2976:
2972:
2968:
2961:
2959:
2955:
2950:
2946:
2942:
2938:
2934:
2930:
2923:
2920:
2915:
2911:
2906:
2901:
2897:
2893:
2889:
2885:
2881:
2874:
2871:
2866:
2862:
2857:
2852:
2848:
2844:
2840:
2836:
2832:
2828:
2824:
2817:
2814:
2809:
2805:
2801:
2797:
2793:
2789:
2782:
2779:
2774:
2770:
2765:
2760:
2756:
2752:
2748:
2741:
2738:
2733:
2729:
2724:
2719:
2715:
2711:
2707:
2703:
2699:
2692:
2689:
2684:
2680:
2675:
2670:
2666:
2662:
2658:
2654:
2650:
2643:
2641:
2637:
2632:
2628:
2623:
2618:
2614:
2610:
2606:
2602:
2598:
2591:
2588:
2583:
2579:
2574:
2569:
2564:
2559:
2555:
2551:
2548:(5): e96070.
2547:
2543:
2539:
2532:
2529:
2524:
2520:
2515:
2510:
2506:
2502:
2498:
2494:
2490:
2483:
2480:
2475:
2471:
2466:
2461:
2457:
2453:
2449:
2445:
2441:
2434:
2431:
2427:
2423:
2420:
2415:
2412:
2407:
2403:
2398:
2393:
2389:
2385:
2381:
2374:
2371:
2366:
2362:
2357:
2352:
2348:
2344:
2337:
2334:
2329:
2325:
2320:
2315:
2311:
2307:
2303:
2296:
2293:
2288:
2284:
2279:
2274:
2270:
2266:
2262:
2258:
2254:
2250:
2246:
2239:
2236:
2231:
2227:
2223:
2219:
2215:
2211:
2207:
2203:
2196:
2193:
2188:
2184:
2179:
2174:
2170:
2166:
2162:
2155:
2152:
2147:
2143:
2138:
2133:
2129:
2125:
2121:
2114:
2111:
2106:
2102:
2097:
2092:
2088:
2084:
2080:
2073:
2070:
2065:
2061:
2056:
2051:
2047:
2043:
2039:
2032:
2029:
2024:
2020:
2015:
2010:
2005:
2000:
1996:
1992:
1988:
1981:
1978:
1973:
1969:
1964:
1959:
1955:
1951:
1947:
1940:
1937:
1932:
1928:
1923:
1918:
1913:
1908:
1904:
1900:
1896:
1889:
1887:
1885:
1883:
1879:
1874:
1870:
1865:
1860:
1855:
1850:
1846:
1842:
1838:
1834:
1830:
1823:
1821:
1817:
1812:
1808:
1803:
1798:
1793:
1788:
1784:
1780:
1776:
1769:
1767:
1765:
1761:
1756:
1752:
1748:
1744:
1740:
1736:
1728:
1725:
1720:
1716:
1711:
1706:
1702:
1698:
1694:
1690:
1689:Cell Research
1686:
1679:
1676:
1671:
1667:
1662:
1657:
1652:
1647:
1643:
1639:
1635:
1628:
1626:
1622:
1617:
1613:
1608:
1603:
1599:
1595:
1591:
1587:
1583:
1576:
1573:
1568:
1562:
1555:
1554:
1546:
1543:
1538:
1534:
1530:
1526:
1522:
1518:
1514:
1510:
1503:
1500:
1495:
1491:
1487:
1483:
1479:
1475:
1471:
1467:
1463:
1459:
1452:
1449:
1444:
1440:
1435:
1430:
1426:
1422:
1418:
1411:
1408:
1403:
1399:
1394:
1389:
1385:
1381:
1377:
1373:
1369:
1362:
1359:
1354:
1350:
1346:
1342:
1338:
1334:
1330:
1326:
1319:
1316:
1309:
1299:
1296:
1290:
1287:
1280:
1276:
1273:
1271:
1268:
1267:
1263:
1261:
1259:
1255:
1251:
1247:
1245:
1241:
1239:
1234:
1230:
1222:
1216:
1211:
1208:
1205:
1201:
1197:
1193:
1189:
1185:
1181:
1177:
1176:
1172:
1170:
1168:
1164:
1160:
1156:
1149:
1147:
1145:
1144:tombusviruses
1141:
1137:
1136:recombination
1133:
1129:
1122:Recombination
1121:
1119:
1116:
1114:
1110:
1109:
1105:and one from
1104:
1103:
1097:
1096:RNA replicase
1093:
1091:
1087:
1080:
1079:
1074:
1070:
1069:
1064:
1062:
1058:
1054:
1053:
1048:
1044:
1041:
1040:
1035:
1031:
1030:
1026:
1022:
1021:
1016:
1015:
1010:
1009:
1004:
1003:
998:
995:
994:
989:
988:
983:
979:
977:
976:
975:Coronaviridae
971:
970:
969:
968:
963:
959:
958:
957:
950:
943:
929:
926:
924:
920:
917:
913:
909:
906:
904:
900:
896:
892:
889:
886:
882:
877:
873:
870:
867:
865:
861:
858:
855:
853:
849:
845:
841:
836:
831:
828:
817:
815:
811:
808:
805:
803:
799:
796:
792:
788:
785:
783:
779:
775:
771:
768:
765:
761:
756:
752:
749:
746:
744:
740:
737:
734:
732:
728:
724:
720:
715:
710:
707:
697:
694:
692:
688:
685:
681:
677:
674:
672:
668:
664:
660:
657:
654:
650:
645:
641:
638:
634:
630:
627:
625:
621:
618:
615:
613:
609:
606:
603:
600:
596:
593:
590:
588:
584:
580:
576:
571:
566:
563:
559:
556:
551:
550:Bacteriophage
547:
546:
540:
534:
529:
526:
522:
518:
517:aspartic acid
514:
510:
509:alpha helices
506:
503:
493:
486:
484:
480:
477:
473:
469:
465:
456:
452:
449:
446:
443:
440:
439:
438:
436:
428:
426:
424:
420:
416:
412:
408:
403:
401:
398:
394:
389:
387:
383:
378:
376:
372:
358:
347:
335:
333:
331:
330:actinomycin D
327:
323:
315:
313:
311:
307:
303:
299:
294:
292:
288:
287:transcription
284:
280:
279:complementary
276:
272:
268:
264:
263:RNA replicase
260:
256:
244:
241:
239:
235:
232:
229:
227:
223:
220:
217:
215:
211:
206:
202:
199:
195:
192:
190:
189:Gene Ontology
186:
183:
180:
177:
174:
171:
167:
164:
161:
159:
155:
152:
149:
147:
143:
140:
137:
135:
131:
128:
127:NiceZyme view
125:
123:
119:
116:
113:
111:
107:
104:
101:
99:
95:
90:
87:
84:
82:
78:
75:
72:
70:
66:
61:
56:
50:
45:
40:
35:
30:
19:
4740:irreversible
4625:Key elements
4522:Key elements
4465:
4436:Genetic code
4426:Introduction
4370:Translocases
4367:
4354:
4341:
4328:
4315:
4305:Transferases
4302:
4289:
4146:Binding site
4048:
4026:
4001:
3781:
3622:
3587:
3345:Class I PI 3
3310:Pantothenate
3181:2.7.1-2.7.4:
3165:Transferases
3072:
3068:
3058:
3023:
3019:
3009:
2974:
2970:
2932:
2928:
2922:
2887:
2883:
2873:
2830:
2826:
2816:
2791:
2787:
2781:
2754:
2750:
2740:
2705:
2701:
2691:
2656:
2652:
2604:
2600:
2590:
2545:
2541:
2531:
2496:
2492:
2482:
2447:
2443:
2433:
2414:
2387:
2383:
2373:
2346:
2336:
2309:
2305:
2295:
2252:
2248:
2238:
2208:(2): 85–98.
2205:
2201:
2195:
2168:
2164:
2154:
2127:
2123:
2113:
2086:
2082:
2072:
2045:
2041:
2031:
1994:
1991:PLOS Biology
1990:
1980:
1953:
1949:
1939:
1902:
1898:
1836:
1832:
1782:
1778:
1741:(1): 29–39.
1738:
1734:
1727:
1692:
1688:
1678:
1641:
1637:
1589:
1585:
1575:
1552:
1545:
1512:
1508:
1502:
1461:
1457:
1451:
1424:
1420:
1410:
1375:
1371:
1361:
1328:
1325:FEBS Letters
1324:
1318:
1298:
1289:
1253:
1242:
1236:
1226:
1158:
1155:Sendai virus
1153:
1140:carmoviruses
1125:
1117:
1106:
1100:
1094:
1086:Flaviviruses
1084:
1076:
1068:Birnaviridae
1066:
1061:bunyaviruses
1050:
1037:
1027:
1018:
1012:
1006:
1000:
993:Cystoviridae
991:
987:Fiersviridae
985:
973:
967:Birnaviridae
965:
962:retroviruses
955:
543:
530:
502:antiparallel
498:
481:
475:
467:
461:
432:
417:and produce
404:
390:
379:
368:
336:Distribution
319:
295:
262:
258:
254:
253:
115:BRENDA entry
29:
4591:Translation
4428:to genetics
4141:Active site
3896:Transposase
3886:Recombinase
3531:-nucleoside
3357:Sphingosine
3069:RNA Biology
2794:: 247–257.
1184:Hepatitis C
1179:functional.
1108:Leviviridae
1090:polyprotein
1055:), such as
1014:Hypoviridae
1008:Totiviridae
838:Identifiers
717:Identifiers
573:Identifiers
531:Eukaryotic
429:Replication
386:endocytosis
382:polio virus
326:polio virus
271:replication
103:IntEnz view
63:Identifiers
57:(PDB 4WTG).
4814:Categories
4735:reversible
4698:lac operon
4674:imprinting
4669:Epigenetic
4661:Regulation
4616:Eukaryotic
4562:5' capping
4513:Eukaryotic
4344:Isomerases
4318:Hydrolases
4185:Regulation
3698:Telomerase
3541:Polymerase
3315:Mevalonate
3278:Riboflavin
3169:phosphorus
2929:Toxicology
1310:References
1254:C. elegans
1238:C. elegans
1218:processes.
1200:remdesivir
1192:Sofosbuvir
1132:poliovirus
1102:Pestivirus
1002:Reoviridae
891:structures
846:Bunya_RdRp
770:structures
659:structures
521:asparagine
505:beta sheet
375:telomerase
322:mengovirus
306:eukaryotes
302:SARS-CoV-2
293:template.
172:structures
139:KEGG entry
86:9026-28-2
55:sofosbuvir
4806:IPR000208
4606:Bacterial
4503:Bacterial
4223:EC number
3890:Integrase
3814:3' to 5'
3459:Guanylate
3454:Uridylate
3444:Adenylate
3288:Thymidine
3283:Shikimate
3026:: e0146.
2255:: 15774.
2042:Structure
1905:(2): 76.
1785:(1): 24.
1210:GS-441524
1196:Ribavirin
1078:IPR007100
1052:IPR007099
1039:IPR016269
869:IPR007322
748:IPR007855
617:IPR001205
507:with two
487:Structure
458:template.
411:microRNAs
402:protein.
371:conserved
92:Databases
4830:EC 2.7.7
4802:InterPro
4718:microRNA
4632:Ribosome
4611:Archaeal
4567:Splicing
4539:Promoter
4508:Archaeal
4452: →
4448: →
4247:Kinetics
4171:Cofactor
4134:Activity
3824:RNase PH
3432:acceptor
3413:Creatine
3406:acceptor
3378:acceptor
3320:Pyruvate
3305:Glycerol
3266:Platelet
3241:Galacto-
3212:acceptor
3129:2.7.7.48
3099:22858672
3050:22303271
3001:20200046
2914:33055231
2865:32358203
2808:23291407
2773:12504565
2751:Virology
2732:14581540
2683:27317698
2582:24816789
2542:PLOS ONE
2523:30482837
2474:26004069
2422:Archived
2406:12069523
2384:Virology
2306:Virology
2287:28585540
2230:30004345
2222:21233849
2187:31103775
2124:Virology
2105:10827187
2023:29953453
1931:29439438
1873:27339134
1811:29300357
1755:19616606
1719:15625012
1670:12553882
1616:25726926
1537:11491076
1486:16163346
1443:14085393
1353:36482110
1264:See also
1188:COVID-19
1167:oligomer
1157:(family
1073:InterPro
1047:InterPro
1034:InterPro
908:RCSB PDB
864:InterPro
787:RCSB PDB
743:InterPro
725:RdRP_euk
676:RCSB PDB
612:InterPro
243:proteins
231:articles
219:articles
176:RCSB PDB
74:2.7.7.48
4790:Biology
4471:RNA→DNA
4466:RNA→RNA
4454:Protein
4357:Ligases
4127:Enzymes
3775:PrimPol
3760:Primase
3234:Hepatic
3229:Fructo-
3090:3551858
3041:3268507
2992:2896523
2905:7566896
2856:7199908
2835:Bibcode
2827:Science
2674:5001610
2631:3021340
2622:7133339
2573:4015915
2550:Bibcode
2514:6282212
2465:4459711
2365:1895057
2328:8607261
2278:5467207
2257:Bibcode
2146:9878607
2064:9309225
2014:6040757
1972:9630251
1963:1369631
1922:5850383
1899:Viruses
1864:4948327
1841:Bibcode
1802:5795437
1779:Viruses
1710:7092015
1607:5653219
1529:2449282
1494:4370363
1466:Bibcode
1402:8709232
1345:2759231
1075::
1049::
1036::
944:Viruses
857:PF04196
736:PF05183
592:PF00680
476:de novo
468:de novo
395:L, and
316:History
304:. Some
298:viruses
198:QuickGO
163:profile
146:MetaCyc
81:CAS no.
4776:Portal
4331:Lyases
4062:2.7.13
4040:2.7.12
4015:2.7.11
3990:2.7.10
3829:PNPase
3797:PNPase
3753:POLRMT
3748:ssRNAP
3261:Muscle
3224:Gluco-
3188:kinase
3122:(MeSH)
3097:
3087:
3048:
3038:
2999:
2989:
2947:
2912:
2902:
2863:
2853:
2806:
2771:
2730:
2723:254248
2720:
2681:
2671:
2629:
2619:
2580:
2570:
2521:
2511:
2472:
2462:
2404:
2363:
2326:
2285:
2275:
2228:
2220:
2185:
2144:
2103:
2062:
2021:
2011:
1970:
1960:
1929:
1919:
1871:
1861:
1809:
1799:
1753:
1717:
1707:
1668:
1661:151600
1658:
1614:
1604:
1563:
1535:
1527:
1492:
1484:
1458:Nature
1441:
1400:
1393:190630
1390:
1351:
1343:
1250:plants
1233:siRNAs
1130:, the
923:PDBsum
897:
887:
843:Symbol
802:PDBsum
776:
766:
722:Symbol
691:PDBsum
665:
655:
637:SUPFAM
605:CL0027
581:RdRP_1
578:Symbol
555:O31945
539:Q9Y7G6
513:motifs
464:primer
421:using
267:enzyme
265:is an
226:PubMed
208:Search
194:AmiGO
182:PDBsum
122:ExPASy
110:BRENDA
98:IntEnz
69:EC no.
4599:Types
4496:Types
4283:Types
3910:2.7.8
3879:Other
3518:2.7.7
3478:2.7.6
3423:2.7.4
3400:2.7.3
3372:2.7.2
3256:Liver
3219:Hexo-
3206:2.7.1
2890:(1).
2499:(6).
2226:S2CID
1644:: 1.
1557:(PDF)
1533:S2CID
1490:S2CID
1349:S2CID
1281:Notes
1202:, an
633:SCOPe
624:SCOP2
542:) in
261:) or
158:PRIAM
4800:and
4798:Pfam
4375:list
4368:EC7
4362:list
4355:EC6
4349:list
4342:EC5
4336:list
4329:EC4
4323:list
4316:EC3
4310:list
4303:EC2
4297:list
4290:EC1
4049:see
4027:see
4002:see
3376:COOH
3175:2.7)
3095:PMID
3046:PMID
2997:PMID
2949:1873
2945:PMID
2910:PMID
2861:PMID
2804:PMID
2769:PMID
2728:PMID
2679:PMID
2627:PMID
2601:Cell
2578:PMID
2519:PMID
2493:mBio
2470:PMID
2444:Cell
2402:PMID
2361:PMID
2324:PMID
2283:PMID
2218:PMID
2183:PMID
2142:PMID
2101:PMID
2060:PMID
2019:PMID
1968:PMID
1927:PMID
1869:PMID
1807:PMID
1751:PMID
1735:Gene
1715:PMID
1666:PMID
1612:PMID
1561:ISBN
1525:PMID
1509:Cell
1482:PMID
1439:PMID
1398:PMID
1341:PMID
1258:RISC
1248:and
1194:and
1186:and
1142:and
1059:and
964:and
916:PDBj
912:PDBe
895:ECOD
885:Pfam
852:Pfam
818:2j7n
795:PDBj
791:PDBe
774:ECOD
764:Pfam
731:Pfam
684:PDBj
680:PDBe
663:ECOD
653:Pfam
629:2jlg
601:clan
599:Pfam
587:Pfam
413:and
400:NS5B
324:and
259:RdRp
238:NCBI
179:PDBe
134:KEGG
4825:RNA
4450:RNA
4446:DNA
3792:PAP
3733:III
3667:/Y
3658:TDT
3639:/X
3631:III
3623:Pfu
3598:/B
3588:Taq
3563:/A
3330:PFP
3300:NAD
3085:PMC
3077:doi
3036:PMC
3028:doi
2987:PMC
2979:doi
2937:doi
2900:PMC
2892:doi
2851:PMC
2843:doi
2831:368
2796:doi
2759:doi
2755:304
2718:PMC
2710:doi
2669:PMC
2661:doi
2617:PMC
2609:doi
2568:PMC
2558:doi
2509:PMC
2501:doi
2460:PMC
2452:doi
2448:161
2392:doi
2388:296
2351:doi
2314:doi
2310:216
2273:PMC
2265:doi
2210:doi
2173:doi
2169:431
2132:doi
2128:252
2091:doi
2087:275
2050:doi
2009:PMC
1999:doi
1958:PMC
1950:RNA
1917:PMC
1907:doi
1859:PMC
1849:doi
1837:113
1797:PMC
1787:doi
1743:doi
1739:447
1705:PMC
1697:doi
1656:PMC
1646:doi
1602:PMC
1594:doi
1590:112
1517:doi
1474:doi
1462:437
1429:doi
1425:238
1388:PMC
1380:doi
1333:doi
1329:252
1204:FDA
903:PDB
814:PDB
782:PDB
671:PDB
291:DNA
275:RNA
273:of
214:PMC
170:PDB
4816::
4804::
4017::
3992::
3977:PO
3738:IV
3728:II
3637:IV
3633:/C
3596:II
3582:T7
3527:PO
3520::
3480::
3427:PO
3425::
3402::
3374::
3210:OH
3208::
3193:PO
3173:EC
3167::
3126:EC
3093:.
3083:.
3071:.
3067:.
3044:.
3034:.
3022:.
3018:.
2995:.
2985:.
2975:38
2973:.
2969:.
2957:^
2943:.
2931:.
2908:.
2898:.
2888:34
2886:.
2882:.
2859:.
2849:.
2841:.
2829:.
2825:.
2802:.
2792:14
2790:.
2767:.
2753:.
2749:.
2726:.
2716:.
2706:77
2704:.
2700:.
2677:.
2667:.
2657:44
2655:.
2651:.
2639:^
2625:.
2615:.
2605:47
2603:.
2599:.
2576:.
2566:.
2556:.
2544:.
2540:.
2517:.
2507:.
2495:.
2491:.
2468:.
2458:.
2446:.
2442:.
2400:.
2386:.
2382:.
2359:.
2345:.
2322:.
2308:.
2304:.
2281:.
2271:.
2263:.
2251:.
2247:.
2224:.
2216:.
2204:.
2181:.
2167:.
2163:.
2140:.
2126:.
2122:.
2099:.
2085:.
2081:.
2058:.
2044:.
2040:.
2017:.
2007:.
1995:16
1993:.
1989:.
1966:.
1952:.
1948:.
1925:.
1915:.
1903:10
1901:.
1897:.
1881:^
1867:.
1857:.
1847:.
1835:.
1831:.
1819:^
1805:.
1795:.
1783:10
1781:.
1777:.
1763:^
1749:.
1737:.
1713:.
1703:.
1693:14
1691:.
1687:.
1664:.
1654:.
1640:.
1636:.
1624:^
1610:.
1600:.
1588:.
1584:.
1531:.
1523:.
1513:52
1511:.
1488:.
1480:.
1472:.
1460:.
1437:.
1423:.
1419:.
1396:.
1386:.
1376:70
1374:.
1370:.
1347:.
1339:.
1327:.
1146:.
1017:,
1011:,
1005:,
914:;
910:;
893:/
793:;
789:;
772:/
682:;
678:;
661:/
635:/
631:/
196:/
4778::
4410:e
4403:t
4396:v
4377:)
4373:(
4364:)
4360:(
4351:)
4347:(
4338:)
4334:(
4325:)
4321:(
4312:)
4308:(
4299:)
4295:(
4119:e
4112:t
4105:v
3979:4
3975:(
3892:)
3888:(
3770:2
3765:1
3743:V
3681:κ
3676:ι
3671:η
3665:V
3653:μ
3648:λ
3643:β
3617:ζ
3612:ε
3607:δ
3602:α
3577:ν
3572:θ
3567:γ
3561:I
3533:)
3529:4
3525:(
3496:)
3493:7
3491:O
3489:2
3487:P
3485:(
3429:4
3404:N
3271:2
3251:1
3198:)
3195:4
3191:(
3186:/
3157:e
3150:t
3143:v
3101:.
3079::
3073:9
3052:.
3030::
3024:9
3003:.
2981::
2951:.
2939::
2933:5
2916:.
2894::
2867:.
2845::
2837::
2810:.
2798::
2775:.
2761::
2734:.
2712::
2685:.
2663::
2633:.
2611::
2584:.
2560::
2552::
2546:9
2525:.
2503::
2497:9
2476:.
2454::
2428:.
2408:.
2394::
2367:.
2353::
2330:.
2316::
2289:.
2267::
2259::
2253:8
2232:.
2212::
2206:9
2189:.
2175::
2148:.
2134::
2107:.
2093::
2066:.
2052::
2046:5
2025:.
2001::
1974:.
1954:4
1933:.
1909::
1875:.
1851::
1843::
1813:.
1789::
1757:.
1745::
1721:.
1699::
1672:.
1648::
1642:3
1618:.
1596::
1569:.
1539:.
1519::
1496:.
1476::
1468::
1445:.
1431::
1404:.
1382::
1355:.
1335::
1246:,
1240:,
1081:)
1071:(
1042:)
536:(
257:(
36:.
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.