1424:
1092:(GFP)-tagged DNA polymerases α. They detected DNA replication of pairs of the tagged loci spaced apart symmetrically from a replication origin and found that the distance between the pairs decreased markedly by time. This finding suggests that the mechanism of DNA replication goes with DNA factories. That is, couples of replication factories are loaded on replication origins and the factories associated with each other. Also, template DNAs move into the factories, which bring extrusion of the template ssDNAs and new DNAs. Meister's finding is the first direct evidence of replication factory model. Subsequent research has shown that DNA helicases form dimers in many eukaryotic cells and bacterial replication machineries stay in single intranuclear location during DNA synthesis.
1416:
1592:
growing strand and not on the free nucleotides, proof-reading by removing a mismatched terminal nucleotide would be problematic: Once a nucleotide is added, the triphosphate is lost and a single phosphate remains on the backbone between the new nucleotide and the rest of the strand. If the added nucleotide were mismatched, removal would result in a DNA strand terminated by a monophosphate at the end of the "growing strand" rather than a high energy triphosphate. So strand would be stuck and wouldn't be able to grow anymore. In actuality, the high energy triphosphates hydrolyzed at each step originate from the free nucleotides, not the polymerized strand, so this issue does not exist.
491:. Sequences used by initiator proteins tend to be "AT-rich" (rich in adenine and thymine bases), because A-T base pairs have two hydrogen bonds (rather than the three formed in a C-G pair) and thus are easier to strand-separate. In eukaryotes, the origin recognition complex catalyzes the assembly of initiator proteins into the pre-replication complex. In addition, a recent report suggests that budding yeast ORC dimerizes in a cell cycle dependent manner to control licensing. In turn, the process of ORC dimerization is mediated by a cell cycle-dependent Noc3p dimerization cycle in vivo, and this role of Noc3p is separable from its role in ribosome biogenesis.
293:, and the different ends of a single strand are called the "3′ (three-prime) end" and the "5′ (five-prime) end". By convention, if the base sequence of a single strand of DNA is given, the left end of the sequence is the 5′ end, while the right end of the sequence is the 3′ end. The strands of the double helix are anti-parallel, with one being 5′ to 3′, and the opposite strand 3′ to 5′. These terms refer to the carbon atom in deoxyribose to which the next phosphate in the chain attaches. Directionality has consequences in DNA synthesis, because DNA polymerase can synthesize DNA in only one direction by adding nucleotides to the 3′ end of a DNA strand.
424:
400:
300:) means that the information contained within each strand is redundant. Phosphodiester (intra-strand) bonds are stronger than hydrogen (inter-strand) bonds. The actual job of the phosphodiester bonds is where in DNA polymers connect the 5' carbon atom of one nucleotide to the 3' carbon atom of another nucleotide, while the hydrogen bonds stabilize DNA double helices across the helix axis but not in the direction of the axis. This makes it possible to separate the strands from one another. The nucleotides on a single strand can therefore be used to reconstruct nucleotides on a newly synthesized partner strand.
1228:, which causes proteolytic destruction of Cdc6. Cdk-dependent phosphorylation of Mcm proteins promotes their export out of the nucleus along with Cdt1 during S phase, preventing the loading of new Mcm complexes at origins during a single cell cycle. Cdk phosphorylation of the origin replication complex also inhibits pre-replication complex assembly. The individual presence of any of these three mechanisms is sufficient to inhibit pre-replication complex assembly. However, mutations of all three proteins in the same cell does trigger reinitiation at many origins of replication within one cell cycle.
1286:
stuck forks are not copied, then the daughter strands get nick nick unreplicated sites. The un-replicated sites on one parent's strand hold the other strand together but not daughter strands. Therefore, the resulting sister chromatids cannot separate from each other and cannot divide into 2 daughter cells. When neighboring origins fire and a fork from one origin is stalled, fork from other origin access on an opposite direction of the stalled fork and duplicate the un-replicated sites. As other mechanism of the rescue there is application of
193:
771:
1080:
replication machineries these components coordinate. In most of the bacteria, all of the factors involved in DNA replication are located on replication forks and the complexes stay on the forks during DNA replication. Replication machineries are also referred to as replisomes, or DNA replication systems. These terms are generic terms for proteins located on replication forks. In eukaryotic and some bacterial cells the replisomes are not formed.
408:
1385:
31:
1495:. Repeating this process through multiple cycles amplifies the targeted DNA region. At the start of each cycle, the mixture of template and primers is heated, separating the newly synthesized molecule and template. Then, as the mixture cools, both of these become templates for annealing of new primers, and the polymerase extends from these. As a result, the number of copies of the target region doubles each round,
758:
5507:
1299:
315:
1174:
835:
738:, the low-processivity enzyme, Pol α, helps to initiate replication because it forms a complex with primase. In eukaryotes, leading strand synthesis is thought to be conducted by Pol ε; however, this view has recently been challenged, suggesting a role for Pol δ. Primer removal is completed Pol δ while repair of DNA during replication is completed by Pol ε.
5519:
1069:
84:. The double helix describes the appearance of a double-stranded DNA which is thus composed of two linear strands that run opposite to each other and twist together to form. During replication, these strands are separated. Each strand of the original DNA molecule then serves as a template for the production of its counterpart, a process referred to as
1344:, chromosome replication takes more time than dividing the cell. The bacteria solve this by initiating a new round of replication before the previous one has been terminated. The new round of replication will form the chromosome of the cell that is born two generations after the dividing cell. This mechanism creates overlapping replication cycles.
1255:. Replication sites can be detected by immunostaining daughter strands and replication enzymes and monitoring GFP-tagged replication factors. By these methods it is found that replication foci of varying size and positions appear in S phase of cell division and their number per nucleus is far smaller than the number of genomic replication forks.
432:
374:
post-replication mismatch repair mechanisms monitor the DNA for errors, being capable of distinguishing mismatches in the newly synthesized DNA Strand from the original strand sequence. Together, these three discrimination steps enable replication fidelity of less than one mistake for every 10 nucleotides added.
705:(RRM). This primase is structurally similar to many viral RNA-dependent RNA polymerases, reverse transcriptases, cyclic nucleotide generating cyclases and DNA polymerases of the A/B/Y families that are involved in DNA replication and repair. In eukaryotic replication, the primase forms a complex with Pol α.
1235:
is a key inhibitor of pre-replication complex assembly. Geminin binds Cdt1, preventing its binding to the origin recognition complex. In G1, levels of geminin are kept low by the APC, which ubiquitinates geminin to target it for degradation. When geminin is destroyed, Cdt1 is released, allowing it to
891:
Clamp proteins act as a sliding clamp on DNA, allowing the DNA polymerase to bind to its template and aid in processivity. The inner face of the clamp enables DNA to be threaded through it. Once the polymerase reaches the end of the template or detects double-stranded DNA, the sliding clamp undergoes
507:
onto the DNA. In eukaryotes, the Mcm complex is the helicase that will split the DNA helix at the replication forks and origins. The Mcm complex is recruited at late G1 phase and loaded by the ORC-Cdc6-Cdt1 complex onto the DNA via ATP-dependent protein remodeling. The loading of the Mcm complex onto
1285:
and favors normal progress of replication forks. Progress of replication forks is inhibited by many factors; collision with proteins or with complexes binding strongly on DNA, deficiency of dNTPs, nicks on template DNAs and so on. If replication forks get stuck and the rest of the sequences from the
1102:
rings, there is the only chance for the disentanglement in DNA replication. Fixing of replication machineries as replication factories can improve the success rate of DNA replication. If replication forks move freely in chromosomes, catenation of nuclei is aggravated and impedes mitotic segregation.
802:
The lagging strand is the strand of new DNA whose direction of synthesis is opposite to the direction of the growing replication fork. Because of its orientation, replication of the lagging strand is more complicated as compared to that of the leading strand. As a consequence, the DNA polymerase on
1336:
ATP builds up when the cell is in a rich medium, triggering DNA replication once the cell has reached a specific size. ATP competes with ADP to bind to DnaA, and the DnaA-ATP complex is able to initiate replication. A certain number of DnaA proteins are also required for DNA replication — each time
1139:
Termination requires that the progress of the DNA replication fork must stop or be blocked. Termination at a specific locus, when it occurs, involves the interaction between two components: (1) a termination site sequence in the DNA, and (2) a protein which binds to this sequence to physically stop
937:
The enzyme responsible for catalyzing the addition of nucleotide substrates to DNA in the 5′ to 3′ direction during DNA replication. Also performs proof-reading and error correction. There exist many different types of DNA Polymerase, each of which perform different functions in different types of
853:
As helicase unwinds DNA at the replication fork, the DNA ahead is forced to rotate. This process results in a build-up of twists in the DNA ahead. This build-up creates a torsional load that would eventually stop the replication fork. Topoisomerases are enzymes that temporarily break the strands of
778:
The replication fork is a structure that forms within the long helical DNA during DNA replication. It is produced by enzymes called helicases that break the hydrogen bonds that hold the DNA strands together in a helix. The resulting structure has two branching "prongs", each one made up of a single
519:
complexes are activated, which stimulate expression of genes that encode components of the DNA synthetic machinery. G1/S-Cdk activation also promotes the expression and activation of S-Cdk complexes, which may play a role in activating replication origins depending on species and cell type. Control
1209:
Once the DNA has gone through the "G1/S" test, it can only be copied once in every cell cycle. When the Mcm complex moves away from the origin, the pre-replication complex is dismantled. Because a new Mcm complex cannot be loaded at an origin until the pre-replication subunits are reactivated, one
1083:
In an alternative figure, DNA factories are similar to projectors and DNAs are like as cinematic films passing constantly into the projectors. In the replication factory model, after both DNA helicases for leading strands and lagging strands are loaded on the template DNAs, the helicases run along
1213:
Activation of S-Cdks in early S phase promotes the destruction or inhibition of individual pre-replication complex components, preventing immediate reassembly. S and M-Cdks continue to block pre-replication complex assembly even after S phase is complete, ensuring that assembly cannot occur again
1111:
Eukaryotes initiate DNA replication at multiple points in the chromosome, so replication forks meet and terminate at many points in the chromosome. Because eukaryotes have linear chromosomes, DNA replication is unable to reach the very end of the chromosomes. Due to this problem, DNA is lost in
1079:
consist of factors involved in DNA replication and appearing on template ssDNAs. Replication machineries include primosotors are replication enzymes; DNA polymerase, DNA helicases, DNA clamps and DNA topoisomerases, and replication proteins; e.g. single-stranded DNA binding proteins (SSB). In the
1591:
of this process may also help explain the directionality of synthesis—if DNA were synthesized in the 3′ to 5′ direction, the energy for the process would come from the 5′ end of the growing strand rather than from free nucleotides. The problem is that if the high energy triphosphates were on the
565:
After α-primase synthesizes the first primers, the primer-template junctions interact with the clamp loader, which loads the sliding clamp onto the DNA to begin DNA synthesis. The components of the preinitiation complex remain associated with replication forks as they move out from the origin.
557:
In early S phase, S-Cdk and Cdc7 activation lead to the assembly of the preinitiation complex, a massive protein complex formed at the origin. Formation of the preinitiation complex displaces Cdc6 and Cdt1 from the origin replication complex, inactivating and disassembling the pre-replication
373:
In general, DNA polymerases are highly accurate, with an intrinsic error rate of less than one mistake for every 10 nucleotides added. Some DNA polymerases can also delete nucleotides from the end of a developing strand in order to fix mismatched bases. This is known as proofreading. Finally,
849:
In all cases the helicase is composed of six polypeptides that wrap around only one strand of the DNA being replicated. The two polymerases are bound to the helicase hexamer. In eukaryotes the helicase wraps around the leading strand, and in prokaryotes it wraps around the lagging strand.
779:
strand of DNA. These two strands serve as the template for the leading and lagging strands, which will be created as DNA polymerase matches complementary nucleotides to the templates; the templates may be properly referred to as the leading strand template and the lagging strand template.
549:, which binds Cdc7 directly and promotes its protein kinase activity. Cdc7 has been found to be a rate-limiting regulator of origin activity. Together, the G1/S-Cdks and/or S-Cdks and Cdc7 collaborate to directly activate the replication origins, leading to initiation of DNA synthesis.
362:. When a nucleotide is being added to a growing DNA strand, the formation of a phosphodiester bond between the proximal phosphate of the nucleotide to the growing chain is accompanied by hydrolysis of a high-energy phosphate bond with release of the two distal phosphate groups as a
1276:
By firing of replication origins, controlled spatially and temporally, the formation of replication foci is regulated. D. A. Jackson et al.(1998) revealed that neighboring origins fire simultaneously in mammalian cells. Spatial juxtaposition of replication sites brings
785:
Since the leading and lagging strand templates are oriented in opposite directions at the replication fork, a major issue is how to achieve synthesis of new lagging strand DNA, whose direction of synthesis is opposite to the direction of the growing replication fork.
443:, it must first replicate its DNA. DNA replication is an all-or-none process; once replication begins, it proceeds to completion. Once replication is complete, it does not occur again in the same cell cycle. This is made possible by the division of initiation of the
70:. This is essential for cell division during growth and repair of damaged tissues, while it also ensures that each of the new cells receives its own copy of the DNA. The cell possesses the distinctive property of division, which makes replication of DNA essential.
1333:, which binds and sequesters the origin sequence; in addition, DnaA (required for initiation of replication) binds less well to hemimethylated DNA. As a result, newly replicated origins are prevented from immediately initiating another round of DNA replication.
544:
to activate replication origins. Cdc7 is not active throughout the cell cycle, and its activation is strictly timed to avoid premature initiation of DNA replication. In late G1, Cdc7 activity rises abruptly as a result of association with the regulatory subunit
666:
removes the primer RNA fragments, and a low processivity DNA polymerase distinct from the replicative polymerase enters to fill the gaps. When this is complete, a single nick on the leading strand and several nicks on the lagging strand can be found.
330:
that carry out all forms of DNA replication. DNA polymerases in general cannot initiate synthesis of new strands but can only extend an existing DNA or RNA strand paired with a template strand. To begin synthesis, a short fragment of RNA, called a
1205:
The G1/S checkpoint (restriction checkpoint) regulates whether eukaryotic cells enter the process of DNA replication and subsequent division. Cells that do not proceed through this checkpoint remain in the G0 stage and do not replicate their DNA.
578:
group before synthesis can be initiated (note: the DNA template is read in 3′ to 5′ direction whereas a new strand is synthesized in the 5′ to 3′ direction—this is often confused). Four distinct mechanisms for DNA synthesis are recognized:
718:
is the polymerase enzyme primarily responsible for DNA replication. It assembles into a replication complex at the replication fork that exhibits extremely high processivity, remaining intact for the entire replication cycle. In contrast,
640:
that use the rolling circle replication (RCR) mechanism, the RCR endonuclease creates a nick in the genome strand (single stranded viruses) or one of the DNA strands (plasmids). The 5′ end of the nicked strand is transferred to a
1095:
Replication
Factories Disentangle Sister Chromatids. The disentanglement is essential for distributing the chromatids into daughter cells after DNA replication. Because sister chromatids after DNA replication hold each other by
318:
DNA polymerases adds nucleotides to the 3′ end of a strand of DNA. If a mismatch is accidentally incorporated, the polymerase is inhibited from further extension. Proofreading removes the mismatched nucleotide and extension
1190:. As the cell grows and divides, it progresses through stages in the cell cycle; DNA replication takes place during the S phase (synthesis phase). The progress of the eukaryotic cell through the cycle is controlled by
653:
adds RNA primers to the template strands. The leading strand receives one RNA primer while the lagging strand receives several. The leading strand is continuously extended from the primer by a DNA polymerase with high
1306:
Most bacteria do not go through a well-defined cell cycle but instead continuously copy their DNA; during rapid growth, this can result in the concurrent occurrence of multiple rounds of replication. In
532:
are primarily responsible for DNA replication. Clb5,6-Cdk1 complexes directly trigger the activation of replication origins and are therefore required throughout S phase to directly activate each origin.
381:. During the period of exponential DNA increase at 37 °C, the rate was 749 nucleotides per second. The mutation rate per base pair per replication during phage T4 DNA synthesis is 1.7 per 10.
4687:
1120:. This shortens the telomeres of the daughter DNA chromosome. As a result, cells can only divide a certain number of times before the DNA loss prevents further division. (This is known as the
621:, the 3′ OH group is provided by the side chain of an amino acid of the genome attached protein (the terminal protein) to which nucleotides are added by the DNA polymerase to form a new strand.
275:, creating the phosphate-deoxyribose backbone of the DNA double helix with the nucleobases pointing inward (i.e., toward the opposing strand). Nucleobases are matched between strands through
958:
Bind to ssDNA and prevent the DNA double helix from re-annealing after DNA helicase unwinds it, thus maintaining the strand separation, and facilitating the synthesis of the new strand.
727:
activity in addition to its polymerase activity, and uses its exonuclease activity to degrade the RNA primers ahead of it as it extends the DNA strand behind it, in a process called
1155:, enable only one direction of replication fork to pass through. As a result, the replication forks are constrained to always meet within the termination region of the chromosome.
892:
a conformational change that releases the DNA polymerase. Clamp-loading proteins are used to initially load the clamp, recognizing the junction between template and RNA primers.
3873:
Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, et al. (January 1988). "Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase".
1311:, the best-characterized bacteria, DNA replication is regulated through several mechanisms, including: the hemimethylation and sequestering of the origin sequence, the ratio of
152:(artificially, outside a cell). DNA polymerases isolated from cells and artificial DNA primers can be used to start DNA synthesis at known sequences in a template DNA molecule.
880:
that play an important role in regulating gene expression so the replicated DNA must be coiled around histones at the same places as the original DNA. To ensure this, histone
1217:
In budding yeast, inhibition of assembly is caused by Cdk-dependent phosphorylation of pre-replication complex components. At the onset of S phase, phosphorylation of Cdc6 by
1147:
Because bacteria have circular chromosomes, termination of replication occurs when the two replication forks meet each other on the opposite end of the parental chromosome.
1423:
2472:
McCarthy D, Minner C, Bernstein H, Bernstein C (October 1976). "DNA elongation rates and growing point distributions of wild-type phage T4 and a DNA-delay amber mutant".
1780:
4680:
415:
DNA replication, like all biological polymerization processes, proceeds in three enzymatically catalyzed and coordinated steps: initiation, elongation and termination.
1415:
1273:. The Heun's results denied the traditional concepts, budding yeasts do not have lamins, and support that replication origins self-assemble and form replication foci.
731:. Pol I is much less processive than Pol III because its primary function in DNA replication is to create many short DNA regions rather than a few very long regions.
794:
The leading strand is the strand of new DNA which is synthesized in the same direction as the growing replication fork. This sort of DNA replication is continuous.
1084:
the DNAs into each other. The helicases remain associated for the remainder of replication process. Peter
Meister et al. observed directly replication sites in
558:
complex. Loading the preinitiation complex onto the origin activates the Mcm helicase, causing unwinding of the DNA helix. The preinitiation complex also loads
1132:
extends the repetitive sequences of the telomere region to prevent degradation. Telomerase can become mistakenly active in somatic cells, sometimes leading to
2219:
4673:
3742:"E. coli SeqA protein binds oriC in two different methyl-modulated reactions appropriate to its roles in DNA replication initiation and origin sequestration"
537:
749:" (resembling the Greek letter theta: θ). In contrast, eukaryotes have longer linear chromosomes and initiate replication at multiple origins within these.
1116:
are regions of repetitive DNA close to the ends and help prevent loss of genes due to this shortening. Shortening of the telomeres is a normal process in
88:. As a result of semi-conservative replication, the new helix will be composed of an original DNA strand as well as a newly synthesized strand. Cellular
3933:
1337:
the origin is copied, the number of binding sites for DnaA doubles, requiring the synthesis of more DnaA to enable another initiation of replication.
690:
protein superfamily which contains a catalytic domain of the TOPRIM fold type. The TOPRIM fold contains an α/β core with four conserved strands in a
1261:,(2001) tracked GFP-tagged replication foci in budding yeast cells and revealed that replication origins move constantly in G1 and S phase and the
4497:
4460:
3484:
3373:
2801:
2522:
2374:
1060:
and DNA-replication. These results lead to the development of kinetic models accounting for the synergetic interactions and their stability.
1014:
649:
Cellular organisms use the first of these pathways since it is the most well-known. In this mechanism, once the two strands are separated,
161:
423:
4580:
1830:
1236:
function in pre-replication complex assembly. At the end of G1, the APC is inactivated, allowing geminin to accumulate and bind Cdt1.
399:
290:
4589:
4155:
3546:
3517:
3240:
3176:
2623:
2547:
2515:
2401:
2349:
2001:
1915:
1872:
1053:
953:
111:
which contains the genetic material of an organism. Unwinding of DNA at the origin and synthesis of new strands, accommodated by an
2868:"Toprim--a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins"
38:
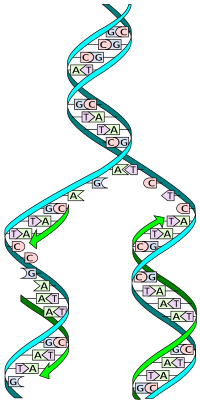
is un'zipped' and unwound, then each separated strand (turquoise) acts as a template for replicating a new partner strand (green).
4642:
1439:
1987:
1858:
888:
before it is replicated and replace the histones in the correct place. Some steps in this reassembly are somewhat speculative.
503:
then associate with the bound origin recognition complex at the origin in order to form a larger complex necessary to load the
745:
with two prongs. In bacteria, which have a single origin of replication on their circular chromosome, this process creates a "
469:, a large complex of initiator proteins assembles into the pre-replication complex at particular points in the DNA, known as "
5341:
4936:
4502:
1329:
GATC DNA sequences, DNA synthesis results in hemimethylated sequences. This hemimethylated DNA is recognized by the protein
1072:
E. coli
Replisome. Notably, the DNA on lagging strand forms a loop. The exact structure of replisome is not well understood.
3535:"Intracellular Control of Cell-Cycle Events: S-Phase Cyclin-Cdk Complexes (S-Cdks) Initiate DNA Replication Once Per Cycle"
2316:
1491:
to span a target region in template DNA, and then polymerizes partner strands in each direction from these primers using a
5477:
4312:
4162:
3926:
870:
5497:
783:
DNA is read by DNA polymerase in the 3′ to 5′ direction, meaning the new strand is synthesized in the 5' to 3' direction.
377:
The rate of DNA replication in a living cell was first measured as the rate of phage T4 DNA elongation in phage-infected
370:
into inorganic phosphate consumes a second high-energy phosphate bond and renders the reaction effectively irreversible.
5125:
85:
3581:
Nguyen VQ, Co C, Li JJ (June 2001). "Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms".
1265:
decreased significantly in S phase. Traditionally, replication sites were fixed on spatial structure of chromosomes by
5549:
5045:
4801:
4303:
3971:
3946:
1492:
500:
390:
3229:"DNA Replication Mechanisms: Special Proteins Help to Open Up the DNA Double Helix in Front of the Replication Fork"
1239:
Replication of chloroplast and mitochondrial genomes occurs independently of the cell cycle, through the process of
5554:
4432:
4408:
4277:
4109:
4056:
4038:
3950:
3789:
Cooper S, Helmstetter CE (February 1968). "Chromosome replication and the division cycle of
Escherichia coli B/r".
3509:
1488:
1140:
DNA replication. In various bacterial species, this is named the DNA replication terminus site-binding protein, or
1033:
504:
488:
394:
332:
900:
At the replication fork, many replication enzymes assemble on the DNA into a complex molecular machine called the
5346:
4486:
1484:
1474:
1225:
1218:
1194:. Progression through checkpoints is controlled through complex interactions between various proteins, including
1089:
645:
residue on the nuclease and the free 3′ OH group is then used by the DNA polymerase to synthesize the new strand.
606:) employ a transfer RNA that primes DNA replication by providing a free 3′ OH that is used for elongation by the
153:
74:
5539:
5193:
5094:
5011:
4647:
3919:
2695:"An Essential and Cell-Cycle-Dependent ORC Dimerization Cycle Regulates Eukaryotic Chromosomal DNA Replication"
1450:
1364:
1319:, and the levels of protein DnaA. All these control the binding of initiator proteins to the origin sequences.
741:
As DNA synthesis continues, the original DNA strands continue to unwind on each side of the bubble, forming a
599:
to synthesize a short RNA primer with a free 3′ OH group which is subsequently elongated by a DNA polymerase.
5220:
4996:
4049:
3980:
1563:
1199:
516:
456:
444:
359:
338:
DNA polymerase adds a new strand of DNA by extending the 3′ end of an existing nucleotide chain, adding new
192:
3501:
2007:
854:
DNA, relieving the tension caused by unwinding the two strands of the DNA helix; topoisomerases (including
5448:
4764:
4546:
4477:
1523:
1375:
1312:
866:
520:
of these Cdks vary depending on cell type and stage of development. This regulation is best understood in
89:
3534:
5208:
5181:
4472:
4241:
3911:
2296:
1316:
1191:
1057:
839:
702:
607:
470:
347:
220:
DNA exists as a double-stranded structure, with both strands coiled together to form the characteristic
169:
157:
104:
3228:
3164:
2630:
2220:"Solving the Chicken-and-the-Egg Problem – "A Step Closer to the Reconstruction of the Origin of Life""
1878:
1002:
Provides a starting point of RNA (or DNA) for DNA polymerase to begin synthesis of the new DNA strand.
164:(TMA) are examples. In March 2021, researchers reported evidence suggesting that a preliminary form of
5152:
5147:
5023:
4519:
4492:
4443:
3882:
3590:
2736:"Noc3p, a bHLH protein, plays an integral role in the initiation of DNA replication in budding yeast"
2408:
2259:
559:
272:
3079:
Huberman JA, Riggs AD (March 1968). "On the mechanism of DNA replication in mammalian chromosomes".
698:
Ia, topoisomerase II, the OLD-family nucleases and DNA repair proteins related to the RecR protein.
493:
An essential Noc3p dimerization cycle mediates ORC double-hexamer formation in replication licensing
492:
5544:
5388:
5230:
5186:
5142:
4880:
4249:
3390:
2535:
1988:"Chapter 27, Section 4: DNA Replication of Both Strands Proceeds Rapidly from Specific Start Sites"
1528:
1360:
1330:
881:
343:
948:
A protein which prevents elongating DNA polymerases from dissociating from the DNA parent strand.
701:
The primase used by archaea and eukaryotes, in contrast, contains a highly derived version of the
5438:
3771:
3614:
3457:
2940:
2224:
1496:
1262:
1240:
822:. The RNA primers are then removed and replaced with DNA, and the fragments of DNA are joined by
55:
2631:
12.1. General
Features of Chromosomal Replication: Three Common Features of Replication Origins
2565:"DnaA protein binding to individual DnaA boxes in the Escherichia coli replication origin, oriC"
873:
bind to the DNA until a second strand is synthesized, preventing secondary structure formation.
2425:"The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases"
1804:
5433:
5137:
4455:
4272:
3898:
3855:
3806:
3763:
3722:
3704:
3665:
3606:
3542:
3513:
3480:
3449:
3369:
3346:
3297:
3279:
3236:
3209:
3172:
3145:
3096:
3038:
2989:
2932:
2897:
2848:
2807:
2797:
2757:
2716:
2675:
2619:
2594:
2543:
2518:
2511:
2489:
2454:
2397:
2370:
2345:
2277:
2200:
2149:
2091:
2073:
1997:
1965:
1911:
1868:
1741:
1692:
1643:
1041:
989:
904:. The following is a list of major DNA replication enzymes that participate in the replisome:
764:
659:
78:
47:
5559:
5523:
5169:
5063:
4696:
4412:
4261:
4256:
4148:
3890:
3845:
3837:
3798:
3753:
3712:
3696:
3655:
3645:
3598:
3439:
3336:
3328:
3287:
3269:
3201:
3135:
3127:
3088:
3061:
3028:
3020:
2979:
2971:
2924:
2887:
2879:
2838:
2747:
2706:
2665:
2655:
2584:
2576:
2481:
2444:
2436:
2267:
2190:
2180:
2139:
2131:
2081:
2065:
1955:
1947:
1731:
1723:
1682:
1674:
1633:
1625:
1568:
1037:
924:
819:
770:
742:
728:
710:
475:
297:
276:
209:
120:
3058:
Distinguishing the pathways of primer removal during
Eukaryotic Okazaki fragment maturation
923:
Also known as helix destabilizing enzyme. Helicase separates the two strands of DNA at the
5426:
5099:
5028:
4831:
4417:
4009:
1538:
1326:
746:
283:. Adenine pairs with thymine (two hydrogen bonds), and guanine pairs with cytosine (three
2825:
Donaldson AD, Raghuraman MK, Friedman KL, Cross FR, Brewer BJ, Fangman WL (August 1998).
180:, could have been a replicator molecule itself in the very early development of life, or
3886:
3594:
3165:"DNA Replication Mechanisms: DNA Topoisomerases Prevent DNA Tangling During Replication"
2263:
1151:
regulates this process through the use of termination sequences that, when bound by the
5511:
5416:
5303:
5174:
5033:
5016:
4821:
4751:
4265:
4245:
3850:
3825:
3717:
3684:
3660:
3633:
3341:
3316:
3292:
3258:"The replication machinery of LUCA: common origin of DNA replication and transcription"
3257:
3140:
3115:
3033:
3008:
2984:
2959:
2670:
2643:
2589:
2564:
2449:
2424:
2195:
2168:
2144:
2115:
2086:
2053:
1960:
1935:
1736:
1711:
1687:
1662:
1638:
1613:
1513:
1266:
1121:
1019:
978:
Relieves strain of unwinding by DNA helicase; this is a specific type of topoisomerase
932:
869:; these structures can interfere with the movement of DNA polymerase. To prevent this,
723:
is the enzyme responsible for replacing RNA primers with DNA. DNA Pol I has a 5′ to 3′
574:
DNA polymerase has 5′–3′ activity. All known DNA replication systems require a free 3′
529:
525:
407:
323:
309:
284:
132:
127:
are associated with the replication fork to help in the initiation and continuation of
100:
4665:
2892:
2867:
2843:
2826:
2752:
2735:
1760:
1384:
806:
The lagging strand is synthesized in short, separated segments. On the lagging strand
763:
a: template, b: leading strand, c: lagging strand, d: replication fork, e: primer, f:
350:(phosphoanhydride) bonds between the three phosphates attached to each unincorporated
139:
that complement each (template) strand. DNA replication occurs during the S-stage of
42:(bases) are matched to synthesize the new partner strands into two new double helices.
30:
5533:
5456:
5421:
5264:
5254:
5225:
4858:
4851:
4467:
4394:
4004:
3802:
3758:
3741:
3092:
2613:
2485:
2391:
1991:
1862:
1588:
1553:
1518:
1427:
Epigenetic consequences of nucleosome reassembly defects at stalled replication forks
1164:
1085:
963:
859:
695:
691:
618:
521:
440:
367:
363:
128:
3775:
3461:
2944:
2928:
2028:
708:
Multiple DNA polymerases take on different roles in the DNA replication process. In
5443:
5286:
5246:
5213:
4985:
4971:
4869:
4606:
3618:
3256:
Koonin, Eugene V.; Krupovic, Mart; Ishino, Sonoko; Ishino, Yoshizumi (2020-06-09).
1117:
918:
655:
221:
197:
177:
165:
35:
1419:
Replication fork restarts by homologous recombination following replication stress
495:
ORC and Noc3p are continuously bound to the chromatin throughout the cell cycle.
3894:
3650:
3024:
2711:
2694:
2660:
1951:
1012:
Lengthens telomeric DNA by adding repetitive nucleotide sequences to the ends of
671:
works to fill these nicks in, thus completing the newly replicated DNA molecule.
346:. The energy for this process of DNA polymerization comes from hydrolysis of the
5467:
5333:
5281:
5276:
4789:
4551:
3700:
2069:
1629:
1543:
1508:
1222:
1152:
1141:
1068:
724:
658:, while the lagging strand is extended discontinuously from each primer forming
629:
229:
181:
5506:
3563:
3444:
3427:
3332:
3274:
3192:
Reece RJ, Maxwell A (26 September 2008). "DNA gyrase: structure and function".
2580:
2135:
1903:
757:
17:
5375:
5370:
5323:
5316:
5311:
5296:
5291:
5132:
5067:
4816:
4758:
4637:
4611:
4399:
4384:
3685:"Principles and concepts of DNA replication in bacteria, archaea, and eukarya"
3205:
1614:"Principles and concepts of DNA replication in bacteria, archaea, and eukarya"
1352:
1298:
1187:
1168:
1129:
1049:
1007:
983:
973:
855:
823:
633:
625:
614:
603:
355:
351:
339:
268:
245:
241:
237:
225:
140:
136:
81:
39:
3708:
3283:
2883:
2693:
Amin A, Wu R, Cheung MH, Scott JF, Wang Z, Zhou Z, et al. (March 2020).
2077:
1202:. Unlike bacteria, eukaryotic DNA replicates in the confines of the nucleus.
5380:
5365:
5352:
5104:
4892:
4811:
4806:
4796:
4784:
4723:
4575:
4389:
4379:
3475:
Watson JD, Baker TA, Bell SP, Gann A, Levine M, Losick R, Inglis CH (2008).
3131:
2811:
2612:
Lodish H, Berk A, Zipursky LS, Matsudaira P, Baltimore D, Darnell J (2000).
1461:
1136:
formation. Increased telomerase activity is one of the hallmarks of cancer.
1125:
1113:
943:
901:
885:
735:
683:
314:
280:
233:
213:
3859:
3726:
3669:
3610:
3453:
3350:
3301:
3149:
3114:
Gao Y, Cui Y, Fox T, Lin S, Wang H, de Val N, et al. (February 2019).
3060:(Ph.D. thesis). School of Medicine and Dentistry, University of Rochester.
3042:
2993:
2936:
2761:
2720:
2679:
2458:
2281:
2204:
2153:
2095:
1969:
1745:
1696:
1678:
1647:
1186:
Within eukaryotes, DNA replication is controlled within the context of the
834:
3902:
3810:
3767:
3213:
3100:
2975:
2901:
2852:
2598:
1173:
814:"reads" the template DNA and initiates synthesis of a short complementary
508:
the origin DNA marks the completion of pre-replication complex formation.
5411:
5357:
5259:
5203:
5198:
5089:
5050:
5006:
4908:
4713:
4652:
4633:
4205:
4196:
3994:
3740:
Slater S, Wold S, Lu M, Boye E, Skarstad K, Kleckner N (September 1995).
2493:
1852:
1456:
1251:
In vertebrate cells, replication sites concentrate into positions called
1045:
1029:
675:
642:
575:
466:
252:
148:
116:
93:
67:
2827:"CLB5-dependent activation of late replication origins in S. cerevisiae"
2272:
2247:
2185:
2052:
Chagin, Vadim O.; Stear, Jeffrey H.; Cardoso, M. Cristina (April 2010).
1727:
1431:
There are many events that contribute to replication stress, including:
511:
If environmental conditions are right in late G1 phase, the G1 and G1/S
5393:
5271:
5055:
5040:
4743:
4718:
4216:
4191:
4186:
4018:
3479:(6th ed.). San Francisco: Pearson/Benjamin Cummings. p. 237.
1356:
1232:
1098:
997:
877:
811:
715:
679:
650:
637:
596:
592:
541:
462:
358:; in particular, bases with three attached phosphate groups are called
260:
256:
248:
173:
124:
2440:
2409:
Chapter 27, Section 2: DNA Polymerases
Require a Template and a Primer
5484:
5462:
5159:
5082:
5077:
4842:
4733:
4728:
3602:
3065:
3009:"Reconsidering DNA Polymerases at the Replication Fork in Eukaryotes"
1548:
1195:
1133:
668:
512:
327:
264:
112:
108:
3841:
1210:
origin of replication can not be used twice in the same cell cycle.
1044:) have found synergetic interactions between the replisome enzymes (
431:
3683:
O'Donnell, Michael; Langston, Lance; Stillman, Bruce (2013-07-01).
3533:
Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2002).
3227:
Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2002).
3163:
Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2002).
2534:
Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2002).
2340:
Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2002).
694:
topology. This structure is also found in the catalytic domains of
342:
matched to the template strand, one at a time, via the creation of
4966:
4958:
4776:
4738:
4566:
4561:
4556:
4539:
4534:
4529:
4524:
4512:
4507:
4225:
4220:
4177:
4172:
4167:
3634:"The obligate human pathogen, Neisseria gonorrhoeae, is polyploid"
2563:
Weigel C, Schmidt A, Rückert B, Lurz R, Messer W (November 1997).
1422:
1414:
1270:
1067:
756:
720:
663:
588:
584:
484:
422:
406:
398:
313:
263:, commonly abbreviated as A, C, G, and T. Adenine and guanine are
201:
191:
5472:
5164:
4705:
4448:
4372:
4367:
4362:
4357:
4352:
4347:
4342:
4337:
4332:
4327:
4322:
4317:
4209:
4139:
4134:
4129:
4124:
4119:
4114:
4102:
4095:
4086:
4081:
4076:
4071:
4066:
4061:
4023:
3999:
3987:
1558:
1533:
1348:
843:
687:
546:
496:
480:
205:
63:
4669:
3915:
674:
The primase used in this process differs significantly between
1379:
865:
Bare single-stranded DNA tends to fold back on itself forming
818:
primer. A DNA polymerase extends the primed segments, forming
815:
354:. Free bases with their attached phosphate groups are called
59:
624:
In the single stranded DNA viruses—a group that includes the
58:
of producing two identical replicas of DNA from one original
1290:
that excess origins do not fire in normal DNA replication.
335:, must be created and paired with the template DNA strand.
3364:
Griffiths AJ, Wessler SR, Lewontin RC, Carroll SB (2008).
2644:"Replication initiation: Implications in genome integrity"
146:
DNA replication (DNA amplification) can also be performed
3317:"Chaperoning histones during DNA replication and repair"
1859:"Chapter 27: DNA Replication, Recombination, and Repair"
1112:
each replication cycle from the end of the chromosome.
224:. Each single strand of DNA is a chain of four types of
2317:"DNA function & structure (with diagram) (article)"
1831:"Semi-Conservative DNA Replication; Meselson and Stahl"
1396:
803:
this strand is seen to "lag behind" the other strand.
3391:"Will the Hayflick limit keep us from living forever?"
3194:
Critical
Reviews in Biochemistry and Molecular Biology
3116:"Structures and operating principles of the replisome"
2120:
single-molecule manipulation data analysis and models"
1302:
Dam methylates adenine of GATC sites after replication
1018:. This allows germ cells and stem cells to avoid the
774:
Many enzymes are involved in the DNA replication fork.
123:
growing bi-directionally from the origin. A number of
5495:
2915:
Frick DN, Richardson CC (July 2001). "DNA primases".
427:
Role of initiators for initiation of DNA replication
5404:
5332:
5245:
5113:
4995:
4984:
4957:
4929:
4879:
4868:
4841:
4830:
4775:
4704:
4624:
4598:
4428:
4299:
4290:
4234:
4034:
3967:
3958:
2344:(4th ed.). Garland Science. pp. 238–240.
1902:Lodish H, Berk A, Zipursky SL, et al. (2000).
1214:until all Cdk activity is reduced in late mitosis.
988:Re-anneals the semi-conservative strands and joins
296:The pairing of complementary bases in DNA (through
103:, DNA replication begins at specific locations, or
2390:Berg JM, Tymoczko JL, Stryer L, Clarke ND (2002).
2124:Computational and Structural Biotechnology Journal
1986:Berg JM, Tymoczko JL, Stryer L, Clarke ND (2002).
1857:Berg JM, Tymoczko JL, Stryer L, Clarke ND (2002).
92:and error-checking mechanisms ensure near perfect
2866:Aravind L, Leipe DD, Koonin EV (September 1998).
1612:O'Donnell M, Langston L, Stillman B (July 2013).
3315:Ransom M, Dennehey BK, Tyler JK (January 2010).
4625:
3826:"Causes and consequences of replication stress"
2418:
2416:
2169:"tRNA sequences can assemble into a replicator"
2167:Kühnlein A, Lanzmich SA, Braun D (March 2021).
2109:
2107:
2105:
1781:"GENETICS / DNA REPLICATION (BASIC) – Pathwayz"
1444:Conflicts between replication and transcription
1128:line, which passes DNA to the next generation,
3959:
2787:
2785:
2783:
2781:
2779:
2777:
2775:
2773:
2771:
1447:Insufficiency of essential replication factors
1056:) and with the DNA replication fork enhancing
968:Relaxes the DNA from its super-coiled nature.
4681:
3927:
3421:
3419:
3417:
3415:
3413:
3411:
2796:. London: New Science Press. pp. 64–75.
2297:"Base Pair: Definition, Rules In DNA And RNA"
1929:
1927:
1455:Overexpression or constitutive activation of
8:
3502:"Chapter 13.2.3. Termination of replication"
3426:Meister P, Taddei A, Gasser SM (June 2006).
1904:"DNA Replication, Repair, and Recombination"
1663:"DNA replication origins-where do we begin?"
4291:
2114:Jarillo J, Ibarra B, Cao-García FJ (2021).
1712:"Mechanisms of DNA replication termination"
4992:
4876:
4838:
4688:
4674:
4666:
4296:
3964:
3934:
3920:
3912:
3689:Cold Spring Harbor Perspectives in Biology
2964:Microbiology and Molecular Biology Reviews
2734:Zhang Y, Yu Z, Fu X, Liang C (June 2002).
2058:Cold Spring Harbor Perspectives in Biology
1981:
1979:
1936:"Origins of DNA replication in eukaryotes"
1618:Cold Spring Harbor Perspectives in Biology
686:. Bacteria use a primase belonging to the
3849:
3757:
3716:
3659:
3649:
3443:
3340:
3291:
3273:
3139:
3032:
2983:
2891:
2842:
2751:
2710:
2669:
2659:
2588:
2448:
2271:
2194:
2184:
2143:
2085:
1959:
1897:
1895:
1735:
1686:
1661:Prioleau MN, MacAlpine DM (August 2016).
1637:
2423:McCulloch SD, Kunkel TA (January 2008).
1761:"What Is DNA Replication And Its Steps?"
1297:
1281:of replication forks. The clustering do
1172:
906:
833:
769:
636:and others—and also the many phages and
562:and other DNA polymerases onto the DNA.
430:
403:Overview of the steps in DNA replication
366:. Enzymatic hydrolysis of the resulting
62:molecule. DNA replication occurs in all
29:
5502:
3428:"In and out of the replication factory"
2536:"Chapter 5: DNA Replication Mechanisms"
1604:
1580:
3824:Zeman MK, Cimprich KA (January 2014).
1716:Nature Reviews. Molecular Cell Biology
267:bases, while cytosine and thymine are
135:synthesizes the new strands by adding
3632:Tobiason DM, Seifert HS (June 2006).
2794:The cell cycle: principles of control
2369:. Blackwell Publishing. p. 112.
1851:Imperfect DNA replication results in
876:Double-stranded DNA is coiled around
583:All cellular life forms and many DNA
208:in the structure are colour-coded by
66:acting as the most essential part of
7:
435:Formation of pre-replication complex
162:transcription-mediated amplification
3570:(2nd ed.). Oxford: Wiley-Liss.
2958:Barry ER, Bell SD (December 2006).
1710:Dewar JM, Walter JC (August 2017).
1479:Researchers commonly replicate DNA
1435:Misincorporation of ribonucleotides
1283:rescue of stalled replication forks
212:and the detailed structures of two
1934:Hu Y, Stillman B (February 2023).
1340:In fast-growing bacteria, such as
1221:causes the binding of Cdc6 to the
1177:The cell cycle of eukaryotic cells
858:) achieve this by adding negative
172:, the biological synthesis of new
25:
4590:Control of chromosome duplication
4156:Autonomously replicating sequence
2642:Lin YC, Prasanth SG (July 2021).
2054:"Organization of DNA replication"
1054:Single-strand DNA-binding protein
954:Single-strand DNA-binding protein
838:The assembled human DNA clamp, a
479:the primary initiator protein is
291:DNA strands have a directionality
5517:
5505:
3564:"Chapter 13: Genome Replication"
3366:Introduction to Genetic Analysis
2960:"DNA replication in the archaea"
2508:The Molecular Basis of Mutation.
1383:
830:Dynamics at the replication fork
2929:10.1146/annurev.biochem.70.1.39
761:Scheme of the replication fork.
228:. Nucleotides in DNA contain a
5342:Last universal common ancestor
4937:Defective interfering particle
871:single-strand binding proteins
216:are shown in the bottom right.
1:
5478:Clonally transmissible cancer
4914:Satellite-like nucleic acids
4313:DNA polymerase III holoenzyme
4163:Single-strand binding protein
3539:Molecular Biology of the Cell
3477:Molecular Biology of the Gene
3368:. W. H. Freeman and Company.
3233:Molecular Biology of the Cell
3169:Molecular Biology of the Cell
2917:Annual Review of Biochemistry
2844:10.1016/s1097-2765(00)80127-6
2753:10.1016/s0092-8674(02)00805-x
2618:. W. H. Freeman and Company.
2540:Molecular Biology of the Cell
2367:Fundamental Molecular Biology
2342:Molecular Biology of the Cell
2246:Friedberg EC (January 2003).
2218:Maximilian L (3 April 2021).
1370:Problems with DNA replication
1231:In animal cells, the protein
602:The retroelements (including
3895:10.1126/science.239.4839.487
3803:10.1016/0022-2836(68)90425-7
3791:Journal of Molecular Biology
3759:10.1016/0092-8674(95)90272-4
3651:10.1371/journal.pbio.0040185
3093:10.1016/0022-2836(68)90013-2
3081:Journal of Molecular Biology
3025:10.1016/j.molcel.2015.07.004
2712:10.1016/j.celrep.2020.02.046
2661:10.1016/j.dnarep.2021.103131
2486:10.1016/0022-2836(76)90346-6
2474:Journal of Molecular Biology
2396:. W.H. Freeman and Company.
1996:. W.H. Freeman and Company.
1952:10.1016/j.molcel.2022.12.024
1910:(4th ed.). WH Freeman.
1867:. W.H. Freeman and Company.
1313:adenosine triphosphate (ATP)
913:Function in DNA replication
86:semiconservative replication
3701:10.1101/cshperspect.a010108
2070:10.1101/cshperspect.a000737
1630:10.1101/cshperspect.a010108
1493:thermostable DNA polymerase
1317:adenosine diphosphate (ADP)
1288:dormant replication origins
1034:single-molecule experiments
391:Prokaryotic DNA replication
168:, a necessary component of
5576:
5034:Class II or DNA transposon
5029:Class I or retrotransposon
4409:Prokaryotic DNA polymerase
4110:Minichromosome maintenance
4057:Origin recognition complex
3510:BIOS Scientific Publishers
3445:10.1016/j.cell.2006.06.014
3333:10.1016/j.cell.2010.01.004
3275:10.1186/s12915-020-00800-9
3056:Rossi ML (February 2009).
2510:Holden-Day, San Francisco
2295:Sabhadiya A (2023-03-13).
2136:10.1016/j.csbj.2021.06.032
1759:Sabhadiya A (2022-03-01).
1487:(PCR). PCR uses a pair of
1472:
1373:
1346:
1162:
927:behind the topoisomerase.
489:origin recognition complex
454:
395:Eukaryotic DNA replication
388:
307:
5347:Earliest known life forms
5221:Repeated sequences in DNA
4487:Eukaryotic DNA polymerase
3206:10.3109/10409239109114072
1809:Learn Science at Scitable
1485:polymerase chain reaction
1475:Polymerase chain reaction
1469:Polymerase chain reaction
1090:green fluorescent protein
540:is also required through
271:. These nucleotides form
196:The structure of the DNA
154:Polymerase chain reaction
5194:Endogenous viral element
5012:Horizontal gene transfer
3007:Stillman B (July 2015).
2581:10.1093/emboj/16.21.6574
1497:increasing exponentially
1365:Plasmid partition system
1226:ubiquitin protein ligase
1200:cyclin-dependent kinases
896:DNA replication proteins
360:nucleoside triphosphates
4891:dsDNA satellite virus (
4050:Pre-replication complex
3981:Pre-replication complex
3132:10.1126/science.aav7003
2248:"DNA damage and repair"
1667:Genes & Development
1564:Replication (computing)
1077:Replication machineries
992:of the lagging strand.
457:Pre-replication complex
451:Pre-replication complex
445:pre-replication complex
244:correspond to the four
176:in accordance with the
5449:Helper dependent virus
4765:Biological dark matter
3389:Clark J (2009-05-11).
2884:10.1093/nar/26.18.4205
2872:Nucleic Acids Research
2615:Molecular Cell Biology
1908:Molecular Cell Biology
1679:10.1101/gad.285114.116
1524:Chromosome segregation
1428:
1420:
1376:DNA replication stress
1303:
1192:cell cycle checkpoints
1178:
1073:
1015:eukaryotic chromosomes
846:
775:
767:
617:and the φ29 family of
524:, where the S cyclins
436:
428:
412:
411:Steps in DNA synthesis
404:
320:
217:
105:origins of replication
68:biological inheritance
43:
5209:Endogenous retrovirus
5182:Origin of replication
4898:ssDNA satellite virus
4888:ssRNA satellite virus
4473:Replication protein A
4242:Origin of replication
2976:10.1128/MMBR.00029-06
1426:
1418:
1301:
1176:
1071:
1064:Replication machinery
837:
773:
760:
703:RNA recognition motif
608:reverse transcriptase
553:Preinitiation complex
536:In a similar manner,
434:
426:
410:
402:
348:high-energy phosphate
317:
195:
158:ligase chain reaction
96:for DNA replication.
34:DNA replication: The
33:
5153:Secondary chromosome
5148:Extrachromosomal DNA
5024:Transposable element
4444:Replication factor C
2705:(10): 3323–3338.e6.
1451:Common fragile sites
867:secondary structures
344:phosphodiester bonds
273:phosphodiester bonds
240:. The four types of
131:. Most prominently,
73:DNA is made up of a
5389:Model lipid bilayer
5231:Interspersed repeat
3887:1988Sci...239..487S
3830:Nature Cell Biology
3595:2001Natur.411.1068N
3589:(6841): 1068–1073.
3541:. Garland Science.
3235:. Garland Science.
3171:. Garland Science.
2542:. Garland Science.
2273:10.1038/nature01408
2264:2003Natur.421..436F
2186:10.7554/eLife.63431
2029:"What is a genome?"
1728:10.1038/nrm.2017.42
1529:Data storage device
1361:Plasmid copy number
385:Replication process
5550:Cellular processes
4699:organic structures
2792:Morgan DO (2007).
2365:Allison L (2007).
2116:"DNA replication:
1811:. Nature Education
1429:
1421:
1395:. You can help by
1304:
1241:D-loop replication
1179:
1074:
1022:on cell division.
862:to the DNA helix.
847:
776:
768:
437:
429:
413:
405:
321:
218:
56:biological process
44:
27:Biological process
5555:Molecular biology
5493:
5492:
5434:Non-cellular life
5241:
5240:
4980:
4979:
4953:
4952:
4907:ssRNA satellite (
4663:
4662:
4620:
4619:
4456:Flap endonuclease
4286:
4285:
4273:Okazaki fragments
3881:(4839): 487–491.
3562:Brown TA (2002).
3500:Brown TA (2002).
3486:978-0-8053-9592-1
3375:978-0-7167-6887-6
2878:(18): 4205–4213.
2803:978-0-19-920610-0
2575:(21): 6574–6583.
2523:978-0-8162-2450-0
2441:10.1038/cr.2008.4
2376:978-1-4051-0379-4
2258:(6921): 436–440.
1673:(15): 1683–1697.
1413:
1412:
1247:Replication focus
1042:magnetic tweezers
1026:
1025:
990:Okazaki Fragments
820:Okazaki fragments
765:Okazaki fragments
660:Okazaki fragments
121:replication forks
48:molecular biology
16:(Redirected from
5567:
5522:
5521:
5520:
5510:
5509:
5501:
5170:Gene duplication
4993:
4989:self-replication
4877:
4839:
4697:Self-replicating
4690:
4683:
4676:
4667:
4413:DNA polymerase I
4297:
4257:Replication fork
4149:Licensing factor
3965:
3936:
3929:
3922:
3913:
3907:
3906:
3870:
3864:
3863:
3853:
3821:
3815:
3814:
3786:
3780:
3779:
3761:
3737:
3731:
3730:
3720:
3680:
3674:
3673:
3663:
3653:
3629:
3623:
3622:
3603:10.1038/35082600
3578:
3572:
3571:
3559:
3553:
3552:
3530:
3524:
3523:
3497:
3491:
3490:
3472:
3466:
3465:
3447:
3423:
3406:
3405:
3403:
3401:
3386:
3380:
3379:
3361:
3355:
3354:
3344:
3312:
3306:
3305:
3295:
3277:
3253:
3247:
3246:
3224:
3218:
3217:
3200:(3–4): 335–375.
3189:
3183:
3182:
3160:
3154:
3153:
3143:
3111:
3105:
3104:
3076:
3070:
3069:
3053:
3047:
3046:
3036:
3004:
2998:
2997:
2987:
2955:
2949:
2948:
2912:
2906:
2905:
2895:
2863:
2857:
2856:
2846:
2822:
2816:
2815:
2789:
2766:
2765:
2755:
2731:
2725:
2724:
2714:
2690:
2684:
2683:
2673:
2663:
2639:
2633:
2629:
2609:
2603:
2602:
2592:
2569:The EMBO Journal
2560:
2554:
2553:
2531:
2525:
2506:Drake JW (1970)
2504:
2498:
2497:
2469:
2463:
2462:
2452:
2420:
2411:
2407:
2387:
2381:
2380:
2362:
2356:
2355:
2337:
2331:
2330:
2328:
2327:
2313:
2307:
2306:
2304:
2303:
2292:
2286:
2285:
2275:
2243:
2237:
2236:
2234:
2232:
2215:
2209:
2208:
2198:
2188:
2164:
2158:
2157:
2147:
2111:
2100:
2099:
2089:
2049:
2043:
2042:
2040:
2039:
2025:
2019:
2018:
2016:
2015:
2006:. Archived from
1983:
1974:
1973:
1963:
1931:
1922:
1921:
1899:
1890:
1889:
1887:
1886:
1877:. Archived from
1849:
1843:
1842:
1835:Nature Education
1829:Pray LA (2008).
1826:
1820:
1819:
1817:
1816:
1801:
1795:
1794:
1792:
1791:
1777:
1771:
1770:
1768:
1767:
1756:
1750:
1749:
1739:
1707:
1701:
1700:
1690:
1658:
1652:
1651:
1641:
1609:
1593:
1585:
1569:Self-replication
1408:
1405:
1387:
1380:
1253:replication foci
1038:optical tweezers
925:Replication Fork
907:
884:disassemble the
753:Replication fork
743:replication fork
729:nick translation
326:are a family of
298:hydrogen bonding
64:living organisms
21:
5575:
5574:
5570:
5569:
5568:
5566:
5565:
5564:
5540:DNA replication
5530:
5529:
5528:
5518:
5516:
5504:
5496:
5494:
5489:
5439:Synthetic virus
5427:Artificial cell
5400:
5328:
5237:
5126:RNA replication
5121:DNA replication
5109:
5100:Group II intron
4998:
4988:
4976:
4967:Mammalian prion
4949:
4925:
4904:dsRNA satellite
4901:ssDNA satellite
4871:
4864:
4833:
4826:
4771:
4700:
4694:
4664:
4659:
4616:
4594:
4434:
4430:
4424:
4418:Klenow fragment
4301:
4282:
4266:leading strands
4230:
4040:
4036:
4030:
3969:
3954:
3943:DNA replication
3940:
3910:
3872:
3871:
3867:
3842:10.1038/ncb2897
3823:
3822:
3818:
3788:
3787:
3783:
3739:
3738:
3734:
3682:
3681:
3677:
3631:
3630:
3626:
3580:
3579:
3575:
3561:
3560:
3556:
3549:
3532:
3531:
3527:
3520:
3499:
3498:
3494:
3487:
3474:
3473:
3469:
3425:
3424:
3409:
3399:
3397:
3388:
3387:
3383:
3376:
3363:
3362:
3358:
3314:
3313:
3309:
3255:
3254:
3250:
3243:
3226:
3225:
3221:
3191:
3190:
3186:
3179:
3162:
3161:
3157:
3113:
3112:
3108:
3078:
3077:
3073:
3055:
3054:
3050:
3006:
3005:
3001:
2957:
2956:
2952:
2914:
2913:
2909:
2865:
2864:
2860:
2824:
2823:
2819:
2804:
2791:
2790:
2769:
2733:
2732:
2728:
2692:
2691:
2687:
2641:
2640:
2636:
2626:
2611:
2610:
2606:
2562:
2561:
2557:
2550:
2533:
2532:
2528:
2505:
2501:
2471:
2470:
2466:
2422:
2421:
2414:
2404:
2389:
2388:
2384:
2377:
2364:
2363:
2359:
2352:
2339:
2338:
2334:
2325:
2323:
2315:
2314:
2310:
2301:
2299:
2294:
2293:
2289:
2245:
2244:
2240:
2230:
2228:
2217:
2216:
2212:
2166:
2165:
2161:
2113:
2112:
2103:
2051:
2050:
2046:
2037:
2035:
2027:
2026:
2022:
2013:
2011:
2004:
1985:
1984:
1977:
1933:
1932:
1925:
1918:
1901:
1900:
1893:
1884:
1882:
1875:
1856:
1850:
1846:
1828:
1827:
1823:
1814:
1812:
1803:
1802:
1798:
1789:
1787:
1779:
1778:
1774:
1765:
1763:
1758:
1757:
1753:
1709:
1708:
1704:
1660:
1659:
1655:
1611:
1610:
1606:
1602:
1597:
1596:
1586:
1582:
1577:
1539:Gene expression
1505:
1477:
1471:
1464:inaccessibility
1409:
1403:
1400:
1393:needs expansion
1378:
1372:
1367:
1296:
1249:
1184:
1171:
1163:Main articles:
1161:
1124:.) Within the
1109:
1093:
1066:
898:
842:of the protein
832:
800:
792:
762:
755:
747:theta structure
572:
555:
459:
453:
421:
397:
389:Main articles:
387:
324:DNA polymerases
312:
306:
190:
52:DNA replication
28:
23:
22:
18:Replication eye
15:
12:
11:
5:
5573:
5571:
5563:
5562:
5557:
5552:
5547:
5542:
5532:
5531:
5527:
5526:
5514:
5491:
5490:
5488:
5487:
5482:
5481:
5480:
5475:
5465:
5459:
5453:
5452:
5451:
5446:
5436:
5431:
5430:
5429:
5424:
5414:
5408:
5406:
5402:
5401:
5399:
5398:
5397:
5396:
5391:
5383:
5378:
5373:
5368:
5362:
5361:
5360:
5349:
5344:
5338:
5336:
5330:
5329:
5327:
5326:
5321:
5320:
5319:
5314:
5306:
5304:Kappa organism
5301:
5300:
5299:
5294:
5289:
5284:
5279:
5269:
5268:
5267:
5262:
5251:
5249:
5243:
5242:
5239:
5238:
5236:
5235:
5234:
5233:
5228:
5218:
5217:
5216:
5211:
5206:
5201:
5191:
5190:
5189:
5179:
5178:
5177:
5175:Non-coding DNA
5172:
5167:
5157:
5156:
5155:
5150:
5145:
5140:
5130:
5129:
5128:
5117:
5115:
5111:
5110:
5108:
5107:
5102:
5097:
5095:Group I intron
5092:
5087:
5086:
5085:
5075:
5074:
5073:
5070:
5061:
5058:
5053:
5048:
5038:
5037:
5036:
5031:
5021:
5020:
5019:
5017:Genomic island
5014:
5003:
5001:
4997:Mobile genetic
4990:
4982:
4981:
4978:
4977:
4975:
4974:
4969:
4963:
4961:
4955:
4954:
4951:
4950:
4948:
4947:
4946:
4945:
4942:
4933:
4931:
4927:
4926:
4924:
4923:
4922:
4921:
4918:
4912:
4905:
4902:
4899:
4896:
4889:
4885:
4883:
4874:
4866:
4865:
4863:
4862:
4855:
4847:
4845:
4836:
4828:
4827:
4825:
4824:
4822:dsDNA-RT virus
4819:
4817:ssRNA-RT virus
4814:
4812:(−)ssRNA virus
4809:
4807:(+)ssRNA virus
4804:
4799:
4794:
4793:
4792:
4781:
4779:
4773:
4772:
4770:
4769:
4768:
4767:
4762:
4752:Incertae sedis
4748:
4747:
4746:
4741:
4736:
4731:
4721:
4716:
4710:
4708:
4702:
4701:
4695:
4693:
4692:
4685:
4678:
4670:
4661:
4660:
4658:
4657:
4656:
4655:
4650:
4645:
4630:
4628:
4622:
4621:
4618:
4617:
4615:
4614:
4609:
4602:
4600:
4596:
4595:
4593:
4592:
4586:
4585:
4584:
4583:
4572:
4571:
4570:
4569:
4564:
4559:
4554:
4544:
4543:
4542:
4537:
4532:
4527:
4517:
4516:
4515:
4510:
4505:
4500:
4490:
4483:
4482:
4481:
4480:
4470:
4465:
4464:
4463:
4453:
4452:
4451:
4440:
4438:
4426:
4425:
4423:
4422:
4421:
4420:
4405:
4404:
4403:
4402:
4392:
4387:
4382:
4377:
4376:
4375:
4370:
4365:
4360:
4355:
4350:
4345:
4340:
4335:
4330:
4325:
4320:
4309:
4307:
4294:
4288:
4287:
4284:
4283:
4281:
4280:
4275:
4270:
4269:
4268:
4253:
4252:
4238:
4236:
4232:
4231:
4229:
4228:
4223:
4213:
4212:
4202:
4201:
4200:
4199:
4194:
4183:
4182:
4181:
4180:
4175:
4170:
4159:
4158:
4152:
4151:
4145:
4144:
4143:
4142:
4137:
4132:
4127:
4122:
4117:
4106:
4105:
4099:
4098:
4092:
4091:
4090:
4089:
4084:
4079:
4074:
4069:
4064:
4053:
4052:
4046:
4044:
4039:preparation in
4032:
4031:
4029:
4028:
4027:
4026:
4015:
4014:
4013:
4012:
4007:
4002:
3991:
3990:
3984:
3983:
3977:
3975:
3962:
3956:
3955:
3941:
3939:
3938:
3931:
3924:
3916:
3909:
3908:
3865:
3816:
3797:(3): 519–540.
3781:
3752:(6): 927–936.
3732:
3695:(7): a010108.
3675:
3624:
3573:
3554:
3547:
3525:
3518:
3492:
3485:
3467:
3407:
3381:
3374:
3356:
3327:(2): 183–195.
3307:
3248:
3241:
3219:
3184:
3177:
3155:
3106:
3087:(2): 327–341.
3071:
3048:
3019:(2): 139–141.
3013:Molecular Cell
2999:
2970:(4): 876–887.
2950:
2907:
2858:
2837:(2): 173–182.
2831:Molecular Cell
2817:
2802:
2767:
2746:(7): 849–860.
2726:
2685:
2634:
2624:
2604:
2555:
2548:
2526:
2499:
2480:(4): 963–981.
2464:
2435:(1): 148–161.
2412:
2402:
2382:
2375:
2357:
2350:
2332:
2308:
2287:
2238:
2210:
2159:
2101:
2064:(4): a000737.
2044:
2020:
2002:
1975:
1946:(3): 352–372.
1940:Molecular Cell
1923:
1916:
1891:
1873:
1844:
1821:
1805:"double helix"
1796:
1772:
1751:
1722:(8): 507–516.
1702:
1653:
1624:(7): a010108.
1603:
1601:
1598:
1595:
1594:
1579:
1578:
1576:
1573:
1572:
1571:
1566:
1561:
1556:
1551:
1546:
1541:
1536:
1531:
1526:
1521:
1516:
1514:Cell (biology)
1511:
1504:
1501:
1473:Main article:
1470:
1467:
1466:
1465:
1459:
1453:
1448:
1445:
1442:
1440:DNA structures
1436:
1411:
1410:
1390:
1388:
1374:Main article:
1371:
1368:
1295:
1292:
1267:nuclear matrix
1259:P. Heun et al.
1248:
1245:
1183:
1180:
1160:
1157:
1122:Hayflick limit
1108:
1105:
1088:by monitoring
1082:
1065:
1062:
1024:
1023:
1020:Hayflick limit
1010:
1004:
1003:
1000:
994:
993:
986:
980:
979:
976:
970:
969:
966:
960:
959:
956:
950:
949:
946:
940:
939:
935:
933:DNA polymerase
929:
928:
921:
915:
914:
911:
897:
894:
831:
828:
799:
798:Lagging strand
796:
791:
790:Leading strand
788:
754:
751:
647:
646:
622:
619:bacteriophages
611:
600:
571:
568:
554:
551:
487:, this is the
455:Main article:
452:
449:
441:cell to divide
420:
417:
386:
383:
310:DNA polymerase
308:Main article:
305:
304:DNA polymerase
302:
285:hydrogen bonds
277:hydrogen bonds
189:
186:
133:DNA polymerase
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
5572:
5561:
5558:
5556:
5553:
5551:
5548:
5546:
5543:
5541:
5538:
5537:
5535:
5525:
5515:
5513:
5508:
5503:
5499:
5486:
5483:
5479:
5476:
5474:
5471:
5470:
5469:
5466:
5464:
5460:
5458:
5457:Nanobacterium
5454:
5450:
5447:
5445:
5442:
5441:
5440:
5437:
5435:
5432:
5428:
5425:
5423:
5422:Cell division
5420:
5419:
5418:
5415:
5413:
5410:
5409:
5407:
5403:
5395:
5392:
5390:
5387:
5386:
5384:
5382:
5379:
5377:
5374:
5372:
5369:
5367:
5363:
5359:
5356:
5355:
5354:
5350:
5348:
5345:
5343:
5340:
5339:
5337:
5335:
5331:
5325:
5322:
5318:
5315:
5313:
5310:
5309:
5307:
5305:
5302:
5298:
5295:
5293:
5290:
5288:
5285:
5283:
5280:
5278:
5275:
5274:
5273:
5270:
5266:
5265:Hydrogenosome
5263:
5261:
5258:
5257:
5256:
5255:Mitochondrion
5253:
5252:
5250:
5248:
5247:Endosymbiosis
5244:
5232:
5229:
5227:
5226:Tandem repeat
5224:
5223:
5222:
5219:
5215:
5212:
5210:
5207:
5205:
5202:
5200:
5197:
5196:
5195:
5192:
5188:
5185:
5184:
5183:
5180:
5176:
5173:
5171:
5168:
5166:
5163:
5162:
5161:
5158:
5154:
5151:
5149:
5146:
5144:
5141:
5139:
5136:
5135:
5134:
5131:
5127:
5124:
5123:
5122:
5119:
5118:
5116:
5114:Other aspects
5112:
5106:
5103:
5101:
5098:
5096:
5093:
5091:
5088:
5084:
5081:
5080:
5079:
5076:
5071:
5069:
5065:
5062:
5059:
5057:
5054:
5052:
5049:
5047:
5044:
5043:
5042:
5039:
5035:
5032:
5030:
5027:
5026:
5025:
5022:
5018:
5015:
5013:
5010:
5009:
5008:
5005:
5004:
5002:
5000:
4994:
4991:
4987:
4983:
4973:
4970:
4968:
4965:
4964:
4962:
4960:
4956:
4943:
4940:
4939:
4938:
4935:
4934:
4932:
4928:
4919:
4916:
4915:
4913:
4910:
4906:
4903:
4900:
4897:
4894:
4890:
4887:
4886:
4884:
4882:
4878:
4875:
4873:
4867:
4861:
4860:
4859:Avsunviroidae
4856:
4854:
4853:
4852:Pospiviroidae
4849:
4848:
4846:
4844:
4840:
4837:
4835:
4829:
4823:
4820:
4818:
4815:
4813:
4810:
4808:
4805:
4803:
4800:
4798:
4795:
4791:
4788:
4787:
4786:
4783:
4782:
4780:
4778:
4774:
4766:
4763:
4761:
4760:
4756:
4755:
4754:
4753:
4749:
4745:
4742:
4740:
4737:
4735:
4732:
4730:
4727:
4726:
4725:
4722:
4720:
4717:
4715:
4712:
4711:
4709:
4707:
4706:Cellular life
4703:
4698:
4691:
4686:
4684:
4679:
4677:
4672:
4671:
4668:
4654:
4651:
4649:
4646:
4644:
4641:
4640:
4639:
4635:
4632:
4631:
4629:
4627:
4623:
4613:
4610:
4608:
4604:
4603:
4601:
4597:
4591:
4588:
4587:
4582:
4579:
4578:
4577:
4574:
4573:
4568:
4565:
4563:
4560:
4558:
4555:
4553:
4550:
4549:
4548:
4545:
4541:
4538:
4536:
4533:
4531:
4528:
4526:
4523:
4522:
4521:
4518:
4514:
4511:
4509:
4506:
4504:
4501:
4499:
4496:
4495:
4494:
4491:
4488:
4485:
4484:
4479:
4476:
4475:
4474:
4471:
4469:
4468:Topoisomerase
4466:
4462:
4459:
4458:
4457:
4454:
4450:
4447:
4446:
4445:
4442:
4441:
4439:
4436:
4427:
4419:
4416:
4415:
4414:
4410:
4407:
4406:
4401:
4398:
4397:
4396:
4395:Topoisomerase
4393:
4391:
4388:
4386:
4383:
4381:
4378:
4374:
4371:
4369:
4366:
4364:
4361:
4359:
4356:
4354:
4351:
4349:
4346:
4344:
4341:
4339:
4336:
4334:
4331:
4329:
4326:
4324:
4321:
4319:
4316:
4315:
4314:
4311:
4310:
4308:
4305:
4298:
4295:
4293:
4289:
4279:
4276:
4274:
4271:
4267:
4263:
4260:
4259:
4258:
4255:
4254:
4251:
4247:
4243:
4240:
4239:
4237:
4233:
4227:
4224:
4222:
4218:
4215:
4214:
4211:
4207:
4204:
4203:
4198:
4195:
4193:
4190:
4189:
4188:
4185:
4184:
4179:
4176:
4174:
4171:
4169:
4166:
4165:
4164:
4161:
4160:
4157:
4154:
4153:
4150:
4147:
4146:
4141:
4138:
4136:
4133:
4131:
4128:
4126:
4123:
4121:
4118:
4116:
4113:
4112:
4111:
4108:
4107:
4104:
4101:
4100:
4097:
4094:
4093:
4088:
4085:
4083:
4080:
4078:
4075:
4073:
4070:
4068:
4065:
4063:
4060:
4059:
4058:
4055:
4054:
4051:
4048:
4047:
4045:
4042:
4033:
4025:
4022:
4021:
4020:
4017:
4016:
4011:
4008:
4006:
4003:
4001:
3998:
3997:
3996:
3993:
3992:
3989:
3986:
3985:
3982:
3979:
3978:
3976:
3973:
3966:
3963:
3961:
3957:
3952:
3948:
3944:
3937:
3932:
3930:
3925:
3923:
3918:
3917:
3914:
3904:
3900:
3896:
3892:
3888:
3884:
3880:
3876:
3869:
3866:
3861:
3857:
3852:
3847:
3843:
3839:
3835:
3831:
3827:
3820:
3817:
3812:
3808:
3804:
3800:
3796:
3792:
3785:
3782:
3777:
3773:
3769:
3765:
3760:
3755:
3751:
3747:
3743:
3736:
3733:
3728:
3724:
3719:
3714:
3710:
3706:
3702:
3698:
3694:
3690:
3686:
3679:
3676:
3671:
3667:
3662:
3657:
3652:
3647:
3643:
3639:
3635:
3628:
3625:
3620:
3616:
3612:
3608:
3604:
3600:
3596:
3592:
3588:
3584:
3577:
3574:
3569:
3565:
3558:
3555:
3550:
3548:0-8153-3218-1
3544:
3540:
3536:
3529:
3526:
3521:
3519:1-85996-228-9
3515:
3511:
3507:
3503:
3496:
3493:
3488:
3482:
3478:
3471:
3468:
3463:
3459:
3455:
3451:
3446:
3441:
3438:(7): 1233–5.
3437:
3433:
3429:
3422:
3420:
3418:
3416:
3414:
3412:
3408:
3396:
3395:Howstuffworks
3392:
3385:
3382:
3377:
3371:
3367:
3360:
3357:
3352:
3348:
3343:
3338:
3334:
3330:
3326:
3322:
3318:
3311:
3308:
3303:
3299:
3294:
3289:
3285:
3281:
3276:
3271:
3267:
3263:
3259:
3252:
3249:
3244:
3242:0-8153-3218-1
3238:
3234:
3230:
3223:
3220:
3215:
3211:
3207:
3203:
3199:
3195:
3188:
3185:
3180:
3178:0-8153-3218-1
3174:
3170:
3166:
3159:
3156:
3151:
3147:
3142:
3137:
3133:
3129:
3126:(6429): 835.
3125:
3121:
3117:
3110:
3107:
3102:
3098:
3094:
3090:
3086:
3082:
3075:
3072:
3067:
3063:
3059:
3052:
3049:
3044:
3040:
3035:
3030:
3026:
3022:
3018:
3014:
3010:
3003:
3000:
2995:
2991:
2986:
2981:
2977:
2973:
2969:
2965:
2961:
2954:
2951:
2946:
2942:
2938:
2934:
2930:
2926:
2922:
2918:
2911:
2908:
2903:
2899:
2894:
2889:
2885:
2881:
2877:
2873:
2869:
2862:
2859:
2854:
2850:
2845:
2840:
2836:
2832:
2828:
2821:
2818:
2813:
2809:
2805:
2799:
2795:
2788:
2786:
2784:
2782:
2780:
2778:
2776:
2774:
2772:
2768:
2763:
2759:
2754:
2749:
2745:
2741:
2737:
2730:
2727:
2722:
2718:
2713:
2708:
2704:
2700:
2696:
2689:
2686:
2681:
2677:
2672:
2667:
2662:
2657:
2653:
2649:
2645:
2638:
2635:
2632:
2627:
2625:0-7167-3136-3
2621:
2617:
2616:
2608:
2605:
2600:
2596:
2591:
2586:
2582:
2578:
2574:
2570:
2566:
2559:
2556:
2551:
2549:0-8153-3218-1
2545:
2541:
2537:
2530:
2527:
2524:
2520:
2517:
2516:0-8162-2450-1
2513:
2509:
2503:
2500:
2495:
2491:
2487:
2483:
2479:
2475:
2468:
2465:
2460:
2456:
2451:
2446:
2442:
2438:
2434:
2430:
2429:Cell Research
2426:
2419:
2417:
2413:
2410:
2405:
2403:0-7167-3051-0
2399:
2395:
2394:
2386:
2383:
2378:
2372:
2368:
2361:
2358:
2353:
2351:0-8153-3218-1
2347:
2343:
2336:
2333:
2322:
2318:
2312:
2309:
2298:
2291:
2288:
2283:
2279:
2274:
2269:
2265:
2261:
2257:
2253:
2249:
2242:
2239:
2227:
2226:
2221:
2214:
2211:
2206:
2202:
2197:
2192:
2187:
2182:
2178:
2174:
2170:
2163:
2160:
2155:
2151:
2146:
2141:
2137:
2133:
2130:: 3765–3778.
2129:
2125:
2121:
2119:
2110:
2108:
2106:
2102:
2097:
2093:
2088:
2083:
2079:
2075:
2071:
2067:
2063:
2059:
2055:
2048:
2045:
2034:
2030:
2024:
2021:
2010:on 2020-03-26
2009:
2005:
2003:0-7167-3051-0
1999:
1995:
1994:
1989:
1982:
1980:
1976:
1971:
1967:
1962:
1957:
1953:
1949:
1945:
1941:
1937:
1930:
1928:
1924:
1919:
1917:0-7167-3136-3
1913:
1909:
1905:
1898:
1896:
1892:
1881:on 2020-03-26
1880:
1876:
1874:0-7167-3051-0
1870:
1866:
1865:
1860:
1854:
1848:
1845:
1840:
1836:
1832:
1825:
1822:
1810:
1806:
1800:
1797:
1786:
1782:
1776:
1773:
1762:
1755:
1752:
1747:
1743:
1738:
1733:
1729:
1725:
1721:
1717:
1713:
1706:
1703:
1698:
1694:
1689:
1684:
1680:
1676:
1672:
1668:
1664:
1657:
1654:
1649:
1645:
1640:
1635:
1631:
1627:
1623:
1619:
1615:
1608:
1605:
1599:
1590:
1584:
1581:
1574:
1570:
1567:
1565:
1562:
1560:
1557:
1555:
1554:Hachimoji DNA
1552:
1550:
1547:
1545:
1542:
1540:
1537:
1535:
1532:
1530:
1527:
1525:
1522:
1520:
1519:Cell division
1517:
1515:
1512:
1510:
1507:
1506:
1502:
1500:
1498:
1494:
1490:
1486:
1482:
1476:
1468:
1463:
1460:
1458:
1454:
1452:
1449:
1446:
1443:
1441:
1437:
1434:
1433:
1432:
1425:
1417:
1407:
1398:
1394:
1391:This section
1389:
1386:
1382:
1381:
1377:
1369:
1366:
1362:
1358:
1354:
1350:
1345:
1343:
1338:
1334:
1332:
1328:
1325:
1320:
1318:
1314:
1310:
1300:
1293:
1291:
1289:
1284:
1280:
1274:
1272:
1268:
1264:
1260:
1256:
1254:
1246:
1244:
1242:
1237:
1234:
1229:
1227:
1224:
1220:
1215:
1211:
1207:
1203:
1201:
1197:
1193:
1189:
1181:
1175:
1170:
1166:
1165:Cell division
1158:
1156:
1154:
1150:
1145:
1143:
1137:
1135:
1131:
1127:
1123:
1119:
1118:somatic cells
1115:
1106:
1104:
1101:
1100:
1091:
1087:
1086:budding yeast
1081:
1078:
1070:
1063:
1061:
1059:
1058:DNA-unwinding
1055:
1051:
1047:
1043:
1039:
1035:
1032:
1031:
1021:
1017:
1016:
1011:
1009:
1006:
1005:
1001:
999:
996:
995:
991:
987:
985:
982:
981:
977:
975:
972:
971:
967:
965:
964:Topoisomerase
962:
961:
957:
955:
952:
951:
947:
945:
942:
941:
936:
934:
931:
930:
926:
922:
920:
917:
916:
912:
909:
908:
905:
903:
895:
893:
889:
887:
883:
879:
874:
872:
868:
863:
861:
857:
851:
845:
841:
836:
829:
827:
825:
821:
817:
813:
809:
804:
797:
795:
789:
787:
784:
780:
772:
766:
759:
752:
750:
748:
744:
739:
737:
732:
730:
726:
722:
717:
713:
712:
706:
704:
699:
697:
696:topoisomerase
693:
692:Rossmann-like
689:
685:
681:
677:
672:
670:
665:
661:
657:
652:
644:
639:
635:
631:
630:geminiviruses
627:
623:
620:
616:
612:
609:
605:
601:
598:
594:
590:
586:
582:
581:
580:
577:
569:
567:
563:
561:
552:
550:
548:
543:
539:
534:
531:
527:
523:
522:budding yeast
518:
514:
509:
506:
502:
498:
494:
490:
486:
482:
478:
477:
472:
468:
464:
458:
450:
448:
446:
442:
433:
425:
418:
416:
409:
401:
396:
392:
384:
382:
380:
375:
371:
369:
368:pyrophosphate
365:
364:pyrophosphate
361:
357:
353:
349:
345:
341:
336:
334:
329:
325:
316:
311:
303:
301:
299:
294:
292:
288:
286:
282:
278:
274:
270:
266:
262:
258:
254:
250:
247:
243:
239:
235:
231:
227:
223:
215:
211:
207:
203:
199:
194:
188:DNA structure
187:
185:
183:
179:
175:
171:
167:
163:
159:
155:
151:
150:
144:
142:
138:
134:
130:
129:DNA synthesis
126:
122:
119:, results in
118:
114:
110:
106:
102:
97:
95:
91:
87:
83:
80:
79:complementary
76:
71:
69:
65:
61:
57:
53:
49:
41:
37:
32:
19:
5444:Viral vector
5287:Gerontoplast
5214:Transpoviron
5120:
4986:Nucleic acid
4972:Fungal prion
4870:Helper-virus
4857:
4850:
4757:
4750:
4607:Processivity
4433:synthesis in
3942:
3878:
3874:
3868:
3833:
3829:
3819:
3794:
3790:
3784:
3749:
3745:
3735:
3692:
3688:
3678:
3641:
3638:PLOS Biology
3637:
3627:
3586:
3582:
3576:
3567:
3557:
3538:
3528:
3505:
3495:
3476:
3470:
3435:
3431:
3398:. Retrieved
3394:
3384:
3365:
3359:
3324:
3320:
3310:
3265:
3261:
3251:
3232:
3222:
3197:
3193:
3187:
3168:
3158:
3123:
3119:
3109:
3084:
3080:
3074:
3057:
3051:
3016:
3012:
3002:
2967:
2963:
2953:
2920:
2916:
2910:
2875:
2871:
2861:
2834:
2830:
2820:
2793:
2743:
2739:
2729:
2702:
2699:Cell Reports
2698:
2688:
2651:
2647:
2637:
2614:
2607:
2572:
2568:
2558:
2539:
2529:
2507:
2502:
2477:
2473:
2467:
2432:
2428:
2393:Biochemistry
2392:
2385:
2366:
2360:
2341:
2335:
2324:. Retrieved
2321:Khan Academy
2320:
2311:
2300:. Retrieved
2290:
2255:
2251:
2241:
2229:. Retrieved
2225:SciTechDaily
2223:
2213:
2176:
2172:
2162:
2127:
2123:
2117:
2061:
2057:
2047:
2036:. Retrieved
2032:
2023:
2012:. Retrieved
2008:the original
1993:Biochemistry
1992:
1943:
1939:
1907:
1883:. Retrieved
1879:the original
1864:Biochemistry
1863:
1847:
1838:
1834:
1824:
1813:. Retrieved
1808:
1799:
1788:. Retrieved
1785:pathwayz.org
1784:
1775:
1764:. Retrieved
1754:
1719:
1715:
1705:
1670:
1666:
1656:
1621:
1617:
1607:
1583:
1480:
1478:
1430:
1401:
1397:adding to it
1392:
1341:
1339:
1335:
1323:
1321:
1308:
1305:
1287:
1282:
1278:
1275:
1258:
1257:
1252:
1250:
1238:
1230:
1216:
1212:
1208:
1204:
1185:
1148:
1146:
1138:
1110:
1097:
1094:
1076:
1075:
1028:
1027:
1013:
919:DNA helicase
899:
890:
875:
864:
852:
848:
807:
805:
801:
793:
782:
781:
777:
740:
733:
709:
707:
700:
673:
656:processivity
648:
634:parvoviruses
626:circoviruses
615:adenoviruses
604:retroviruses
573:
564:
556:
535:
510:
474:
460:
438:
414:
378:
376:
372:
337:
322:
295:
289:
222:double helix
219:
198:double helix
178:genetic code
166:transfer RNA
147:
145:
98:
90:proofreading
75:double helix
72:
51:
45:
36:double helix
5468:Cancer cell
5334:Abiogenesis
5282:Chromoplast
5277:Chloroplast
5060:Degradative
4802:dsRNA virus
4797:ssDNA virus
4790:Giant virus
4785:dsDNA virus
4626:Termination
4300:Prokaryotic
4292:Replication
3968:Prokaryotic
3947:prokaryotic
3945:(comparing
3644:(6): e185.
3400:January 20,
3262:BMC Biology
1544:Epigenetics
1509:Autopoiesis
1153:Tus protein
1142:Ter protein
1107:Termination
725:exonuclease
716:DNA Pol III
505:Mcm complex
356:nucleotides
340:nucleotides
269:pyrimidines
246:nucleobases
230:deoxyribose
226:nucleotides
182:abiogenesis
170:translation
160:(LCR), and
137:nucleotides
40:Nucleotides
5545:Senescence
5534:Categories
5376:Proteinoid
5371:Coacervate
5324:Nitroplast
5317:Trophosome
5312:Bacteriome
5297:Apicoplast
5292:Leucoplast
5133:Chromosome
5051:Resistance
4759:Parakaryon
4638:Telomerase
4612:DNA ligase
4605:Movement:
4429:Eukaryotic
4400:DNA gyrase
4385:DNA ligase
4304:elongation
4035:Eukaryotic
3972:initiation
3960:Initiation
3951:eukaryotic
3836:(1): 2–9.
2654:: 103131.
2648:DNA Repair
2326:2020-12-10
2302:2023-08-04
2179:: e63431.
2038:2020-12-10
2033:yourgenome
2014:2019-08-09
1885:2019-08-09
1815:2020-12-10
1790:2020-12-10
1766:2023-08-04
1600:References
1589:energetics
1483:using the
1353:Min System
1347:See also:
1327:methylates
1279:clustering
1188:cell cycle
1182:Eukaryotes
1169:Cell cycle
1159:Regulation
1130:telomerase
1050:polymerase
1008:Telomerase
984:DNA ligase
974:DNA gyrase
882:chaperones
860:supercoils
856:DNA gyrase
824:DNA ligase
736:eukaryotes
684:eukaryotes
570:Elongation
465:and early
419:Initiation
319:continues.
281:base pairs
242:nucleotide
238:nucleobase
214:base pairs
141:interphase
5524:Astronomy
5385:Research
5366:Protocell
5105:Retrozyme
5064:Virulence
5046:Fertility
4893:Virophage
4881:Satellite
4872:dependent
4724:Eukaryota
4576:DNA clamp
4390:DNA clamp
4380:Replisome
3709:1943-0264
3284:1741-7007
3268:(1): 61.
3066:1802/6537
2923:: 39–80.
2078:1943-0264
1853:mutations
1462:Chromatin
1457:oncogenes
1126:germ cell
1114:Telomeres
944:DNA clamp
902:replisome
886:chromatin
721:DNA Pol I
560:α-primase
234:phosphate
232:sugar, a
115:known as
107:, in the
5412:Organism
5405:See also
5381:Sulphobe
5358:Ribozyme
5353:RNA life
5260:Mitosome
5204:Prophage
5199:Provirus
5187:Replicon
5143:Circular
5090:Phagemid
5007:Mobilome
4999:elements
4909:Virusoid
4832:Subviral
4744:Protista
4729:Animalia
4714:Bacteria
4634:Telomere
4250:Replicon
4206:Helicase
4197:RNASEH2A
4041:G1 phase
3995:Helicase
3860:24366029
3776:14652024
3727:23818497
3670:16719561
3611:11429609
3462:15397410
3454:16814710
3351:20141833
3302:32517760
3150:30679383
3043:26186286
2994:17158702
2945:33197061
2937:11395402
2812:70173205
2762:12110182
2721:32160540
2680:33992866
2459:18166979
2282:12540918
2205:33648631
2154:34285777
2118:In vitro
2096:20452942
1970:36640769
1841:(1): 98.
1746:28537574
1697:27542827
1648:23818497
1503:See also
1481:in vitro
1438:Unusual
1404:May 2020
1322:Because
1294:Bacteria
1263:dynamics
1046:helicase
1030:In vitro
878:histones
808:template
676:bacteria
643:tyrosine
638:plasmids
593:plasmids
576:hydroxyl
467:G1 phase
461:In late
279:to form
253:cytosine
236:, and a
174:proteins
149:in vitro
125:proteins
117:helicase
94:fidelity
5560:Copying
5512:Biology
5498:Portals
5394:Jeewanu
5308:Organs
5272:Plastid
5072:Cryptic
5041:Plasmid
4739:Plantae
4719:Archaea
4547:epsilon
4435:S phase
4262:Lagging
4217:Primase
4192:RNASEH1
4187:RNase H
4019:Primase
3903:2448875
3883:Bibcode
3875:Science
3851:4354890
3811:4866337
3768:7553853
3718:3685895
3661:1470461
3619:4393812
3591:Bibcode
3568:Genomes
3506:Genomes
3342:3433953
3293:7281927
3214:1657531
3141:6681829
3120:Science
3101:5689363
3034:4636199
2985:1698513
2902:9722641
2853:9734354
2671:8296962
2599:9351837
2590:1170261
2450:3639319
2260:Bibcode
2231:3 April
2196:7924937
2145:8267548
2087:2845211
1961:9898300
1737:6386472
1688:5002974
1639:3685895
1489:primers
1357:Plasmid
1342:E. coli
1324:E. coli
1309:E. coli
1233:geminin
1196:cyclins
1149:E. coli
1099:Cohesin
1036:(using
998:Primase
938:cells.
812:primase
711:E. coli
680:archaea
651:primase
613:In the
597:primase
585:viruses
542:S phase
476:E. coli
471:origins
463:mitosis
379:E. coli
328:enzymes
261:thymine
257:guanine
249:adenine
210:element
204:). The
156:(PCR),
82:strands
77:of two
54:is the
5485:Virome
5463:Nanobe
5160:Genome
5138:Linear
5083:Fosmid
5078:Cosmid
4843:Viroid
4834:agents
4278:Primer
3901:
3858:
3848:
3809:
3774:
3766:
3725:
3715:
3707:
3668:
3658:
3617:
3609:
3583:Nature
3545:
3516:
3483:
3460:
3452:
3372:
3349:
3339:
3300:
3290:
3282:
3239:
3212:
3175:
3148:
3138:
3099:
3041:
3031:
2992:
2982:
2943:
2935:
2900:
2893:147817
2890:
2851:
2810:
2800:
2760:
2719:
2678:
2668:
2622:
2597:
2587:
2546:
2521:
2514:
2494:789903
2492:
2457:
2447:
2400:
2373:
2348:
2280:
2252:Nature
2203:
2193:
2152:
2142:
2094:
2084:
2076:
2000:
1968:
1958:
1914:
1871:
1744:
1734:
1695:
1685:
1646:
1636:
1549:Genome
1363:, and
1271:lamins
1134:cancer
1052:, and
910:Enzyme
840:trimer
669:Ligase
632:, the
628:, the
595:use a
589:phages
513:cyclin
473:". In
439:For a
333:primer
265:purine
259:, and
200:(type
113:enzyme
109:genome
4959:Prion
4930:Other
4777:Virus
4734:Fungi
4567:POLE4
4562:POLE3
4557:POLE2
4540:POLD4
4535:POLD3
4530:POLD2
4525:POLD1
4520:delta
4513:PRIM2
4508:PRIM1
4503:POLA2
4498:POLA1
4493:alpha
4226:PRIM2
4221:PRIM1
4178:SSBP4
4173:SSBP3
4168:SSBP2
3772:S2CID
3615:S2CID
3458:S2CID
2941:S2CID
2173:eLife
1575:Notes
664:RNase
485:yeast
483:; in
481:Dna A
206:atoms
202:B-DNA
99:In a
5473:HeLa
5417:Cell
5165:Gene
4653:DKC1
4648:TERC
4643:TERT
4599:Both
4581:PCNA
4552:POLE
4478:RPA1
4461:FEN1
4449:RFC1
4373:holE
4368:holD
4363:holC
4358:holB
4353:holA
4348:dnaX
4343:dnaT
4338:dnaQ
4333:dnaN
4328:dnaH
4323:dnaE
4318:dnaC
4264:and
4235:Both
4210:HFM1
4140:MCM7
4135:MCM6
4130:MCM5
4125:MCM4
4120:MCM3
4115:MCM2
4103:Cdt1
4096:Cdc6
4087:ORC6
4082:ORC5
4077:ORC4
4072:ORC3
4067:ORC2
4062:ORC1
4024:dnaG
4005:dnaB
4000:dnaA
3988:dnaC
3899:PMID
3856:PMID
3807:PMID
3764:PMID
3746:Cell
3723:PMID
3705:ISSN
3666:PMID
3607:PMID
3543:ISBN
3514:ISBN
3481:ISBN
3450:PMID
3432:Cell
3402:2015
3370:ISBN
3347:PMID
3321:Cell
3298:PMID
3280:ISSN
3237:ISBN
3210:PMID
3173:ISBN
3146:PMID
3097:PMID
3039:PMID
2990:PMID
2933:PMID
2898:PMID
2849:PMID
2808:OCLC
2798:ISBN
2758:PMID
2740:Cell
2717:PMID
2676:PMID
2620:ISBN
2595:PMID
2544:ISBN
2519:ISBN
2512:ISBN
2490:PMID
2455:PMID
2398:ISBN
2371:ISBN
2346:ISBN
2278:PMID
2233:2021
2201:PMID
2150:PMID
2092:PMID
2074:ISSN
1998:ISBN
1966:PMID
1912:ISBN
1869:ISBN
1742:PMID
1693:PMID
1644:PMID
1587:The
1559:Life
1534:Gene
1349:FtsZ
1331:SeqA
1219:Cdk1
1198:and
1167:and
1040:and
844:PCNA
810:, a
688:DnaG
678:and
591:and
547:DBF4
538:Cdc7
530:Clb6
528:and
526:Clb5
501:Cdt1
499:and
497:Cdc6
393:and
352:base
101:cell
5056:Col
4944:DNA
4941:RNA
4920:DNA
4917:RNA
4246:Ori
3949:to
3891:doi
3879:239
3846:PMC
3838:doi
3799:doi
3754:doi
3713:PMC
3697:doi
3656:PMC
3646:doi
3599:doi
3587:411
3440:doi
3436:125
3337:PMC
3329:doi
3325:140
3288:PMC
3270:doi
3202:doi
3136:PMC
3128:doi
3124:363
3089:doi
3062:hdl
3029:PMC
3021:doi
2980:PMC
2972:doi
2925:doi
2888:PMC
2880:doi
2839:doi
2748:doi
2744:109
2707:doi
2666:PMC
2656:doi
2652:103
2585:PMC
2577:doi
2482:doi
2478:106
2445:PMC
2437:doi
2268:doi
2256:421
2191:PMC
2181:doi
2140:PMC
2132:doi
2082:PMC
2066:doi
1956:PMC
1948:doi
1732:PMC
1724:doi
1683:PMC
1675:doi
1634:PMC
1626:doi
1399:.
1315:to
1269:or
1223:SCF
816:RNA
734:In
517:Cdk
287:).
60:DNA
46:In
5536::
5068:Ti
4636::
4411::
4219::
4208::
4010:T7
3897:.
3889:.
3877:.
3854:.
3844:.
3834:16
3832:.
3828:.
3805:.
3795:31
3793:.
3770:.
3762:.
3750:82
3748:.
3744:.
3721:.
3711:.
3703:.
3691:.
3687:.
3664:.
3654:.
3640:.
3636:.
3613:.
3605:.
3597:.
3585:.
3566:.
3537:.
3512:.
3508:.
3504:.
3456:.
3448:.
3434:.
3430:.
3410:^
3393:.
3345:.
3335:.
3323:.
3319:.
3296:.
3286:.
3278:.
3266:18
3264:.
3260:.
3231:.
3208:.
3198:26
3196:.
3167:.
3144:.
3134:.
3122:.
3118:.
3095:.
3085:32
3083:.
3037:.
3027:.
3017:59
3015:.
3011:.
2988:.
2978:.
2968:70
2966:.
2962:.
2939:.
2931:.
2921:70
2919:.
2896:.
2886:.
2876:26
2874:.
2870:.
2847:.
2833:.
2829:.
2806:.
2770:^
2756:.
2742:.
2738:.
2715:.
2703:30
2701:.
2697:.
2674:.
2664:.
2650:.
2646:.
2593:.
2583:.
2573:16
2571:.
2567:.
2538:.
2488:.
2476:.
2453:.
2443:.
2433:18
2431:.
2427:.
2415:^
2319:.
2276:.
2266:.
2254:.
2250:.
2222:.
2199:.
2189:.
2177:10
2175:.
2171:.
2148:.
2138:.
2128:19
2126:.
2122:.
2104:^
2090:.
2080:.
2072:.
2060:.
2056:.
2031:.
1990:.
1978:^
1964:.
1954:.
1944:83
1942:.
1938:.
1926:^
1906:.
1894:^
1861:.
1855:.
1837:.
1833:.
1807:.
1783:.
1740:.
1730:.
1720:18
1718:.
1714:.
1691:.
1681:.
1671:30
1669:.
1665:.
1642:.
1632:.
1620:.
1616:.
1499:.
1359:,
1355:,
1351:,
1243:.
1144:.
1048:,
826:.
714:,
662:.
587:,
447:.
255:,
251:,
184:.
143:.
50:,
5500::
5461:?
5455:?
5364:†
5351:?
5066:/
4911:)
4895:)
4689:e
4682:t
4675:v
4489::
4437:)
4431:(
4306:)
4302:(
4248:/
4244:/
4043:)
4037:(
3974:)
3970:(
3953:)
3935:e
3928:t
3921:v
3905:.
3893::
3885::
3862:.
3840::
3813:.
3801::
3778:.
3756::
3729:.
3699::
3693:5
3672:.
3648::
3642:4
3621:.
3601::
3593::
3551:.
3522:.
3489:.
3464:.
3442::
3404:.
3378:.
3353:.
3331::
3304:.
3272::
3245:.
3216:.
3204::
3181:.
3152:.
3130::
3103:.
3091::
3068:.
3064::
3045:.
3023::
2996:.
2974::
2947:.
2927::
2904:.
2882::
2855:.
2841::
2835:2
2814:.
2764:.
2750::
2723:.
2709::
2682:.
2658::
2628:.
2601:.
2579::
2552:.
2496:.
2484::
2461:.
2439::
2406:.
2379:.
2354:.
2329:.
2305:.
2284:.
2270::
2262::
2235:.
2207:.
2183::
2156:.
2134::
2098:.
2068::
2062:2
2041:.
2017:.
1972:.
1950::
1920:.
1888:.
1839:1
1818:.
1793:.
1769:.
1748:.
1726::
1699:.
1677::
1650:.
1628::
1622:5
1406:)
1402:(
682:/
610:.
515:-
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.