954:. After infection, reverse transcription is accompanied by template switching between the two genome copies (copy choice recombination). There are two models that suggest why RNA transcriptase switches templates. The first, the forced copy-choice model, proposes that reverse transcriptase changes the RNA template when it encounters a nick, implying that recombination is obligatory to maintaining virus genome integrity. The second, the dynamic choice model, suggests that reverse transcriptase changes templates when the RNAse function and the polymerase function are not in sync rate-wise, implying that recombination occurs at random and is not in response to genomic damage. A study by Rawson et al. supported both models of recombination. From 5 to 14 recombination events per genome occur at each replication cycle. Template switching (recombination) appears to be necessary for maintaining genome integrity and as a repair mechanism for salvaging damaged genomes.
612:
42:
686:
1072:
963:
1086:
480:(RNase H), and DNA-dependent DNA polymerase activity. Collectively, these activities enable the enzyme to convert single-stranded RNA into double-stranded cDNA. In retroviruses and retrotransposons, this cDNA can then integrate into the host genome, from which new RNA copies can be made via host-cell
790:
and forms a base-paired duplex with the viral RNA at PBS. The fact that the PBS is located near the 5’ terminus of viral RNA is unusual because reverse transcriptase synthesize DNA from 3’ end of the primer in the 5’ to 3’ direction (with respect to the newly synthesized DNA strand). Therefore, the
888:
There are three different replication systems during the life cycle of a retrovirus. The first process is the reverse transcriptase synthesis of viral DNA from viral RNA, which then forms newly made complementary DNA strands. The second replication process occurs when host cellular DNA polymerase
778:, in which there is a translocation of short DNA product from initial RNA-dependent DNA synthesis to acceptor template regions at the other end of the genome, which are later reached and processed by the reverse transcriptase for its DNA-dependent DNA activity.
651:, can allow RNA to serve as a template in assembling and making DNA strands. HIV infects humans with the use of this enzyme. Without reverse transcriptase, the viral genome would not be able to incorporate into the host cell, resulting in failure to replicate.
989:
uses reverse transcriptase to copy its genetic material and generate new viruses (part of a retrovirus proliferation circle), specific drugs have been designed to disrupt the process and thereby suppress its growth. Collectively, these drugs are known as
4919:
785:
is annealed to viral RNA is called the primer-binding site (PBS). The RNA 5’end to the PBS site is called U5, and the RNA 3’ end to the PBS is called the leader. The tRNA primer is unwound between 14 and 22
889:
replicates the integrated viral DNA. Lastly, RNA polymerase II transcribes the proviral DNA into RNA, which will be packed into virions. Mutation can occur during one or all of these replication steps.
860:
Valerian Dolja of Oregon State argues that viruses, due to their diversity, have played an evolutionary role in the development of cellular life, with reverse transcriptase playing a central role.
876:
family, which is vital to their replication. By degrading the RNA template, it allows the other strand of DNA to be synthesized. Some fragments from the digestion also serve as the primer for the
4929:
673:
The process of reverse transcription, also called retrotranscription or retrotras, is extremely error-prone, and it is during this step that mutations may occur. Such mutations may cause
4924:
1833:
766:
Both strands are extended to form a complete double-stranded DNA copy of the original viral RNA genome, which can then be incorporated into the host's genome by the enzyme
5044:
5022:
830:
utilize reverse transcriptase to move from one position in the genome to another via an RNA intermediate. They are found abundantly in the genomes of plants and animals.
2720:
2715:
4840:
377:
182:
2365:
1033:
1019:
2772:
791:
primer and reverse transcriptase must be relocated to 3’ end of viral RNA. In order to accomplish this reposition, multiple steps and various enzymes including
662:
In virus species with reverse transcriptase lacking DNA-dependent DNA polymerase activity, creation of double-stranded DNA can possibly be done by host-encoded
396:
4950:
1764:
1114:
751:
The tRNA primer then "jumps" to the 3’ end of the viral genome, and the newly synthesised DNA strands hybridizes to the complementary R region on the RNA.
3300:
3295:
3290:
3285:
3247:
3242:
1654:
5129:
4845:
3622:
3280:
3275:
814:
DNA to form a double-stranded viral DNA intermediate (vDNA). The HIV viral RNA structural elements regulate the progression of reverse transcription.
2751:
2746:
2741:
5012:
2465:
2115:"Evidence that BmTXK beta-BmKCT cDNA from Chinese scorpion Buthus martensii Karsch is an artifact generated in the reverse transcription process"
4003:
1158:
Tu X, Das K, Han Q, Bauman JD, Clark AD, Hou X, Frenkel YV, Gaffney BL, Jones RA, Boyer PL, Hughes SH, Sarafianos SG, Arnold E (October 2010).
904:
to accumulate at an accelerated rate relative to proofread forms of replication. The commercially available reverse transcriptases produced by
4651:
3766:
2014:
1746:
1571:
539:
118:
1762:
Romashchenko AG, et al. (1977). "Otdelenie ot preparatov DNK-polimeraz I RNK-zavisimoy DNK-polimeraz; oshistka i svoystva fermenta".
849:
et al., 1971a, 1972) and a few years later in the USSR (Romashchenko 1977). These have since been broadly described as part of bacterial
513:
1056:. The commercial availability of reverse transcriptase greatly improved knowledge in the area of molecular biology, as, along with other
631:
The enzymes are encoded and used by viruses that use reverse transcription as a step in the process of replication. Reverse-transcribing
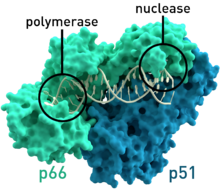
50:-1 reverse transcriptase where the two subunits p51 and p66 are colored and the active sites of polymerase and nuclease are highlighted.
2030:
1841:
4785:
2578:
763:
The tRNA primer leaves and a "jump" happens. The PBS from the second strand hybridizes with the complementary PBS on the first strand.
466:
4355:
4149:
3736:
3670:
2897:
4496:
3364:
3032:
588:
389:
465:. Contrary to a widely held belief, the process does not violate the flows of genetic information as described by the classical
991:
980:
2370:
1456:
912:
908:
are quoted by their manuals as having error rates in the range of 1 in 17,000 bases for AMV and 1 in 30,000 bases for M-MLV.
340:
316:
1457:"Improving the thermal stability of avian myeloblastosis virus reverse transcriptase α-subunit by site-directed mutagenesis"
202:
5069:
5064:
4624:
4442:
3883:
2425:
935:, may have been one of the causes for finding several thousand unannotated transcripts in the genomes of model organisms.
854:
674:
1562:
Telesnitsky A, Goff SP (1993). "Strong-stop strand transfer during reverse transcription". In Skalka MA, Goff SP (eds.).
4934:
4822:
4790:
4775:
3357:
3352:
2458:
869:
857:. In order to initiate synthesis of DNA, a primer is needed. In bacteria, the primer is synthesized during replication.
803:
4457:
3985:
3920:
3592:
3568:
3375:
3260:
3166:
3092:
3017:
2912:
2796:
2731:
2651:
2617:
2568:
1275:"Epigenetically facilitated mutational assimilation: epigenetics as a hub within the inclusive evolutionary synthesis"
734:
acts as a primer and hybridizes to a complementary part of the virus RNA genome called the primer binding site or PBS.
611:
741:
to the U5 (non-coding region) and R region (a direct repeat found at both ends of the RNA molecule) of the viral RNA.
5109:
2661:
2588:
2553:
782:
667:
5055:
5033:
5008:
4983:
4903:
4511:
4471:
4416:
4393:
4365:
4333:
4199:
3543:
3347:
2548:
1045:
1037:
1025:
807:
701:
700:
viruses, are RNA reverse-transcribing viruses with a DNA intermediate. Their genomes consist of two molecules of
493:
138:
4286:
334:
5124:
4520:
4501:
4432:
4166:
4042:
3930:
3856:
3794:
3746:
3698:
3650:
3602:
3481:
3330:
3225:
2986:
2892:
2872:
2855:
2705:
2593:
2528:
2359:
2005:
Bbenek K, Kunkel AT (1993). "The fidelity of retroviral reverse transcriptases". In Skalka MA, Goff PS (eds.).
666:, mistaking the viral DNA-RNA for a primer and synthesizing a double-stranded DNA by a similar mechanism as in
227:
1868:"Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition"
872:. In addition to the transcription function, retroviral reverse transcriptases have a domain belonging to the
190:
1662:
1602:
Moelling K, Broecker F (April 2015). "The reverse transcriptase-RNase H: from viruses to antiviral defense".
321:
4378:
2451:
2433:
481:
2695:
1785:"RNA-dependent DNA polymerase activity of RNA tumor viruses. I. Directing influence of DNA in the reaction"
643:
into DNA, which is then integrated into the host genome and replicated along with it. Reverse-transcribing
5114:
4832:
4605:
4515:
4105:
3941:
3826:
2666:
1129:
897:
2440:
about the discovery in 1970 of retroviral reverse transcriptase and its impact on life sciences research.
2214:"Recombination is required for efficient HIV-1 replication and the maintenance of viral genome integrity"
401:
4641:
4343:
4328:
4264:
4244:
4142:
4075:
3846:
3718:
3578:
728:
575:
531:
309:
916:
186:
4127:
41:
4600:
4595:
4475:
4338:
4318:
4227:
3877:
3815:
3502:
2946:
2937:
2859:
2806:
2627:
2518:
2319:
2067:
1611:
1216:
244:
131:
892:
Reverse transcriptase has a high error rate when transcribing RNA into DNA since, unlike most other
760:
Synthesis of the second DNA strand begins, using the remaining PP fragment of viral RNA as a primer.
143:
4863:
4855:
4636:
4303:
4239:
4203:
3548:
2942:
2932:
2851:
1324:
Temin HM, Mizutani S (June 1970). "RNA-dependent DNA polymerase in virions of Rous sarcoma virus".
1044:
strands, but, with the help of reverse transcriptase, RNA can be transcribed into DNA, thus making
950:
are packaged into each retrovirus particle, but, after an infection, each virus generates only one
834:
is another reverse transcriptase found in many eukaryotes, including humans, which carries its own
663:
484:. The same sequence of reactions is widely used in the laboratory to convert RNA to DNA for use in
337:
239:
1515:
Autexier C, Lue NF (June 2006). "The structure and function of telomerase reverse transcriptase".
261:
5119:
4726:
4664:
4615:
4350:
4177:
4070:
3947:
2421:
2292:
2144:
1715:
1635:
1497:
1392:
1349:
1306:
1240:
535:
518:
1733:
Krieger M, Scott MP, Matsudaira PT, Lodish HF, Darnell JE, Zipursky L, Kaiser C, Berk A (2004).
5104:
5099:
4741:
4731:
4721:
4646:
4630:
4589:
4575:
4570:
4308:
4271:
3522:
3270:
3209:
2974:
2398:
2335:
2284:
2243:
2136:
2095:
2010:
1987:
1938:
1897:
1814:
1742:
1707:
1627:
1567:
1532:
1489:
1434:
1384:
1341:
1232:
1189:
1148:
738:
559:
485:
446:
328:
177:
2355:
1866:
Sarafianos SG, Marchand B, Das K, Himmel DM, Parniak MA, Hughes SH, Arnold E (January 2009).
1549:
1528:
5074:
4736:
4716:
4658:
4554:
4452:
4437:
4383:
4281:
4276:
4135:
3784:
3390:
3127:
3122:
3117:
3112:
3107:
3102:
3081:
2327:
2274:
2233:
2225:
2185:
2175:
2126:
2085:
2075:
2037:
1977:
1969:
1928:
1887:
1879:
1804:
1796:
1697:
1689:
1619:
1524:
1479:
1471:
1426:
1376:
1333:
1296:
1286:
1224:
1179:
1171:
880:(either the same enzyme or a host protein), responsible for making the other (plus) strand.
845:
Initial reports of reverse transcriptase in prokaryotes came as far back as 1971 in France (
737:
Reverse transcriptase then adds DNA nucleotides onto the 3' end of the primer, synthesizing
853:, distinct sequences that code for reverse transcriptase, and are used in the synthesis of
297:
169:
4997:
4987:
4809:
4780:
4406:
4313:
4298:
4014:
3471:
3461:
3214:
2429:
1367:
Baltimore D (June 1970). "RNA-dependent DNA polymerase in virions of RNA tumour viruses".
868:
The reverse transcriptase employs a "right hand" structure similar to that found in other
846:
839:
827:
709:
582:
543:
523:
450:
1410:
Ferris AL, Hizi A, Showalter SD, Pichuantes S, Babe L, Craik CS, Hughes SH (April 1990).
273:
2323:
2071:
1615:
1220:
232:
4965:
4804:
4711:
4703:
4580:
4543:
4088:
3971:
3516:
3341:
3236:
2842:
2762:
2310:
Hu WS, Temin HM (November 1990). "Retroviral recombination and reverse transcription".
2238:
2213:
2190:
2163:
2090:
2055:
1982:
1957:
1892:
1867:
1702:
1677:
1301:
1274:
1184:
1159:
1119:
1104:
1091:
1077:
920:
893:
877:
792:
615:
Reverse transcriptase is shown with its finger, palm, and thumb regions. The catalytic
497:
489:
477:
473:
372:
2131:
2114:
2056:"Apparent non-canonical trans-splicing is generated by reverse transcriptase in vitro"
1933:
1916:
1809:
1784:
685:
352:
5093:
4234:
4037:
3688:
3149:
2786:
2503:
2478:
2474:
2437:
2407:
2386:. Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB)
1735:
1719:
1501:
1430:
347:
123:
17:
2366:
animation of reverse transcriptase action and three reverse transcriptase inhibitors
2296:
2148:
1639:
1310:
74:
4323:
4293:
4222:
3757:
3709:
3661:
3613:
3534:
3319:
3155:
2832:
2641:
2508:
2482:
1973:
1396:
1353:
1244:
1099:
1049:
799:
693:
648:
509:
165:
2443:
915:, reverse transcriptases have also been shown to be involved in processes such as
87:
2279:
2262:
2180:
2080:
1958:"RNase H activity: structure, specificity, and function in reverse transcription"
1800:
1048:
analysis of RNA molecules possible. Reverse transcriptase is used also to create
99:
4889:
4879:
4480:
4217:
4158:
4097:
3640:
2960:
2923:
2377:
HIV Replication 3D Medical
Animation. (Nov 2008). Video by Boehringer Ingelheim.
787:
599:
567:) has two subunits, which have respective molecular weights of 66 and 51
356:
2383:
1412:"Immunologic and proteolytic analysis of HIV-1 reverse transcriptase structure"
1071:
757:
The majority of viral RNA is degraded by RNAse H, leaving only the PP sequence.
4691:
4534:
4447:
4212:
4162:
3979:
3911:
3508:
3314:
3145:
3141:
2376:
1883:
1693:
1475:
1411:
1124:
1067:
1007:
999:
995:
967:
831:
823:
802:
activity that degrades the viral RNA during the synthesis of cDNA, as well as
636:
624:
616:
596:
568:
462:
426:
781:
Retroviral RNA is arranged in 5’ terminus to 3’ terminus. The site where the
4970:
4883:
4420:
4186:
4025:
4020:
3961:
3955:
3199:
3076:
3066:
3054:
2827:
2491:
2331:
1003:
924:
767:
644:
632:
469:, as transfers of information from RNA to DNA are explicitly held possible.
458:
454:
2288:
2247:
2140:
2099:
1991:
1901:
1711:
1631:
1536:
1493:
1193:
705:
2339:
1942:
1818:
1438:
1388:
1345:
1236:
689:
Mechanism of reverse transcription in HIV. Step numbers will not match up.
127:
4817:
4397:
2607:
2229:
1678:"Advances in understanding the initiation of HIV-1 reverse transcription"
951:
901:
697:
592:
472:
Retroviral RT has three sequential biochemical activities: RNA-dependent
94:
2418:
2212:
Rawson JM, Nikolaitchik OA, Keele BF, Pathak VK, Hu WS (November 2018).
1591:(2nd ed.). Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory.
931:
activity of reverse transcriptase, which can be demonstrated completely
659:
Reverse transcriptase creates double-stranded DNA from an RNA template.
285:
4768:
4753:
4369:
2412:
2402:
1623:
1484:
1057:
962:
905:
873:
745:
721:
670:, where the newly synthesized DNA displaces the original RNA template.
620:
304:
106:
2263:"Estimating the in-vivo HIV template switching and recombination rate"
1380:
1337:
1291:
1175:
1152:
1085:
563:
453:
mobile genetic elements to proliferate within the host genome, and by
4746:
4181:
3867:
3805:
3553:
3492:
3385:
2883:
2539:
1228:
947:
850:
712:. Examples of retroviruses include the human immunodeficiency virus (
640:
422:
384:
280:
268:
256:
197:
111:
2261:
Cromer D, Grimm AJ, Schlub TE, Mak J, Davenport MP (January 2016).
795:, ribonuclease H(RNase H) and polynucleotide unwinding are needed.
4763:
4758:
4610:
3438:
3428:
3423:
3413:
3408:
3403:
3398:
3005:
3000:
2995:
1109:
1060:, it allowed scientists to clone, sequence, and characterise RNA.
610:
438:
4920:
CDP-diacylglycerol—glycerol-3-phosphate 3-phosphatidyltransferase
1260:
The
Philosophy and History of Molecular Biology: New Perspectives
4674:
4669:
4565:
4560:
4259:
4254:
4249:
3990:
3821:
3433:
3418:
2847:
2583:
1053:
1024:
Reverse transcriptase is commonly used in research to apply the
731:
717:
292:
159:
81:
69:
4131:
2447:
4051:
4047:
4030:
3176:
3027:
1041:
1029:
986:
944:
835:
754:
The complementary DNA (cDNA) added in (2) is further extended.
713:
554:
527:
442:
430:
47:
2009:. New York: Cold Spring Harbor Laboratory Press. p. 85.
1917:"Structure of the RNA-dependent RNA polymerase of poliovirus"
1207:
Crick F (August 1970). "Central dogma of molecular biology".
585:
also has two subunits, a 63 kDa subunit and a 95 kDa subunit.
2411:(Human immunodeficiency virus Reverse transcriptase) at the
2397:
Overview of all the structural information available in the
1273:
Danchin É, Pocheville A, Rey O, Pujol B, Blanchet S (2019).
1587:
Bernstein A, Weiss R, Tooze J (1985). "RNA tumor viruses".
1566:(1st ed.). New York: Cold Spring Harbor. p. 49.
1262:. Dordrecht: Kluwer Academic Publishers. pp. 187–232.
2384:"Molecule of the Month: Reverse Transcriptase (Sep 2002)"
1160:"Structural basis of HIV-1 resistance to AZT by excision"
1006:(Viread), as well as non-nucleoside inhibitors, such as
748:
degrades the U5 and R regions on the 5’ end of the RNA.
2164:"Response to "The Reality of Pervasive Transcription""
2162:
van Bakel H, Nislow C, Blencowe BJ, Hughes TR (2011).
1661:. Community College of Baltimore Count. Archived from
655:
Process of reverse transcription or retrotranscription
4930:
CDP-diacylglycerol—inositol 3-phosphatidyltransferase
1834:"Could Giant Viruses Be the Origin of Life on Earth?"
4935:
CDP-diacylglycerol—choline O-phosphatidyltransferase
1676:
Krupkin M, Jackson LN, Ha B, Puglisi EV (Dec 2020).
994:
and include the nucleoside and nucleotide analogues
970:(AZT), a drug used to inhibit reverse transcriptase
744:
A domain on the reverse transcriptase enzyme called
5054:
5032:
5007:
4982:
4963:
4943:
4925:
CDP-diacylglycerol—serine O-phosphatidyltransferase
4912:
4902:
4872:
4854:
4831:
4803:
4702:
4542:
4533:
4510:
4470:
4415:
4392:
4364:
4198:
4174:
4096:
4087:
4064:
4001:
3928:
3919:
3910:
3866:
3854:
3845:
3804:
3792:
3783:
3756:
3744:
3735:
3708:
3696:
3687:
3660:
3648:
3639:
3612:
3600:
3591:
3566:
3533:
3491:
3479:
3470:
3455:
3373:
3328:
3313:
3258:
3223:
3208:
3193:
3164:
3140:
3090:
3075:
3062:
3053:
3015:
2984:
2973:
2922:
2910:
2882:
2870:
2841:
2823:
2794:
2785:
2760:
2729:
2703:
2694:
2683:
2649:
2640:
2615:
2606:
2566:
2538:
2526:
2517:
2499:
2490:
395:
383:
371:
366:
346:
327:
315:
303:
291:
279:
267:
255:
250:
238:
226:
221:
216:
196:
176:
158:
153:
137:
117:
105:
93:
80:
68:
60:
55:
32:
1734:
720:). Creation of double-stranded DNA occurs in the
639:, use the enzyme to reverse-transcribe their RNA
1861:
1859:
1034:reverse transcription polymerase chain reaction
1020:Reverse transcription polymerase chain reaction
538:. For their achievements, they shared the 1975
1915:Hansen JL, Long AM, Schultz SC (August 1997).
1450:
1448:
927:transcripts. It has been speculated that this
774:Creation of double-stranded DNA also involves
4143:
2459:
2207:
2205:
2203:
2201:
1455:Konishi A, Yasukawa K, Inouye K (July 2012).
838:template; this RNA is used as a template for
627:active site are shown in ball-and-stick form.
549:Well-studied reverse transcriptases include:
8:
4951:N-acetylglucosamine-1-phosphate transferase
4841:UTP—glucose-1-phosphate uridylyltransferase
2419:TWiV 904: 50 years of reverse transcriptase
1765:Proceedings of the USSR Academy of Sciences
4979:
4909:
4539:
4530:
4195:
4150:
4136:
4128:
4093:
4084:
3925:
3916:
3863:
3851:
3801:
3789:
3753:
3741:
3705:
3693:
3657:
3645:
3609:
3597:
3488:
3476:
3467:
3325:
3220:
3205:
3161:
3087:
3072:
3059:
2981:
2919:
2879:
2867:
2838:
2791:
2700:
2691:
2646:
2612:
2535:
2523:
2514:
2496:
2466:
2452:
2444:
1604:Annals of the New York Academy of Sciences
508:Reverse transcriptases were discovered by
363:
150:
40:
5045:serine/threonine-specific protein kinases
5023:serine/threonine-specific protein kinases
4846:Galactose-1-phosphate uridylyltransferase
3623:Parainfluenza hemagglutinin-neuraminidase
2358:at the U.S. National Library of Medicine
2278:
2237:
2189:
2179:
2130:
2089:
2079:
1981:
1932:
1891:
1808:
1701:
1483:
1300:
1290:
1183:
1164:Nature Structural & Molecular Biology
1529:10.1146/annurev.biochem.75.103004.142412
961:
684:
2054:Houseley J, Tollervey D (August 2010).
1140:
798:The HIV reverse transcriptase also has
716:) and the human T-lymphotropic virus (
522:virions and independently isolated by
213:
29:
3767:Respiratory syncytial virus G protein
1956:Schultz SJ, Champoux JJ (June 2008).
900:ability. This high error rate allows
574:M-MLV reverse transcriptase from the
540:Nobel Prize in Physiology or Medicine
437:. Reverse transcriptases are used by
7:
1783:Hurwitz J, Leis JP (January 1972).
1659:Doc Kaiser's Microbiology Home Page
581:AMV reverse transcriptase from the
1589:Molecular Biology of Tumor Viruses
25:
4356:Glucose-1,6-bisphosphate synthase
3671:Mumps hemagglutinin-neuraminidase
2318:(4985). New York, N.Y.: 1227–33.
1741:. New York: W.H. Freeman and CO.
1040:technique can be applied only to
553:HIV-1 reverse transcriptase from
5130:RNA reverse-transcribing viruses
4497:Ribose-phosphate diphosphokinase
3033:Hepatitis B virus DNA polymerase
2373:(September 2002) at the RCSB PDB
2125:(1–3): 183–4, author reply 185.
2031:"Promega kit instruction manual"
1084:
1070:
992:reverse-transcriptase inhibitors
681:Retroviral reverse transcription
589:Telomerase reverse transcriptase
449:to replicate their genomes, by
2864:(non-enveloped circular ds-DNA)
981:Reverse-transcriptase inhibitor
913:single-nucleotide polymorphisms
696:, also referred to as class VI
514:University of Wisconsin–Madison
2113:Zeng XC, Wang SX (June 2002).
1974:10.1016/j.virusres.2007.12.007
870:viral nucleic acid polymerases
822:Self-replicating stretches of
46:Crystallographic structure of
35:(RNA-dependent DNA polymerase)
1:
5070:Protein-histidine tele-kinase
5065:Protein-histidine pros-kinase
4944:Glycosyl-1-phosphotransferase
3884:Vesiculovirus matrix proteins
2426:Cold Spring Harbor Laboratory
2132:10.1016/S0014-5793(02)02812-0
1934:10.1016/S0969-2126(97)00261-X
1550:Bio-Medicine.org - Retrovirus
1517:Annual Review of Biochemistry
576:Moloney murine leukemia virus
530:from two RNA tumour viruses:
154:Available protein structures:
4776:RNA-dependent RNA polymerase
2280:10.1097/QAD.0000000000000936
2181:10.1371/journal.pbio.1001102
2081:10.1371/journal.pone.0012271
1872:Journal of Molecular Biology
1801:10.1128/JVI.9.1.116-129.1972
1431:10.1016/0042-6822(90)90430-y
804:DNA-dependent DNA polymerase
724:as a series of these steps:
555:human immunodeficiency virus
4683:RNA-directed DNA polymerase
4551:DNA-directed DNA polymerase
3921:Structure and genome of HIV
2978:(circular partially ds-DNA)
966:The molecular structure of
704:single-stranded RNA with a
578:is a single 75 kDa monomer.
217:RNA-directed DNA polymerase
5146:
5036:: protein-dual-specificity
2554:Herpesvirus glycoprotein B
1653:Kaiser GE (January 2008).
1115:Reverse transcribing virus
1017:
978:
583:avian myeloblastosis virus
27:Enzyme which generates DNA
2549:HHV capsid portal protein
1884:10.1016/j.jmb.2008.10.071
1832:Arnold C (17 July 2014).
1694:10.1016/j.sbi.2020.07.005
1552:Retrieved on 17 Feb, 2009
1476:10.1007/s10529-012-0904-9
1036:(RT-PCR). The classical
1026:polymerase chain reaction
806:activity that copies the
494:polymerase chain reaction
362:
149:
39:
5013:protein-serine/threonine
4913:Phosphatidyltransferases
4502:Thiamine diphosphokinase
2360:Medical Subject Headings
923:and creating artificial
3544:Influenza hemagglutinin
2434:John Coffin (scientist)
2332:10.1126/science.1700865
1655:"The Life Cycle of HIV"
998:(trade name Retrovir),
4833:Nucleotidyltransferase
4516:nucleotidyltransferase
4443:Nucleoside-diphosphate
4106:Gag-onc fusion protein
2218:Nucleic Acids Research
2036:. 1999. Archived from
1737:Molecular cell biology
1130:Retrotransposon marker
1032:in a technique called
971:
710:3' polyadenylated tail
690:
628:
4687:Reverse transcriptase
4076:murine leukemia virus
3967:Reverse transcriptase
3847:Indiana vesiculovirus
3719:Measles hemagglutinin
2371:Molecule of the month
2007:Reverse transcriptase
1682:Curr Opin Struct Biol
1564:Reverse transcriptase
1464:Biotechnology Letters
1018:Further information:
979:Further information:
965:
688:
614:
532:murine leukemia virus
461:at the ends of their
435:reverse transcription
415:reverse transcriptase
33:Reverse transcriptase
18:Reverse-transcriptase
4476:diphosphotransferase
4458:Thiamine-diphosphate
4165:-containing groups (
2807:Early 35 kDa protein
911:Other than creating
884:Replication fidelity
810:cDNA strand into an
623:active site and the
457:cells to extend the
5058:: protein-histidine
4976:; protein acceptor)
4864:mRNA capping enzyme
4856:Guanylyltransferase
2324:1990Sci...250.1227H
2224:(20): 10535–10545.
2072:2010PLoSO...512271H
1838:National Geographic
1789:Journal of Virology
1616:2015NYASA1341..126M
1221:1970Natur.227..561C
607:Function in viruses
591:that maintains the
433:, a process termed
4334:Phosphoinositide 3
4178:phosphotransferase
4071:Rous sarcoma virus
2422:Vincent Racaniello
2230:10.1093/nar/gky910
1624:10.1111/nyas.12668
1279:Biological Reviews
972:
939:Template switching
929:template switching
917:transcript fusions
691:
629:
536:Rous sarcoma virus
463:linear chromosomes
5110:Molecular biology
5087:
5086:
5083:
5082:
4959:
4958:
4898:
4897:
4799:
4798:
4712:Template-directed
4466:
4465:
4433:Phosphomevalonate
4125:
4124:
4121:
4120:
4117:
4116:
4113:
4112:
4060:
4059:
3906:
3905:
3902:
3901:
3898:
3897:
3894:
3893:
3841:
3840:
3837:
3836:
3779:
3778:
3775:
3774:
3731:
3730:
3727:
3726:
3683:
3682:
3679:
3678:
3635:
3634:
3631:
3630:
3587:
3586:
3562:
3561:
3451:
3450:
3447:
3446:
3309:
3308:
3189:
3188:
3185:
3184:
3136:
3135:
3049:
3048:
3045:
3044:
3041:
3040:
2969:
2968:
2956:
2955:
2906:
2905:
2819:
2818:
2815:
2814:
2781:
2780:
2679:
2678:
2675:
2674:
2636:
2635:
2602:
2601:
2562:
2561:
2356:RNA+Transcriptase
2016:978-0-87969-382-4
1748:978-0-7167-4366-8
1573:978-0-87969-382-4
1381:10.1038/2261209a0
1375:(5252): 1209–11.
1338:10.1038/2261211a0
1292:10.1111/brv.12453
1258:Sarkar S (1996).
1176:10.1038/nsmb.1908
1014:Molecular biology
826:genomes known as
739:DNA complementary
486:molecular cloning
411:
410:
407:
406:
310:metabolic pathway
212:
211:
208:
207:
203:structure summary
16:(Redirected from
5137:
5075:Histidine kinase
4998:tyrosine kinases
4988:protein-tyrosine
4980:
4910:
4717:RNA polymerase I
4540:
4531:
4384:Aspartate kinase
4379:Phosphoglycerate
4196:
4152:
4145:
4138:
4129:
4094:
4085:
3926:
3917:
3864:
3852:
3802:
3790:
3785:Zaire ebolavirus
3754:
3742:
3706:
3694:
3658:
3646:
3610:
3598:
3489:
3477:
3468:
3391:3C-like protease
3326:
3221:
3206:
3162:
3088:
3082:Duplornaviricota
3073:
3060:
2982:
2920:
2880:
2868:
2839:
2792:
2701:
2692:
2647:
2613:
2536:
2524:
2515:
2497:
2468:
2461:
2454:
2445:
2394:
2392:
2391:
2344:
2343:
2307:
2301:
2300:
2282:
2258:
2252:
2251:
2241:
2209:
2196:
2195:
2193:
2183:
2159:
2153:
2152:
2134:
2110:
2104:
2103:
2093:
2083:
2051:
2045:
2044:
2042:
2035:
2027:
2021:
2020:
2002:
1996:
1995:
1985:
1953:
1947:
1946:
1936:
1912:
1906:
1905:
1895:
1863:
1854:
1853:
1851:
1849:
1844:on July 18, 2014
1840:. Archived from
1829:
1823:
1822:
1812:
1780:
1774:
1773:
1759:
1753:
1752:
1740:
1730:
1724:
1723:
1705:
1673:
1667:
1666:
1650:
1644:
1643:
1599:
1593:
1592:
1584:
1578:
1577:
1559:
1553:
1547:
1541:
1540:
1512:
1506:
1505:
1487:
1461:
1452:
1443:
1442:
1416:
1407:
1401:
1400:
1364:
1358:
1357:
1332:(5252): 1211–3.
1321:
1315:
1314:
1304:
1294:
1270:
1264:
1263:
1255:
1249:
1248:
1229:10.1038/227561a0
1204:
1198:
1197:
1187:
1155:
1145:
1094:
1089:
1088:
1080:
1075:
1074:
828:retrotransposons
818:In cellular life
664:DNA polymerase δ
566:
425:used to convert
364:
214:
151:
44:
30:
21:
5145:
5144:
5140:
5139:
5138:
5136:
5135:
5134:
5125:1970 in biology
5090:
5089:
5088:
5079:
5050:
5028:
5003:
4974:
4968:
4964:2.7.10-2.7.13:
4955:
4939:
4906:: miscellaneous
4894:
4868:
4850:
4827:
4810:exoribonuclease
4807:
4795:
4781:Polyadenylation
4698:
4524:
4518:
4506:
4488:
4484:
4478:
4462:
4424:
4411:
4388:
4360:
4190:
4184:
4176:
4170:
4156:
4126:
4109:
4056:
4015:transactivators
3997:
3890:
3833:
3771:
3723:
3675:
3627:
3583:
3558:
3529:
3472:Influenza virus
3462:Negarnaviricota
3459:
3457:
3443:
3369:
3317:
3305:
3254:
3215:Kitrinoviricota
3212:
3197:
3195:
3181:
3153:
3132:
3079:
3064:
3037:
3011:
2977:
2965:
2952:
2902:
2863:
2845:
2831:
2825:
2811:
2777:
2756:
2725:
2687:
2686:(Duplodnaviria,
2685:
2684:circular ds-DNA
2671:
2632:
2598:
2558:
2507:
2501:
2486:
2472:
2430:David Baltimore
2389:
2387:
2381:
2352:
2347:
2309:
2308:
2304:
2260:
2259:
2255:
2211:
2210:
2199:
2174:(7): e1001102.
2161:
2160:
2156:
2112:
2111:
2107:
2053:
2052:
2048:
2040:
2033:
2029:
2028:
2024:
2017:
2004:
2003:
1999:
1968:(1–2): 86–103.
1955:
1954:
1950:
1914:
1913:
1909:
1865:
1864:
1857:
1847:
1845:
1831:
1830:
1826:
1782:
1781:
1777:
1761:
1760:
1756:
1749:
1732:
1731:
1727:
1675:
1674:
1670:
1652:
1651:
1647:
1601:
1600:
1596:
1586:
1585:
1581:
1574:
1561:
1560:
1556:
1548:
1544:
1514:
1513:
1509:
1459:
1454:
1453:
1446:
1414:
1409:
1408:
1404:
1366:
1365:
1361:
1323:
1322:
1318:
1272:
1271:
1267:
1257:
1256:
1252:
1215:(5258): 561–3.
1206:
1205:
1201:
1157:
1147:
1146:
1142:
1138:
1090:
1083:
1076:
1069:
1066:
1022:
1016:
983:
977:
975:Antiviral drugs
960:
941:
894:DNA polymerases
886:
866:
840:DNA replication
820:
776:strand transfer
683:
675:drug resistance
657:
609:
558:
544:Renato Dulbecco
524:David Baltimore
506:
498:genome analysis
451:retrotransposon
51:
34:
28:
23:
22:
15:
12:
11:
5:
5143:
5141:
5133:
5132:
5127:
5122:
5117:
5112:
5107:
5102:
5092:
5091:
5085:
5084:
5081:
5080:
5078:
5077:
5072:
5067:
5061:
5059:
5052:
5051:
5049:
5048:
5039:
5037:
5030:
5029:
5027:
5026:
5017:
5015:
5005:
5004:
5002:
5001:
4992:
4990:
4977:
4972:
4966:protein kinase
4961:
4960:
4957:
4956:
4954:
4953:
4947:
4945:
4941:
4940:
4938:
4937:
4932:
4927:
4922:
4916:
4914:
4907:
4900:
4899:
4896:
4895:
4893:
4892:
4887:
4876:
4874:
4870:
4869:
4867:
4866:
4860:
4858:
4852:
4851:
4849:
4848:
4843:
4837:
4835:
4829:
4828:
4826:
4825:
4820:
4814:
4812:
4805:Phosphorolytic
4801:
4800:
4797:
4796:
4794:
4793:
4788:
4783:
4778:
4773:
4772:
4771:
4766:
4761:
4751:
4750:
4749:
4739:
4734:
4729:
4724:
4719:
4714:
4708:
4706:
4704:RNA polymerase
4700:
4699:
4697:
4696:
4695:
4694:
4684:
4680:
4679:
4678:
4677:
4672:
4667:
4656:
4655:
4654:
4649:
4644:
4639:
4628:
4622:
4621:
4620:
4613:
4608:
4603:
4598:
4587:
4586:
4585:
4578:
4573:
4568:
4563:
4552:
4548:
4546:
4544:DNA polymerase
4537:
4528:
4522:
4508:
4507:
4505:
4504:
4499:
4493:
4491:
4486:
4482:
4468:
4467:
4464:
4463:
4461:
4460:
4455:
4450:
4445:
4440:
4435:
4429:
4427:
4422:
4413:
4412:
4410:
4409:
4403:
4401:
4390:
4389:
4387:
4386:
4381:
4375:
4373:
4362:
4361:
4359:
4358:
4353:
4348:
4347:
4346:
4341:
4331:
4329:Diacylglycerol
4326:
4321:
4316:
4311:
4306:
4301:
4296:
4291:
4290:
4289:
4279:
4274:
4269:
4268:
4267:
4262:
4257:
4252:
4247:
4240:Phosphofructo-
4237:
4232:
4231:
4230:
4220:
4215:
4209:
4207:
4193:
4188:
4172:
4171:
4157:
4155:
4154:
4147:
4140:
4132:
4123:
4122:
4119:
4118:
4115:
4114:
4111:
4110:
4108:
4103:
4101:
4091:
4089:Fusion protein
4082:
4081:
4080:
4079:
4078:
4073:
4062:
4061:
4058:
4057:
4055:
4054:
4045:
4040:
4035:
4034:
4033:
4028:
4023:
4010:
4008:
3999:
3998:
3996:
3995:
3994:
3993:
3988:
3976:
3975:
3974:
3972:HIV-1 protease
3969:
3964:
3952:
3951:
3950:
3937:
3935:
3923:
3914:
3908:
3907:
3904:
3903:
3900:
3899:
3896:
3895:
3892:
3891:
3889:
3888:
3887:
3886:
3878:matrix protein
3873:
3871:
3861:
3849:
3843:
3842:
3839:
3838:
3835:
3834:
3832:
3831:
3830:
3829:
3824:
3816:matrix protein
3811:
3809:
3799:
3787:
3781:
3780:
3777:
3776:
3773:
3772:
3770:
3769:
3763:
3761:
3751:
3739:
3733:
3732:
3729:
3728:
3725:
3724:
3722:
3721:
3715:
3713:
3703:
3691:
3685:
3684:
3681:
3680:
3677:
3676:
3674:
3673:
3667:
3665:
3655:
3643:
3637:
3636:
3633:
3632:
3629:
3628:
3626:
3625:
3619:
3617:
3607:
3595:
3589:
3588:
3585:
3584:
3582:
3581:
3575:
3573:
3564:
3563:
3560:
3559:
3557:
3556:
3551:
3546:
3540:
3538:
3531:
3530:
3528:
3527:
3526:
3525:
3517:viral envelope
3513:
3512:
3511:
3503:matrix protein
3498:
3496:
3486:
3474:
3465:
3458:negative-sense
3453:
3452:
3449:
3448:
3445:
3444:
3442:
3441:
3436:
3431:
3426:
3421:
3416:
3411:
3406:
3401:
3396:
3395:
3394:
3382:
3380:
3371:
3370:
3368:
3367:
3362:
3361:
3360:
3355:
3350:
3342:viral envelope
3337:
3335:
3323:
3311:
3310:
3307:
3306:
3304:
3303:
3298:
3293:
3288:
3283:
3278:
3273:
3267:
3265:
3256:
3255:
3253:
3252:
3251:
3250:
3245:
3237:viral envelope
3232:
3230:
3218:
3203:
3196:positive-sense
3191:
3190:
3187:
3186:
3183:
3182:
3180:
3179:
3173:
3171:
3159:
3138:
3137:
3134:
3133:
3131:
3130:
3125:
3120:
3115:
3110:
3105:
3099:
3097:
3085:
3070:
3057:
3051:
3050:
3047:
3046:
3043:
3042:
3039:
3038:
3036:
3035:
3030:
3024:
3022:
3013:
3012:
3010:
3009:
3008:
2998:
2993:
2991:
2979:
2971:
2970:
2967:
2966:
2964:
2963:
2957:
2954:
2953:
2951:
2950:
2940:
2935:
2929:
2927:
2917:
2908:
2907:
2904:
2903:
2901:
2900:
2895:
2889:
2887:
2877:
2865:
2843:Polyomaviridae
2836:
2821:
2820:
2817:
2816:
2813:
2812:
2810:
2809:
2803:
2801:
2789:
2783:
2782:
2779:
2778:
2776:
2775:
2769:
2767:
2758:
2757:
2755:
2754:
2749:
2744:
2738:
2736:
2727:
2726:
2724:
2723:
2718:
2712:
2710:
2698:
2689:
2688:Varidnaviria?)
2681:
2680:
2677:
2676:
2673:
2672:
2670:
2669:
2664:
2658:
2656:
2644:
2638:
2637:
2634:
2633:
2631:
2630:
2624:
2622:
2610:
2604:
2603:
2600:
2599:
2597:
2596:
2591:
2586:
2581:
2575:
2573:
2564:
2563:
2560:
2559:
2557:
2556:
2551:
2545:
2543:
2533:
2521:
2519:Herpes simplex
2512:
2494:
2488:
2487:
2475:Viral proteins
2473:
2471:
2470:
2463:
2456:
2448:
2442:
2441:
2428:to speak with
2416:
2395:
2379:
2374:
2368:
2363:
2351:
2350:External links
2348:
2346:
2345:
2302:
2253:
2197:
2154:
2105:
2046:
2043:on 2006-11-21.
2022:
2015:
1997:
1962:Virus Research
1948:
1927:(8): 1109–22.
1907:
1878:(3): 693–713.
1855:
1824:
1775:
1754:
1747:
1725:
1668:
1665:on 2010-07-26.
1645:
1594:
1579:
1572:
1554:
1542:
1523:(1): 493–517.
1507:
1470:(7): 1209–15.
1444:
1402:
1359:
1316:
1285:(1): 259–282.
1265:
1250:
1199:
1170:(10): 1202–9.
1139:
1137:
1134:
1133:
1132:
1127:
1122:
1120:RNA polymerase
1117:
1112:
1107:
1105:DNA polymerase
1102:
1096:
1095:
1092:Viruses portal
1081:
1078:Biology portal
1065:
1062:
1050:cDNA libraries
1015:
1012:
976:
973:
959:
956:
940:
937:
921:exon shuffling
885:
882:
878:DNA polymerase
865:
862:
819:
816:
793:DNA polymerase
772:
771:
764:
761:
758:
755:
752:
749:
742:
735:
702:positive-sense
682:
679:
668:primer removal
656:
653:
649:hepadnaviruses
647:, such as the
608:
605:
604:
603:
586:
579:
572:
505:
502:
490:RNA sequencing
478:ribonuclease H
474:DNA polymerase
409:
408:
405:
404:
399:
393:
392:
387:
381:
380:
375:
369:
368:
360:
359:
350:
344:
343:
332:
325:
324:
319:
313:
312:
307:
301:
300:
295:
289:
288:
283:
277:
276:
271:
265:
264:
259:
253:
252:
248:
247:
242:
236:
235:
230:
224:
223:
219:
218:
210:
209:
206:
205:
200:
194:
193:
180:
174:
173:
163:
156:
155:
147:
146:
141:
135:
134:
121:
115:
114:
109:
103:
102:
97:
91:
90:
85:
78:
77:
72:
66:
65:
62:
58:
57:
53:
52:
45:
37:
36:
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
5142:
5131:
5128:
5126:
5123:
5121:
5118:
5116:
5115:Viral enzymes
5113:
5111:
5108:
5106:
5103:
5101:
5098:
5097:
5095:
5076:
5073:
5071:
5068:
5066:
5063:
5062:
5060:
5057:
5053:
5047:
5046:
5041:
5040:
5038:
5035:
5031:
5025:
5024:
5019:
5018:
5016:
5014:
5010:
5006:
5000:
4999:
4994:
4993:
4991:
4989:
4985:
4981:
4978:
4975:
4967:
4962:
4952:
4949:
4948:
4946:
4942:
4936:
4933:
4931:
4928:
4926:
4923:
4921:
4918:
4917:
4915:
4911:
4908:
4905:
4901:
4891:
4888:
4885:
4881:
4878:
4877:
4875:
4871:
4865:
4862:
4861:
4859:
4857:
4853:
4847:
4844:
4842:
4839:
4838:
4836:
4834:
4830:
4824:
4821:
4819:
4816:
4815:
4813:
4811:
4806:
4802:
4792:
4789:
4787:
4784:
4782:
4779:
4777:
4774:
4770:
4767:
4765:
4762:
4760:
4757:
4756:
4755:
4752:
4748:
4745:
4744:
4743:
4740:
4738:
4735:
4733:
4730:
4728:
4725:
4723:
4720:
4718:
4715:
4713:
4710:
4709:
4707:
4705:
4701:
4693:
4690:
4689:
4688:
4685:
4682:
4681:
4676:
4673:
4671:
4668:
4666:
4663:
4662:
4660:
4657:
4653:
4650:
4648:
4645:
4643:
4640:
4638:
4635:
4634:
4632:
4629:
4626:
4623:
4619:
4618:
4614:
4612:
4609:
4607:
4604:
4602:
4599:
4597:
4594:
4593:
4591:
4588:
4584:
4583:
4579:
4577:
4574:
4572:
4569:
4567:
4564:
4562:
4559:
4558:
4556:
4553:
4550:
4549:
4547:
4545:
4541:
4538:
4536:
4532:
4529:
4526:
4517:
4513:
4509:
4503:
4500:
4498:
4495:
4494:
4492:
4489:
4477:
4473:
4469:
4459:
4456:
4454:
4451:
4449:
4446:
4444:
4441:
4439:
4436:
4434:
4431:
4430:
4428:
4425:
4418:
4414:
4408:
4405:
4404:
4402:
4399:
4395:
4391:
4385:
4382:
4380:
4377:
4376:
4374:
4371:
4367:
4363:
4357:
4354:
4352:
4349:
4345:
4344:Class II PI 3
4342:
4340:
4337:
4336:
4335:
4332:
4330:
4327:
4325:
4322:
4320:
4319:Deoxycytidine
4317:
4315:
4312:
4310:
4307:
4305:
4302:
4300:
4297:
4295:
4292:
4288:
4287:ADP-thymidine
4285:
4284:
4283:
4280:
4278:
4275:
4273:
4270:
4266:
4263:
4261:
4258:
4256:
4253:
4251:
4248:
4246:
4243:
4242:
4241:
4238:
4236:
4233:
4229:
4226:
4225:
4224:
4221:
4219:
4216:
4214:
4211:
4210:
4208:
4205:
4201:
4197:
4194:
4191:
4183:
4179:
4173:
4168:
4164:
4160:
4153:
4148:
4146:
4141:
4139:
4134:
4133:
4130:
4107:
4104:
4102:
4099:
4095:
4092:
4090:
4086:
4083:
4077:
4074:
4072:
4069:
4068:
4067:
4066:
4063:
4053:
4049:
4046:
4044:
4041:
4039:
4036:
4032:
4029:
4027:
4024:
4022:
4019:
4018:
4017:
4016:
4012:
4011:
4009:
4007:
4005:
4000:
3992:
3989:
3987:
3984:
3983:
3982:
3981:
3977:
3973:
3970:
3968:
3965:
3963:
3960:
3959:
3958:
3957:
3953:
3949:
3946:
3945:
3944:
3943:
3939:
3938:
3936:
3934:
3932:
3927:
3924:
3922:
3918:
3915:
3913:
3909:
3885:
3882:
3881:
3880:
3879:
3875:
3874:
3872:
3869:
3865:
3862:
3860:
3858:
3853:
3850:
3848:
3844:
3828:
3825:
3823:
3820:
3819:
3818:
3817:
3813:
3812:
3810:
3807:
3803:
3800:
3798:
3796:
3791:
3788:
3786:
3782:
3768:
3765:
3764:
3762:
3759:
3755:
3752:
3750:
3748:
3743:
3740:
3738:
3734:
3720:
3717:
3716:
3714:
3711:
3707:
3704:
3702:
3700:
3695:
3692:
3690:
3686:
3672:
3669:
3668:
3666:
3663:
3659:
3656:
3654:
3652:
3647:
3644:
3642:
3638:
3624:
3621:
3620:
3618:
3615:
3611:
3608:
3606:
3604:
3599:
3596:
3594:
3593:Parainfluenza
3590:
3580:
3577:
3576:
3574:
3572:
3570:
3565:
3555:
3552:
3550:
3549:Neuraminidase
3547:
3545:
3542:
3541:
3539:
3536:
3532:
3524:
3521:
3520:
3519:
3518:
3514:
3510:
3507:
3506:
3505:
3504:
3500:
3499:
3497:
3494:
3490:
3487:
3485:
3483:
3478:
3475:
3473:
3469:
3466:
3463:
3454:
3440:
3437:
3435:
3432:
3430:
3427:
3425:
3422:
3420:
3417:
3415:
3412:
3410:
3407:
3405:
3402:
3400:
3397:
3392:
3389:
3388:
3387:
3384:
3383:
3381:
3379:
3377:
3372:
3366:
3363:
3359:
3356:
3354:
3351:
3349:
3346:
3345:
3344:
3343:
3339:
3338:
3336:
3334:
3332:
3327:
3324:
3321:
3316:
3312:
3302:
3299:
3297:
3294:
3292:
3289:
3287:
3284:
3282:
3279:
3277:
3274:
3272:
3269:
3268:
3266:
3264:
3262:
3257:
3249:
3246:
3244:
3241:
3240:
3239:
3238:
3234:
3233:
3231:
3229:
3227:
3222:
3219:
3216:
3211:
3207:
3204:
3201:
3192:
3178:
3175:
3174:
3172:
3170:
3168:
3163:
3160:
3157:
3151:
3147:
3143:
3139:
3129:
3126:
3124:
3121:
3119:
3116:
3114:
3111:
3109:
3106:
3104:
3101:
3100:
3098:
3096:
3094:
3089:
3086:
3083:
3078:
3074:
3071:
3068:
3061:
3058:
3056:
3052:
3034:
3031:
3029:
3026:
3025:
3023:
3021:
3019:
3014:
3007:
3004:
3003:
3002:
2999:
2997:
2994:
2992:
2990:
2988:
2983:
2980:
2976:
2972:
2962:
2959:
2958:
2948:
2944:
2941:
2939:
2936:
2934:
2931:
2930:
2928:
2925:
2921:
2918:
2916:
2914:
2909:
2899:
2896:
2894:
2891:
2890:
2888:
2885:
2881:
2878:
2876:
2874:
2869:
2866:
2861:
2857:
2853:
2849:
2844:
2840:
2837:
2834:
2829:
2822:
2808:
2805:
2804:
2802:
2800:
2798:
2793:
2790:
2788:
2787:Baculoviridae
2784:
2774:
2771:
2770:
2768:
2766:
2764:
2759:
2753:
2750:
2748:
2745:
2743:
2740:
2739:
2737:
2735:
2733:
2728:
2722:
2719:
2717:
2714:
2713:
2711:
2709:
2707:
2702:
2699:
2697:
2693:
2690:
2682:
2668:
2665:
2663:
2660:
2659:
2657:
2655:
2653:
2648:
2645:
2643:
2639:
2629:
2626:
2625:
2623:
2621:
2619:
2614:
2611:
2609:
2605:
2595:
2592:
2590:
2587:
2585:
2582:
2580:
2577:
2576:
2574:
2572:
2570:
2565:
2555:
2552:
2550:
2547:
2546:
2544:
2541:
2537:
2534:
2532:
2530:
2525:
2522:
2520:
2516:
2513:
2510:
2505:
2504:Duplodnaviria
2500:linear ds-DNA
2498:
2495:
2493:
2489:
2484:
2480:
2476:
2469:
2464:
2462:
2457:
2455:
2450:
2449:
2446:
2439:
2438:Harold Varmus
2435:
2431:
2427:
2423:
2420:
2417:
2414:
2410:
2409:
2404:
2400:
2396:
2385:
2382:Goodsell DS.
2380:
2378:
2375:
2372:
2369:
2367:
2364:
2361:
2357:
2354:
2353:
2349:
2341:
2337:
2333:
2329:
2325:
2321:
2317:
2313:
2306:
2303:
2298:
2294:
2290:
2286:
2281:
2276:
2273:(2): 185–92.
2272:
2268:
2264:
2257:
2254:
2249:
2245:
2240:
2235:
2231:
2227:
2223:
2219:
2215:
2208:
2206:
2204:
2202:
2198:
2192:
2187:
2182:
2177:
2173:
2169:
2165:
2158:
2155:
2150:
2146:
2142:
2138:
2133:
2128:
2124:
2120:
2116:
2109:
2106:
2101:
2097:
2092:
2087:
2082:
2077:
2073:
2069:
2066:(8): e12271.
2065:
2061:
2057:
2050:
2047:
2039:
2032:
2026:
2023:
2018:
2012:
2008:
2001:
1998:
1993:
1989:
1984:
1979:
1975:
1971:
1967:
1963:
1959:
1952:
1949:
1944:
1940:
1935:
1930:
1926:
1922:
1918:
1911:
1908:
1903:
1899:
1894:
1889:
1885:
1881:
1877:
1873:
1869:
1862:
1860:
1856:
1843:
1839:
1835:
1828:
1825:
1820:
1816:
1811:
1806:
1802:
1798:
1795:(1): 116–29.
1794:
1790:
1786:
1779:
1776:
1771:
1767:
1766:
1758:
1755:
1750:
1744:
1739:
1738:
1729:
1726:
1721:
1717:
1713:
1709:
1704:
1699:
1695:
1691:
1687:
1683:
1679:
1672:
1669:
1664:
1660:
1656:
1649:
1646:
1641:
1637:
1633:
1629:
1625:
1621:
1617:
1613:
1610:(1): 126–35.
1609:
1605:
1598:
1595:
1590:
1583:
1580:
1575:
1569:
1565:
1558:
1555:
1551:
1546:
1543:
1538:
1534:
1530:
1526:
1522:
1518:
1511:
1508:
1503:
1499:
1495:
1491:
1486:
1481:
1477:
1473:
1469:
1465:
1458:
1451:
1449:
1445:
1440:
1436:
1432:
1428:
1425:(2): 456–64.
1424:
1420:
1413:
1406:
1403:
1398:
1394:
1390:
1386:
1382:
1378:
1374:
1370:
1363:
1360:
1355:
1351:
1347:
1343:
1339:
1335:
1331:
1327:
1320:
1317:
1312:
1308:
1303:
1298:
1293:
1288:
1284:
1280:
1276:
1269:
1266:
1261:
1254:
1251:
1246:
1242:
1238:
1234:
1230:
1226:
1222:
1218:
1214:
1210:
1203:
1200:
1195:
1191:
1186:
1181:
1177:
1173:
1169:
1165:
1161:
1154:
1150:
1144:
1141:
1135:
1131:
1128:
1126:
1123:
1121:
1118:
1116:
1113:
1111:
1108:
1106:
1103:
1101:
1098:
1097:
1093:
1087:
1082:
1079:
1073:
1068:
1063:
1061:
1059:
1055:
1051:
1047:
1043:
1039:
1035:
1031:
1028:technique to
1027:
1021:
1013:
1011:
1009:
1005:
1002:(Epivir) and
1001:
997:
993:
988:
982:
974:
969:
964:
957:
955:
953:
949:
946:
938:
936:
934:
930:
926:
922:
918:
914:
909:
907:
903:
899:
895:
890:
883:
881:
879:
875:
871:
863:
861:
858:
856:
852:
848:
843:
841:
837:
833:
829:
825:
817:
815:
813:
809:
805:
801:
796:
794:
789:
784:
779:
777:
769:
765:
762:
759:
756:
753:
750:
747:
743:
740:
736:
733:
730:
727:
726:
725:
723:
719:
715:
711:
707:
703:
699:
695:
687:
680:
678:
676:
671:
669:
665:
660:
654:
652:
650:
646:
642:
638:
634:
626:
622:
618:
613:
606:
601:
598:
594:
590:
587:
584:
580:
577:
573:
570:
565:
561:
556:
552:
551:
550:
547:
545:
541:
537:
533:
529:
525:
521:
520:
515:
511:
503:
501:
499:
495:
491:
487:
483:
482:transcription
479:
475:
470:
468:
467:central dogma
464:
460:
456:
452:
448:
444:
440:
436:
432:
428:
424:
420:
416:
403:
400:
398:
394:
391:
388:
386:
382:
379:
376:
374:
370:
365:
361:
358:
354:
351:
349:
348:Gene Ontology
345:
342:
339:
336:
333:
330:
326:
323:
320:
318:
314:
311:
308:
306:
302:
299:
296:
294:
290:
287:
286:NiceZyme view
284:
282:
278:
275:
272:
270:
266:
263:
260:
258:
254:
249:
246:
243:
241:
237:
234:
231:
229:
225:
220:
215:
204:
201:
199:
195:
192:
188:
184:
181:
179:
175:
171:
167:
164:
161:
157:
152:
148:
145:
142:
140:
136:
133:
129:
125:
122:
120:
116:
113:
110:
108:
104:
101:
98:
96:
92:
89:
86:
83:
79:
76:
73:
71:
67:
63:
59:
54:
49:
43:
38:
31:
19:
5042:
5020:
4995:
4686:
4616:
4581:
4339:Class I PI 3
4304:Pantothenate
4175:2.7.1-2.7.4:
4159:Transferases
4013:
4002:
3978:
3966:
3954:
3940:
3929:
3876:
3855:
3814:
3793:
3758:glycoprotein
3745:
3710:glycoprotein
3697:
3662:glycoprotein
3649:
3614:glycoprotein
3601:
3567:
3535:glycoprotein
3515:
3501:
3480:
3374:
3365:Nucleocapsid
3340:
3329:
3320:Pisuviricota
3259:
3235:
3224:
3165:
3156:Pisuviricota
3091:
3016:
2985:
2911:
2871:
2833:Monodnaviria
2795:
2761:
2730:
2704:
2696:Epstein–Barr
2650:
2642:Adenoviridae
2616:
2567:
2527:
2509:Varidnaviria
2406:
2388:. Retrieved
2315:
2311:
2305:
2270:
2266:
2256:
2221:
2217:
2171:
2168:PLOS Biology
2167:
2157:
2122:
2119:FEBS Letters
2118:
2108:
2063:
2059:
2049:
2038:the original
2025:
2006:
2000:
1965:
1961:
1951:
1924:
1920:
1910:
1875:
1871:
1846:. Retrieved
1842:the original
1837:
1827:
1792:
1788:
1778:
1769:
1763:
1757:
1736:
1728:
1685:
1681:
1671:
1663:the original
1658:
1648:
1607:
1603:
1597:
1588:
1582:
1563:
1557:
1545:
1520:
1516:
1510:
1467:
1463:
1422:
1418:
1405:
1372:
1368:
1362:
1329:
1325:
1319:
1282:
1278:
1268:
1259:
1253:
1212:
1208:
1202:
1167:
1163:
1143:
1100:cDNA library
1023:
1010:(Viramune).
984:
958:Applications
942:
932:
928:
910:
898:proofreading
896:, it has no
891:
887:
867:
859:
844:
821:
811:
800:ribonuclease
797:
780:
775:
773:
694:Retroviruses
692:
672:
661:
658:
637:retroviruses
630:
548:
519:Rous sarcoma
517:
510:Howard Temin
507:
471:
434:
418:
414:
412:
274:BRENDA entry
4890:Transposase
4880:Recombinase
4525:-nucleoside
4351:Sphingosine
4098:oncoprotein
3210:Hepatitis C
2975:Hepatitis B
2961:Agnoprotein
2924:oncoprotein
2898:VP2 and VP3
2424:travels to
1688:: 175–183.
1485:2433/157247
788:nucleotides
645:DNA viruses
633:RNA viruses
617:amino acids
600:chromosomes
526:in 1970 at
447:hepatitis B
262:IntEnz view
222:Identifiers
56:Identifiers
5094:Categories
4692:Telomerase
4535:Polymerase
4309:Mevalonate
4272:Riboflavin
4163:phosphorus
3523:M2 protein
3509:M1 protein
3315:SARS-CoV-2
2390:2013-01-13
1772:: 734–737.
1136:References
1125:Telomerase
1008:nevirapine
1000:lamivudine
996:zidovudine
968:zidovudine
832:Telomerase
824:eukaryotic
635:, such as
625:polymerase
597:eukaryotic
534:and again
496:(PCR), or
476:activity,
455:eukaryotic
427:RNA genome
331:structures
298:KEGG entry
245:9068-38-6
166:structures
5120:Telomeres
4884:Integrase
4808:3' to 5'
4453:Guanylate
4448:Uridylate
4438:Adenylate
4282:Thymidine
4277:Shikimate
4065:Multiple
3962:Integrase
3200:Riboviria
3077:Rotavirus
3067:Riboviria
2828:Riboviria
1921:Structure
1720:221636459
1502:207096569
1156:;
1004:tenofovir
925:antisense
902:mutations
864:Structure
847:Beljanski
812:antisense
768:integrase
593:telomeres
459:telomeres
251:Databases
100:IPR000477
5105:EC 2.7.7
5100:Genetics
4818:RNase PH
4426:acceptor
4407:Creatine
4400:acceptor
4372:acceptor
4314:Pyruvate
4299:Glycerol
4260:Platelet
4235:Galacto-
4206:acceptor
3358:Membrane
3353:Envelope
2947:SV40 Tag
2608:Vaccinia
2297:20086739
2289:26691546
2248:30307534
2149:24619868
2141:12044895
2100:20805885
2060:PLOS ONE
1992:18261820
1902:19022262
1712:32916568
1640:42378727
1632:25703292
1537:16756500
1494:22426840
1419:Virology
1311:67861162
1194:20852643
1064:See also
952:provirus
698:ssRNA-RT
557:type 1 (
441:such as
421:) is an
402:proteins
390:articles
378:articles
335:RCSB PDB
233:2.7.7.49
183:RCSB PDB
95:InterPro
4769:PrimPol
4754:Primase
4228:Hepatic
4223:Fructo-
3689:Measles
3142:Rhinov.
2589:ICP34.5
2413:PDBe-KB
2403:UniProt
2340:1700865
2320:Bibcode
2312:Science
2239:6237782
2191:3134445
2091:2923612
2068:Bibcode
1983:2464458
1943:9309225
1893:2881421
1819:4333538
1703:9973426
1612:Bibcode
1439:1691562
1397:4222378
1389:4316300
1354:4187764
1346:4316301
1302:6378602
1245:4164029
1237:4913914
1217:Bibcode
1185:2987654
1058:enzymes
948:genomes
933:in vivo
906:Promega
874:RNase H
851:Retrons
746:RNAse H
722:cytosol
641:genomes
621:RNase H
619:of the
512:at the
504:History
439:viruses
357:QuickGO
322:profile
305:MetaCyc
240:CAS no.
144:cd00304
112:PS50878
107:PROSITE
75:PF00078
5056:2.7.13
5034:2.7.12
5009:2.7.11
4984:2.7.10
4823:PNPase
4791:PNPase
4747:POLRMT
4742:ssRNAP
4255:Muscle
4218:Gluco-
4182:kinase
3868:capsid
3806:capsid
3554:HA-tag
3493:capsid
3456:ss-RNA
3386:ORF1ab
3194:ss-RNA
3154:etc. (
3063:ds-RNA
2884:capsid
2752:EBNA-3
2747:EBNA-2
2742:EBNA-1
2540:capsid
2436:, and
2408:P03366
2362:(MeSH)
2338:
2295:
2287:
2246:
2236:
2188:
2147:
2139:
2098:
2088:
2013:
1990:
1980:
1941:
1900:
1890:
1848:29 May
1817:
1810:356270
1807:
1745:
1718:
1710:
1700:
1638:
1630:
1570:
1535:
1500:
1492:
1437:
1395:
1387:
1369:Nature
1352:
1344:
1326:Nature
1309:
1299:
1243:
1235:
1209:Nature
1192:
1182:
783:primer
706:5' cap
542:(with
423:enzyme
385:PubMed
367:Search
353:AmiGO
341:PDBsum
281:ExPASy
269:BRENDA
257:IntEnz
228:EC no.
198:PDBsum
172:
162:
132:SUPFAM
88:CL0027
61:Symbol
4904:2.7.8
4873:Other
4512:2.7.7
4472:2.7.6
4417:2.7.4
4394:2.7.3
4366:2.7.2
4250:Liver
4213:Hexo-
4200:2.7.1
4004:VRAPs
3986:gp120
3641:Mumps
3439:ORF9b
3429:ORF7b
3424:ORF7a
3414:ORF3d
3409:ORF3c
3404:ORF3b
3399:ORF3a
3393:(NS5)
3348:Spike
3150:Hep A
3146:Polio
3006:HBeAg
3001:HBcAg
2996:HBsAg
2860:HaPyV
2856:MCPyV
2824:other
2763:ncRNA
2721:LMP-2
2716:LMP-1
2594:ICP47
2579:vmw65
2479:early
2293:S2CID
2145:S2CID
2041:(PDF)
2034:(PDF)
1716:S2CID
1636:S2CID
1498:S2CID
1460:(PDF)
1415:(PDF)
1393:S2CID
1350:S2CID
1307:S2CID
1241:S2CID
1110:msDNA
1052:from
855:msDNA
808:sense
729:Lysyl
317:PRIAM
128:SCOPe
119:SCOP2
64:RVT_1
5043:see
5021:see
4996:see
4370:COOH
4169:2.7)
3991:gp41
3931:VSPs
3857:VSPs
3827:VP24
3822:VP40
3795:VSPs
3747:VSPs
3699:VSPs
3651:VSPs
3603:VSPs
3569:VNPs
3482:VSPs
3434:ORF8
3419:ORF6
3376:VNPs
3331:VSPs
3301:NS5B
3296:NS5A
3291:NS4B
3286:NS4A
3261:VNPs
3226:VSPs
3167:VNPs
3128:NSP6
3123:NSP5
3118:NSP4
3113:NSP3
3108:NSP2
3103:NSP1
3093:VNPs
3018:VNPs
2987:VSPs
2943:LTag
2938:MTag
2933:STag
2913:VNPs
2873:VSPs
2852:MPyV
2848:SV40
2797:VNPs
2773:EBER
2732:VNPs
2706:VSPs
2652:VNPs
2628:B13R
2618:VNPs
2584:ICP8
2569:VNPs
2529:VSPs
2483:late
2481:and
2401:for
2336:PMID
2285:PMID
2267:AIDS
2244:PMID
2137:PMID
2096:PMID
2011:ISBN
1988:PMID
1939:PMID
1898:PMID
1850:2016
1815:PMID
1743:ISBN
1708:PMID
1628:PMID
1608:1341
1568:ISBN
1533:PMID
1490:PMID
1435:PMID
1385:PMID
1342:PMID
1233:PMID
1190:PMID
1153:3KLF
1054:mRNA
943:Two
732:tRNA
718:HTLV
708:and
569:kDas
564:1HMV
445:and
397:NCBI
338:PDBe
293:KEGG
191:PDBj
187:PDBe
170:ECOD
160:Pfam
124:1hmv
84:clan
82:Pfam
70:Pfam
4786:PAP
4727:III
4661:/Y
4652:TDT
4633:/X
4625:III
4617:Pfu
4592:/B
4582:Taq
4557:/A
4324:PFP
4294:NAD
4052:Vpx
4050:or
4048:Vpu
4043:Vif
4038:Nef
4031:Vpr
4026:Rev
4021:Tat
3980:env
3956:pol
3948:p24
3942:gag
3737:RSV
3579:NS1
3281:NS3
3276:NS2
3177:VPg
3055:RNA
3028:HBx
2893:VP1
2667:E1B
2662:E1A
2492:DNA
2399:PDB
2328:doi
2316:250
2275:doi
2234:PMC
2226:doi
2186:PMC
2176:doi
2127:doi
2123:520
2086:PMC
2076:doi
1978:PMC
1970:doi
1966:134
1929:doi
1888:PMC
1880:doi
1876:385
1805:PMC
1797:doi
1770:233
1698:PMC
1690:doi
1620:doi
1525:doi
1480:hdl
1472:doi
1427:doi
1423:175
1377:doi
1373:226
1334:doi
1330:226
1297:PMC
1287:doi
1225:doi
1213:227
1180:PMC
1172:doi
1149:PDB
1046:PCR
1042:DNA
1038:PCR
1030:RNA
987:HIV
985:As
945:RNA
836:RNA
714:HIV
595:of
560:PDB
546:).
528:MIT
516:in
443:HIV
431:DNA
429:to
373:PMC
329:PDB
178:PDB
139:CDD
48:HIV
5096::
5011::
4986::
4971:PO
4732:IV
4722:II
4631:IV
4627:/C
4590:II
4576:T7
4521:PO
4514::
4474::
4421:PO
4419::
4396::
4368::
4204:OH
4202::
4187:PO
4167:EC
4161::
3912:RT
3271:P7
3248:E2
3243:E1
3148:,
3144:,
2858:,
2854:,
2850:,
2432:,
2405::
2334:.
2326:.
2314:.
2291:.
2283:.
2271:30
2269:.
2265:.
2242:.
2232:.
2222:46
2220:.
2216:.
2200:^
2184:.
2170:.
2166:.
2143:.
2135:.
2121:.
2117:.
2094:.
2084:.
2074:.
2062:.
2058:.
1986:.
1976:.
1964:.
1960:.
1937:.
1923:.
1919:.
1896:.
1886:.
1874:.
1870:.
1858:^
1836:.
1813:.
1803:.
1791:.
1787:.
1768:.
1714:.
1706:.
1696:.
1686:65
1684:.
1680:.
1657:.
1634:.
1626:.
1618:.
1606:.
1531:.
1521:75
1519:.
1496:.
1488:.
1478:.
1468:34
1466:.
1462:.
1447:^
1433:.
1421:.
1417:.
1391:.
1383:.
1371:.
1348:.
1340:.
1328:.
1305:.
1295:.
1283:94
1281:.
1277:.
1239:.
1231:.
1223:.
1211:.
1188:.
1178:.
1168:17
1166:.
1162:.
1151::
919:,
842:.
677:.
562::
500:.
492:,
488:,
419:RT
413:A
355:/
189:;
185:;
168:/
130:/
126:/
4973:4
4969:(
4886:)
4882:(
4764:2
4759:1
4737:V
4675:κ
4670:ι
4665:η
4659:V
4647:μ
4642:λ
4637:β
4611:ζ
4606:ε
4601:δ
4596:α
4571:ν
4566:θ
4561:γ
4555:I
4527:)
4523:4
4519:(
4490:)
4487:7
4485:O
4483:2
4481:P
4479:(
4423:4
4398:N
4265:2
4245:1
4192:)
4189:4
4185:(
4180:/
4151:e
4144:t
4137:v
4100::
4006::
3933::
3870::
3859::
3808::
3797::
3760::
3749::
3712::
3701::
3664::
3653::
3616::
3605::
3571::
3537::
3495::
3484::
3464:)
3460:(
3378::
3333::
3322:)
3318:(
3263::
3228::
3217:)
3213:(
3202:)
3198:(
3169::
3158:)
3152:,
3095::
3084:)
3080:(
3069:)
3065:(
3020::
2989::
2949:)
2945:(
2926::
2915::
2886::
2875::
2862:)
2846:(
2835:)
2830:,
2826:(
2799::
2765::
2734::
2708::
2654::
2620::
2571::
2542::
2531::
2511:)
2506:,
2502:(
2485:)
2477:(
2467:e
2460:t
2453:v
2415:.
2393:.
2342:.
2330::
2322::
2299:.
2277::
2250:.
2228::
2194:.
2178::
2172:9
2151:.
2129::
2102:.
2078::
2070::
2064:5
2019:.
1994:.
1972::
1945:.
1931::
1925:5
1904:.
1882::
1852:.
1821:.
1799::
1793:9
1751:.
1722:.
1692::
1642:.
1622::
1614::
1576:.
1539:.
1527::
1504:.
1482::
1474::
1441:.
1429::
1399:.
1379::
1356:.
1336::
1313:.
1289::
1247:.
1227::
1219::
1196:.
1174::
770:.
602:.
571:.
417:(
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.