382:(a Crenarchaeota TopR2 reverse gyrase) initiates an unwinding of approximately 20 base pairs upon binding to a DNA structure. Upon initial binding to the DNA, the helicase domain is in an open conformation, while the topoisomerase IA domain is in a closed conformation. After the binding of ATP to the reverse gyrase structure, the helicase domain closes, and the topoisomerase IA domain opens. This triggers a rewinding of 10 of the 20 base pairs in the unwound bubble, and the topoisomerase IA domain can introduce positive supercoiling during strand passage. As the strand passage occurs, reverse gyrase's topoisomerase IA domain is able to increase the linking number (how many times a strand of DNA is wrapped around the other strand) of the DNA strand as they are renatured. Following ATP hydrolysis-induced rewinding, the reverse gyrase enzyme domains return to their original state (open helicase domain and closed topoisomerase IA domain) and the reverse gyrase is released, ready to bind to a new region of DNA and repeat the process.
26:
229:. These positive supercoils can be introduced to DNA that is either negatively supercoiled or fully relaxed. Where DNA gyrase forms a tetramer and is capable of cleaving a double-stranded region of DNA, reverse gyrase can only cleave single stranded DNA. More specifically, reverse gyrase is a member of the type IA topoisomerase class; along with the ability to relax negatively or positively supercoiled DNA (which does not require ATP), type IA enzymes also tend to have RNA-topoisomerase activities. These RNA topoisomerases help keep longer RNA strands from becoming tangled in what are referred to as "pseudoknots." Due to their ability to interact with RNA, it is thought that this is one of the most ancient class of enzymes found to date.
347:
334:(organisms that can live in temperatures ranging from 40 °C up to as high as 122 °C) are thought to maintain several positive supercoils in their DNA in order to assist with maintaining structural integrity of the DNA under the denaturing capabilities of these high temperatures. Positive supercoiling, which is referred to as overwinding, results in the clockwise twisting of the strand. As previously discovered, one of the biggest benefits to maintaining positive supercoils in DNA strands is preventing separation of the strands in high temperatures.
279:
263:
under the topoisomerase umbrella. Furthermore, the 5.6 number designates this molecule as an isomerase that is capable of changing conformation in cellular molecules. 5.6.2 designates the enzyme further as being capable of altering nucleic acid, or DNA, conformations. Lastly, the full designation of 5.6.2.2 characterizes this enzyme as an ATP-dependent DNA topoisomerase.
337:
While positive supercoiling is certainly more common in thermophiles, positive supercoiling has been found in mesophilic organisms. For example, telomeres and condensins can both utilize positive supercoiling as a means for contributing to chromosomal structure. Furthermore, the reverse gyrase enzyme
249:
reverse gyrase being one of the first to be characterized. Additionally, it has been found that all thermophilic bacteria and archaea contain at least one reverse gyrase enzyme. Some organisms, such as members of the
Crenarchaeota phylum, even have two reverse gyrase enzymes: TopR1, which tends to be
241:
domain, which is responsible for the actual introduction of coils into DNA. However, mechanistic studies have shown that these two domains tend to exhibit weak activities separately and can only perform efficient DNA positive supercoiling activity when working in tandem. Other studies have also shown
309:
The reverse gyrase enzyme contains a zinc finger domain, where two zinc ions help to coordinate enzymatic function. The first zinc ion is kept in place by interactions with four cysteine residues. The second zinc ion is not always found in reverse gyrase enzymes. However, when present, both ions are
262:
As seen in the information box above, reverse gyrase is designated under the EC number 5.6.2.2. The first number of this code (5) designates the enzymes identity as an isomerase. While the enzyme itself does have both a topoisomerase and helicase-like domain, as a gyrase, it is primarily classified
291:
Reverse gyrases have helicase and topoisomerase domains. The active site, where nucleotides are bound by the enzyme, is characterized by Asp78, Phe75, Gln83, Lys106, Asp203, and Thr107 residues. It is hypothesized that the H1 and H2 subdomains also contain nucleotide-binding abilities, and the DNA
300:
The latch domain appears to be variable across species, with domain size ranging from as small as 10 amino acids to as large as 120 amino acids. The latch is thought to function as a control mechanism to prevent the topoisomerase domain from creating negative supercoils and relaxing the DNA, and
326:
in their DNA strands. This helps to condense the genetic material so that it fits within the host cells (or in the case of eukaryotes, within the cell's nuclear region). Negative supercoiling, also referred to as underwinding, results in the counterclockwise twisting of the DNA strand. Negative
242:
that reverse gyrase enzymes tend to favorably attack regions of single-stranded DNA versus double-stranded DNA, which suggests that this enzyme's critical biological function is to ensure the constant renaturation of melted DNA strands, especially in organisms that grow at high temperatures.
232:
Reverse gyrase is an ATP-dependent topoisomerase in terms of its positive supercoiling activity, however, reverse gyrase can also relax DNA strands without introducing positive supercoils through interaction with ADP. The structure of the enzyme includes both a
354:
It is suspected that the helicase and topoisomerase domains of the reverse gyrase enzyme work together to promote positive supercoiling in DNA. However, the exact mechanisms of action appear to differ between organisms. For example,
459:
Fogg JM, Catanese DJ, Randall GL, Swick MC, Zechiedrich L (2009). Benham CJ, Harvey S, Olson WK, Sumners WL, Swigon D (eds.). "Differences
Between Positively and Negatively Supercoiled DNA that Topoisomerases May Distinguish".
327:
supercoiling leaves the DNA strands available for various cellular processes, like genome replication and transcription, as DNA typically needs to be underwound in order to be denatured and accessed by the proper enzymes.
338:
is not exclusive to thermophiles. Some reverse gyrase enzymes even function outside of thermophilic temperature ranges, suggesting that there may be some organisms at mesophilic temperatures that utilize this enzyme.
310:
found near the binding site for nucleic acids. It is thought that these zinc fingers play a role in initial binding of DNA and strand passage, but their exact mechanisms of action appear to vary between organisms.
385:
Regardless of the differences in interactions between the topoisomerase and helicase domains, in general, reverse gyrase enzymes all undergo conformational changes when nucleotides are bound to the active site.
378:
When bound to the DNA, reverse gyrase induces a change in structure via a left-handed wrapping, which more or less functions as an unwinding. Specifically, the reverse gyrase found in
30:
Molecular depictions of the different type I and type II topoisomerase classes. Reverse gyrase is indicated by the 'RG' tag under the type IA topoisomerase section of the diagram.
254:, whose reverse gyrase enzyme tends to naturally exist as two separate peptides versus the typical monomeric polypeptide with a topoisomerase IA domain and a helicase domain.
175:
194:
292:
strand is able to be grabbed by these subdomains and are subsequently fed up through the topoisomerase domain of the enzyme to complete positive supercoiling.
782:"Reverse gyrase functions as a DNA renaturase: annealing of complementary single-stranded circles and positive supercoiling of a bubble substrate"
1105:"Cell proliferation at 122 degrees C and isotopically heavy CH4 production by a hyperthermophilic methanogen under high-pressure cultivation"
481:
271:
The crystal structure of reverse gyrase has been characterized fully, and a crystal structure has been produced based on the enzyme found in
1154:
Valenti A, Perugino G, Rossi M, Ciaramella M (January 2011). "Positive supercoiling in thermophiles and mesophiles: of the good and evil".
301:
instead allows the enzyme to create positive supercoils in an ATP-dependent manner during the strand passage step of the helicase domain.
250:
active in increased temperatures, and TopR2, which shows activity in both low and high temperatures. Other exceptional organisms include
187:
130:
154:
499:"Reverse gyrase binding to DNA alters the double helix structure and produces single-strand cleavage in the absence of ATP"
403:
Kikuchi A, Asai K (21 June 1984). "Reverse gyrase--a topoisomerase which introduces positive superhelical turns into DNA".
148:
41:
357:
135:
223:
219:
199:
25:
123:
58:
350:
Visual depiction of the proposed mechanism of action of the reverse gyrase enzyme with ATP hydrolysis.
1202:
632:
412:
215:
935:"Reverse gyrase--recent advances and current mechanistic understanding of positive DNA supercoiling"
363:
278:
53:
867:"Separate and combined biochemical activities of the subunits of a naturally split reverse gyrase"
151:
1254:
436:
75:
1230:
1171:
1136:
1085:
964:
898:
844:
803:
757:
713:
660:
592:
528:
477:
428:
142:
1220:
1210:
1163:
1126:
1116:
1103:
Takai K, Nakamura K, Toki T, Tsunogai U, Miyazaki M, Miyazaki J, et al. (August 2008).
1075:
1067:
954:
946:
888:
878:
834:
793:
747:
705:
650:
640:
582:
572:
518:
510:
469:
420:
1191:"Reverse gyrase: a helicase-like domain and a type I topoisomerase in the same polypeptide"
375:
ATP hydrolysis ability via the helicase appears to be reduced by the topoisomerase domain.
371:
helicase's ability to hydrolyze ATP appears to be activated by the topoisomerase, whereas
346:
111:
1206:
709:
636:
416:
87:
1131:
1104:
959:
934:
893:
866:
823:"Reverse gyrase, the two domains intimately cooperate to promote positive supercoiling"
655:
620:
587:
560:
514:
170:
46:
1080:
1055:
752:
735:
523:
498:
1248:
1225:
1190:
1056:"Crystal structure of reverse gyrase: insights into the positive supercoiling of DNA"
323:
238:
440:
1036:
678:
497:
Jaxel C, Nadal M, Mirambeau G, Forterre P, Takahashi M, Duguet M (October 1989).
1189:
Confalonieri F, Elie C, Nadal M, de La Tour C, Forterre P, Duguet M (May 1993).
473:
331:
1195:
Proceedings of the
National Academy of Sciences of the United States of America
1109:
Proceedings of the
National Academy of Sciences of the United States of America
625:
Proceedings of the
National Academy of Sciences of the United States of America
322:(living in temperature ranges between 20 °C and 40 °C), tend to have
577:
226:
1071:
821:
Déclais AC, Marsault J, Confalonieri F, de La Tour CB, Duguet M (June 2000).
621:"Direct observation of helicase-topoisomerase coupling within reverse gyrase"
1215:
1121:
883:
645:
319:
245:
This enzyme has been extensively characterized across several
Archaea, with
1175:
1140:
1089:
1018:
968:
902:
848:
839:
822:
807:
798:
781:
664:
596:
318:
Organisms that live under standard temperature and pressure conditions, or
1234:
761:
717:
532:
432:
1000:
950:
234:
222:
into DNA, contrary to the typical negative supercoils introduced by the
1167:
982:
118:
99:
424:
182:
94:
82:
70:
237:
domain, which is responsible for separating nucleic acids, and a
106:
367:
experience opposite phenomena in terms of helicase activity:
1043:. International Union of Biochemistry and Molecular Biology.
1025:. International Union of Biochemistry and Molecular Biology.
1007:. International Union of Biochemistry and Molecular Biology.
989:. International Union of Biochemistry and Molecular Biology.
685:. International Union of Biochemistry and Molecular Biology.
865:
Capp C, Qian Y, Sage H, Huber H, Hsieh TS (December 2010).
736:"Intrinsic DNA-dependent ATPase activity of reverse gyrase"
619:
Yang X, Garnier F, Débat H, Strick TR, Nadal M (May 2020).
559:
Garnier F, Couturier M, Débat H, Nadal M (25 May 2021).
734:
Shibata T, Nakasu S, Yasui K, Kikuchi A (August 1987).
1019:"EC 5.6.2 Enzymes altering nucleic acid conformation"
282:
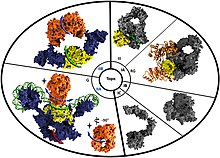
Reverse gyrase linear domains and crystal structure.
462:
193:
181:
169:
164:
141:
129:
117:
105:
93:
81:
69:
64:
52:
40:
35:
18:
561:"Archaea: A Gold Mine for Topoisomerase Diversity"
8:
161:
24:
1224:
1214:
1130:
1120:
1079:
958:
892:
882:
838:
797:
751:
654:
644:
586:
576:
522:
696:Gellert M (1981). "DNA topoisomerases".
345:
277:
1054:Rodríguez AC, Stock D (February 2002).
933:Lulchev P, Klostermeier D (July 2014).
395:
15:
928:
926:
924:
922:
920:
918:
916:
914:
912:
860:
858:
7:
775:
773:
771:
729:
727:
614:
612:
610:
608:
606:
554:
552:
550:
548:
546:
544:
542:
454:
452:
450:
871:The Journal of Biological Chemistry
827:The Journal of Biological Chemistry
786:The Journal of Biological Chemistry
740:The Journal of Biological Chemistry
710:10.1146/annurev.bi.50.070181.004311
515:10.1002/j.1460-2075.1989.tb08466.x
14:
780:Hsieh TS, Plank JL (March 2006).
1156:Biochemical Society Transactions
1:
753:10.1016/S0021-9258(18)60974-3
698:Annual Review of Biochemistry
474:10.1007/978-1-4419-0670-0_5
1271:
1041:IUBMB Enzyme Nomenclature
1023:IUBMB Enzyme Nomenclature
1005:IUBMB Enzyme Nomenclature
987:IUBMB Enzyme Nomenclature
683:IUBMB Enzyme Nomenclature
578:10.3389/fmicb.2021.661411
565:Frontiers in Microbiology
247:Sulfolobus acidocaldarius
160:
23:
314:Thermophile significance
1216:10.1073/pnas.90.10.4753
1122:10.1073/pnas.0712334105
884:10.1074/jbc.M110.173989
646:10.1073/pnas.1921848117
369:Sulfolobus solfataricus
358:Sulfolobus solfataricus
1072:10.1093/emboj/21.3.418
939:Nucleic Acids Research
840:10.1074/jbc.M910091199
799:10.1074/jbc.M513252200
351:
342:Supercoiling mechanism
283:
349:
324:negative supercoiling
281:
252:Nanoarchaeum equitans
224:type II topoisomerase
216:type I topoisomerase
1207:1993PNAS...90.4753C
1115:(31): 10949–10954.
877:(51): 39637–39645.
833:(26): 19498–19504.
746:(22): 10419–10421.
637:2020PNAS..11710856Y
631:(20): 10856–10864.
417:1984Natur.309..677K
373:Thermotoga maritima
364:Thermotoga maritima
330:On the other hand,
273:Thermotoga maritima
220:positive supercoils
1168:10.1042/BST0390058
983:"EC 5. Isomerases"
951:10.1093/nar/gku589
352:
284:
1201:(10): 4753–4757.
945:(13): 8200–8213.
509:(10): 3135–3139.
483:978-1-4419-0669-4
411:(5970): 677–681.
209:
208:
205:
204:
124:metabolic pathway
1262:
1239:
1238:
1228:
1218:
1186:
1180:
1179:
1151:
1145:
1144:
1134:
1124:
1100:
1094:
1093:
1083:
1060:The EMBO Journal
1051:
1045:
1044:
1033:
1027:
1026:
1015:
1009:
1008:
997:
991:
990:
979:
973:
972:
962:
930:
907:
906:
896:
886:
862:
853:
852:
842:
818:
812:
811:
801:
792:(9): 5640–5647.
777:
766:
765:
755:
731:
722:
721:
693:
687:
686:
675:
669:
668:
658:
648:
616:
601:
600:
590:
580:
556:
537:
536:
526:
503:The EMBO Journal
494:
488:
487:
456:
445:
444:
425:10.1038/309677a0
400:
218:that introduces
162:
28:
16:
1270:
1269:
1265:
1264:
1263:
1261:
1260:
1259:
1245:
1244:
1243:
1242:
1188:
1187:
1183:
1153:
1152:
1148:
1102:
1101:
1097:
1053:
1052:
1048:
1035:
1034:
1030:
1017:
1016:
1012:
999:
998:
994:
981:
980:
976:
932:
931:
910:
864:
863:
856:
820:
819:
815:
779:
778:
769:
733:
732:
725:
695:
694:
690:
677:
676:
672:
618:
617:
604:
558:
557:
540:
496:
495:
491:
484:
458:
457:
448:
402:
401:
397:
392:
380:S. solfataricus
344:
316:
307:
298:
289:
269:
260:
31:
12:
11:
5:
1268:
1266:
1258:
1257:
1247:
1246:
1241:
1240:
1181:
1146:
1095:
1066:(3): 418–426.
1046:
1028:
1010:
992:
974:
908:
854:
813:
767:
723:
688:
670:
602:
538:
489:
482:
446:
394:
393:
391:
388:
343:
340:
315:
312:
306:
303:
297:
294:
288:
285:
268:
265:
259:
258:Classification
256:
212:Reverse gyrase
207:
206:
203:
202:
197:
191:
190:
185:
179:
178:
173:
167:
166:
158:
157:
146:
139:
138:
133:
127:
126:
121:
115:
114:
109:
103:
102:
97:
91:
90:
85:
79:
78:
73:
67:
66:
62:
61:
56:
50:
49:
44:
38:
37:
33:
32:
29:
21:
20:
19:Reverse gyrase
13:
10:
9:
6:
4:
3:
2:
1267:
1256:
1253:
1252:
1250:
1236:
1232:
1227:
1222:
1217:
1212:
1208:
1204:
1200:
1196:
1192:
1185:
1182:
1177:
1173:
1169:
1165:
1161:
1157:
1150:
1147:
1142:
1138:
1133:
1128:
1123:
1118:
1114:
1110:
1106:
1099:
1096:
1091:
1087:
1082:
1077:
1073:
1069:
1065:
1061:
1057:
1050:
1047:
1042:
1038:
1032:
1029:
1024:
1020:
1014:
1011:
1006:
1002:
996:
993:
988:
984:
978:
975:
970:
966:
961:
956:
952:
948:
944:
940:
936:
929:
927:
925:
923:
921:
919:
917:
915:
913:
909:
904:
900:
895:
890:
885:
880:
876:
872:
868:
861:
859:
855:
850:
846:
841:
836:
832:
828:
824:
817:
814:
809:
805:
800:
795:
791:
787:
783:
776:
774:
772:
768:
763:
759:
754:
749:
745:
741:
737:
730:
728:
724:
719:
715:
711:
707:
703:
699:
692:
689:
684:
680:
674:
671:
666:
662:
657:
652:
647:
642:
638:
634:
630:
626:
622:
615:
613:
611:
609:
607:
603:
598:
594:
589:
584:
579:
574:
570:
566:
562:
555:
553:
551:
549:
547:
545:
543:
539:
534:
530:
525:
520:
516:
512:
508:
504:
500:
493:
490:
485:
479:
475:
471:
467:
463:
455:
453:
451:
447:
442:
438:
434:
430:
426:
422:
418:
414:
410:
406:
399:
396:
389:
387:
383:
381:
376:
374:
370:
366:
365:
360:
359:
348:
341:
339:
335:
333:
328:
325:
321:
313:
311:
304:
302:
295:
293:
286:
280:
276:
274:
266:
264:
257:
255:
253:
248:
243:
240:
239:topoisomerase
236:
230:
228:
225:
221:
217:
213:
201:
198:
196:
192:
189:
186:
184:
180:
177:
174:
172:
168:
163:
159:
156:
153:
150:
147:
144:
140:
137:
134:
132:
128:
125:
122:
120:
116:
113:
110:
108:
104:
101:
100:NiceZyme view
98:
96:
92:
89:
86:
84:
80:
77:
74:
72:
68:
63:
60:
57:
55:
51:
48:
45:
43:
39:
34:
27:
22:
17:
1198:
1194:
1184:
1162:(1): 58–63.
1159:
1155:
1149:
1112:
1108:
1098:
1063:
1059:
1049:
1040:
1031:
1022:
1013:
1004:
995:
986:
977:
942:
938:
874:
870:
830:
826:
816:
789:
785:
743:
739:
701:
697:
691:
682:
679:"EC 5.6.2.1"
673:
628:
624:
568:
564:
506:
502:
492:
465:
461:
408:
404:
398:
384:
379:
377:
372:
368:
362:
356:
353:
336:
332:thermophiles
329:
317:
308:
305:Zinc Fingers
299:
296:Latch Domain
290:
272:
270:
261:
251:
246:
244:
231:
211:
210:
88:BRENDA entry
59:143180-75-0
704:: 879–910.
287:Active Site
76:IntEnz view
36:Identifiers
571:: 661411.
468:: 73–121.
390:References
320:mesophiles
227:DNA gyrase
145:structures
112:KEGG entry
1255:EC 5.99.1
1037:"5.6.2.2"
267:Structure
65:Databases
1249:Category
1176:21265747
1141:18664583
1090:11823434
1001:"EC 5.6"
969:25013168
903:20929866
849:10748189
808:16407212
665:32371489
597:34113328
235:helicase
200:proteins
188:articles
176:articles
149:RCSB PDB
1235:8389456
1203:Bibcode
1132:2490668
960:4117796
894:3000944
762:3038879
718:6267993
656:7245102
633:Bibcode
588:8185306
533:2555155
441:4242694
433:6328327
413:Bibcode
136:profile
119:MetaCyc
54:CAS no.
47:5.6.2.2
1233:
1223:
1174:
1139:
1129:
1088:
1081:125824
1078:
967:
957:
901:
891:
847:
806:
760:
716:
663:
653:
595:
585:
531:
524:401394
521:
480:
439:
431:
405:Nature
183:PubMed
165:Search
155:PDBsum
95:ExPASy
83:BRENDA
71:IntEnz
42:EC no.
1226:46591
437:S2CID
214:is a
131:PRIAM
1231:PMID
1172:PMID
1137:PMID
1086:PMID
965:PMID
899:PMID
845:PMID
804:PMID
758:PMID
714:PMID
661:PMID
593:PMID
529:PMID
478:ISBN
429:PMID
361:and
195:NCBI
152:PDBe
107:KEGG
1221:PMC
1211:doi
1164:doi
1127:PMC
1117:doi
1113:105
1076:PMC
1068:doi
955:PMC
947:doi
889:PMC
879:doi
875:285
835:doi
831:275
794:doi
790:281
748:doi
744:262
706:doi
651:PMC
641:doi
629:117
583:PMC
573:doi
519:PMC
511:doi
470:doi
466:150
421:doi
409:309
171:PMC
143:PDB
1251::
1229:.
1219:.
1209:.
1199:90
1197:.
1193:.
1170:.
1160:39
1158:.
1135:.
1125:.
1111:.
1107:.
1084:.
1074:.
1064:21
1062:.
1058:.
1039:.
1021:.
1003:.
985:.
963:.
953:.
943:42
941:.
937:.
911:^
897:.
887:.
873:.
869:.
857:^
843:.
829:.
825:.
802:.
788:.
784:.
770:^
756:.
742:.
738:.
726:^
712:.
702:50
700:.
681:.
659:.
649:.
639:.
627:.
623:.
605:^
591:.
581:.
569:12
567:.
563:.
541:^
527:.
517:.
505:.
501:.
476:.
464:.
449:^
435:.
427:.
419:.
407:.
1237:.
1213::
1205::
1178:.
1166::
1143:.
1119::
1092:.
1070::
971:.
949::
905:.
881::
851:.
837::
810:.
796::
764:.
750::
720:.
708::
667:.
643::
635::
599:.
575::
535:.
513::
507:8
486:.
472::
443:.
423::
415::
275:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.