231:) (target Cell, viral entry), the receptor for SARS-CoV-2 ACE2 traffics SARS-CoV-2 to GM1 lipid rafts where it is endocytosed and exposed to cathepsin for cleavage and optimal cells fusion. In low cholesterol ACE2 traffics the virus to TMPRSS2 which also cleaves and allows viral entry but through a putative surface mechanism that is much less efficient. The sensitivity of ACE2 to cholesterol is thought to contribute to less severe
17:
162:
272:. For proteins that are both palmitoylated and bind PIP2, increasing the concentration of PIP2 favors trafficking of the enzyme out of lipid rafts to PIP2. PIP2 is primarily polyunsaturated which causes the lipid to localize away from lipid rafts and allows the PIP2 to oppose palmitate mediated localization.
314:
Mechanical force (shear or swell) can independently disrupt the packing and resultant affinity of palmitate to lipid rafts. This disruption also causes PLD2 to favor trafficking to PIP2 domains. The mechanosensitive ion channel TREK-1 is released from cholesterol dependent lipid rafts in response to
260:
Sequestration can both elevate and reduce the concentration of a protein in proximity to its substrate. When the substrate is present within a lipid raft, sequestration leads to an increased concentration of the protein near the substrate. Conversely, if the substrate is excluded from a lipid raft,
200:
are cell surface receptors that bind to various polypeptide growth factors, cytokines, and hormones. Activation of RTKs is driven by palmitoylation and dimerization, a process facilitated by cholesterol within lipid rafts. Once dimerized, the receptor undergoes autophosphorylation, which triggers a
300:
PUFAs may also increase the concentration of signaling lipids. The arachidonic acid, a very common PUFA in the brain, incorporates into PC and PIP2. Arachidonyl PC is a preferred substrate of PLD likely increasing the amount of PA in a cell. Regulation of raft function by cholesterol effectively
208:(PKC) is a class of enzymes that phosphorylates proteins. Its substrates are typically on the membrane surface where the enzyme is recruited by the lipid diacylglycerol. Thus a portion of PKC activation is through substrate presentation, i.e., by localization with its substrate on the membrane.
94:
enzymes are regulated by substrate presentation. The substrate APP is palmitoylated and moves in and out of GM1 lipid rafts in response to astrocyte cholesterol. Cholesterol delivered by apolipoprotein E (ApoE) drives APP to associate with GM1 lipid rafts. When cholesterol is low, the protein
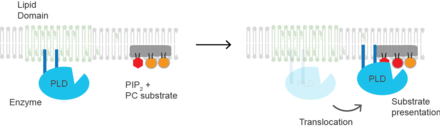
172:. PLD also binds PIP2(red hexagon) domains (grey shading) located separate from GM1 clusters in the plasma membrane and near phosphatidylcholine (PC). When PIP2 increases in the cell PLD translocates to PIP2 where it is exposed to and hydrolyzes PC to phosphatidic acid (red spherical lipid).
301:
regulates substrate presentation and the many palmitoylated proteins that utilize substrate presentation as a mechanism of activation. While speculative, the profound effect of cholesterol and PUFAs on human health is likely through physiological regulation of lipid raft function in cells.
292:(PUFAs) regulate lipid raft formation, hence the biological function of rafts. When saturated lipids and cholesterol increase in the membrane, lipid rafts increase their affinity for palmitoylated proteins. PUFAs have the opposite effect, they fluidize the membrane.
248:
Sequestration is the process of moving a molecule to a lipid raft. Within the plasma membrane, sequestration is primarily driven by packing of saturated lipid with cholesterol or phase separation at very small distances (< 100 nm). At a macroscopic level,
1066:
Petersen, E. Nicholas; Gudheti, Manasa; Pavel, Mahmud Arif; Murphy, Keith R.; Ja, William W.; Jorgensen, Erik M.; Hansen, Scott B. (5 September 2019). "Phospholipase D Transduces Force to TREK-1 Channels in a
Biological Membrane".
23:; A substrate (purple rectangle) is shown sequestered into a lipid domain (green lipids). The substrate's translocation to the disordered region (grey lipids) presents it to its enzyme (blue oval) where it is hydrolyzed.
350:
of PLD2 to lipid rafts. Activation of PLD then activates TREK-1 channels. The membrane mediated PLD2 activation could be transferred to an anesthetic insensitive homolog TRAAK, rending the channel anesthetic sensitive.
157:
where it then gains access to its substrate PC and commences catalysis based on substrate presentation. Presumably, the enzyme is capable of catalyzing a reaction in a lipid raft but lacks a substrate for activity.
701:
Tellier, Edwige; Canault, Matthias; Rebsomen, Laure; Bonardo, Bernadette; Juhan-Vague, Irène; Nalbone, Gilles; Peiretti, Franck (10 December 2006). "The shedding activity of ADAM17 is sequestered in lipid rafts".
1139:
Petersen, E. Nicholas; Pavel, Mahmud Arif; Hansen, Samuel S.; Gudheti, Manasa; Wang, Hao; Yuan, Zixuan; Murphy, Keith R.; Ja, William; Ferris, Heather A.; Jorgensen, Erik; Hansen, Scott B. (26 February 2024).
46:
binds. The substrate is the material acted upon. In the case of an interaction with an enzyme, the protein or organic substrate typically changes chemical form. Substrate presentation differs from
95:
traffics to the disordered region and is cleaved by alpha secretase to produce a non-amylogenic product. The enzymes do not appear to respond to cholesterol, only the substrate moves.
1242:
Pavel, Mahmud Arif; Petersen, E. Nicholas; Wang, Hao; Lerner, Richard A.; Hansen, Scott B. (19 June 2019). "Studies on the mechanism of membrane mediated general anesthesia".
220:) (producing cell, replication). When cells are loaded with cholesterol furin traffics to GM1 lipid rafts where it is localized with the palmitoylated spike protein of
221:
141:(PC) which is unsaturated and is of low abundance in lipid rafts. PC localizes to the disordered region of the cell along with the polyunsaturated lipid
142:
50:
in that the enzyme need not change its conformation to begin catalysis. Substrate presentation is best described for domain partitioning at
189:(mTNF). Cholesterol causes mTNF to cluster with ADAM17 in lipid rafts and shed soluble TNF (sTNF) which is an inflammatory cytokine.
34:. The protein is sequestered away from its substrate and then activated by release and exposure of the protein to its substrate. A
347:
265:
254:
228:
1269:
201:
subsequent phosphorylation cascade. This is a specific case where the substrate and the enzyme are the same molecule.
323:
1193:"Disruption of palmitate-mediated localization; a shared pathway of force and anesthetic activation of TREK-1 channels"
1090:"Disruption of palmitate-mediated localization; a shared pathway of force and anesthetic activation of TREK-1 channels"
370:"Disruption of palmitate-mediated localization; a shared pathway of force and anesthetic activation of TREK-1 channels"
289:
101:
drives the partitioning of molecules. In the cell, this gives rise to compartmentalization within the cell and within
1142:"Mechanical activation of TWIK-related potassium channel by nanoscopic movement and rapid second messenger signaling"
519:
Wang, Hao; Kulas, Joshua A.; Wang, Chao; Holtzman, David M.; Ferris, Heather A.; Hansen, Scott B. (17 August 2021).
67:
232:
197:
87:
261:
sequestration results in decreased interaction between the protein and the substrate, as seen with PLD2.
269:
186:
110:
47:
35:
1243:
1068:
1020:
657:
599:
532:
153:. When PIP2 concentration in the membrane increases, PLD2 leaves the GM1 domains and associates with
109:
regulates raft affinity for the majority of integral raft proteins. Raft regulation is regulated by
138:
1224:
1173:
1121:
1048:
989:
940:
895:
846:
801:
760:
719:
683:
627:
568:
550:
501:
450:
401:
185:), also called TACE, is sequestered into lipid rafts away from its substrate, membrane bound
1214:
1204:
1163:
1153:
1111:
1101:
1038:
1028:
979:
971:
930:
922:
885:
877:
836:
828:
791:
750:
711:
673:
665:
617:
607:
558:
540:
491:
481:
470:"Cholesterol Regulation of Membrane Proteins Revealed by Two-Color Super-Resolution Imaging"
440:
432:
391:
381:
205:
168:; PLD (blue oval) is sequestered into cholesterol-dependent lipid domains (green lipids) by
125:) is a well-defined example of an enzyme activated by substrate presentation. The enzyme is
51:
134:
75:
1168:
1141:
1024:
890:
865:
661:
603:
536:
16:
1219:
1192:
1116:
1089:
1043:
1008:
984:
959:
935:
841:
678:
646:
622:
587:
563:
520:
496:
469:
445:
420:
396:
369:
169:
150:
126:
106:
98:
83:
71:
796:
779:
1263:
343:
264:
Either the substrate of the enzyme can move. Movement is typically the disruption of
102:
161:
1009:"Palmitoylation regulates raft affinity for the majority of integral raft proteins"
881:
588:"Palmitoylation regulates raft affinity for the majority of integral raft proteins"
521:"Regulation of beta-amyloid production in neurons by astrocyte-derived cholesterol"
1209:
1106:
975:
755:
738:
386:
715:
285:
154:
130:
1013:
Proceedings of the
National Academy of Sciences of the United States of America
1007:
Levental, I; Lingwood, D; Grzybek, M; Coskun, U; Simons, K (21 December 2010).
778:
Paige, LA; Nadler, MJ; Harrison, ML; Cassady, JM; Geahlen, RL (25 April 1993).
592:
Proceedings of the
National Academy of Sciences of the United States of America
586:
Levental, I; Lingwood, D; Grzybek, M; Coskun, U; Simons, K (21 December 2010).
436:
926:
832:
486:
339:
335:
250:
913:
Wang, Hao; Yuan, Zixuan; Pavel, Mahmud Arif; Hansen, Scott B. (29 May 2020).
819:
Wang, Hao; Yuan, Zixuan; Pavel, Mahmud Arif; Hansen, Scott B. (29 May 2020).
554:
1033:
914:
820:
612:
545:
91:
1228:
1177:
1125:
1052:
993:
944:
899:
850:
764:
723:
687:
647:"Kinetic disruption of lipid rafts is a mechanosensor for phospholipase D."
631:
572:
505:
454:
405:
964:
Biochimica et
Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids
805:
327:
1158:
669:
645:
Petersen, EN; Chung, HW; Nayebosadri, A; Hansen, SB (15 December 2016).
79:
31:
739:"Growth factor receptors, lipid rafts and caveolae: an evolving story"
866:"Getting in on the action: New tools to see SARS-CoV-2 infect a cell"
182:
43:
39:
421:"Tools for Understanding Nanoscale Lipid Regulation of Ion Channels"
1248:
1073:
331:
217:
160:
15:
780:"Reversible palmitoylation of the protein-tyrosine kinase p56lck"
1088:
Petersen, EN; Pavel, MA; Wang, H; Hansen, SB (28 October 2019).
368:
Petersen, EN; Pavel, MA; Wang, H; Hansen, SB (28 October 2019).
146:
122:
1191:
Petersen, EN; Pavel, MA; Wang, H; Hansen, SB (1 January 2020).
960:"Lipid agonism: The PIP2 paradigm of ligand-gated ion channels"
915:"The role of high cholesterol in age-related COVID19 lethality"
821:"The role of high cholesterol in age-related COVID19 lethality"
743:
Biochimica et
Biophysica Acta (BBA) - Molecular Cell Research
315:
mechanical force. This has the effect of dampening pain.
129:
causing the enzyme to traffic to GM1 lipid domains or "
419:
Robinson, CV; Rohacs, T; Hansen, SB (September 2019).
326:
employs substrate presentation. General anesthetics
468:Yuan, Zixuan; Hansen, Scott B. (20 February 2023).
1197:Biochimica et Biophysica Acta (BBA) - Biomembranes
1094:Biochimica et Biophysica Acta (BBA) - Biomembranes
374:Biochimica et Biophysica Acta (BBA) - Biomembranes
42:acts but can also be a protein surface to which a
257:can limit access of an enzyme with to substrate.
525:Proceedings of the National Academy of Sciences
864:Hansen, Scott B.; Yuan, Zixuan (March 2023).
8:
1247:
1218:
1208:
1167:
1157:
1115:
1105:
1072:
1042:
1032:
983:
934:
889:
840:
795:
754:
677:
621:
611:
562:
544:
495:
485:
444:
395:
385:
30:is a biological process that activates a
360:
38:is typically the substance on which an
143:phosphatidylinositol 4,5-bisphosphate
7:
784:The Journal of Biological Chemistry
14:
346:disrupt lipid raft function and
348:palmitate mediated localization
266:palmitate mediated localization
224:and primes it for viral entry.
882:10.1016/j.chembiol.2023.02.010
425:Trends in Biochemical Sciences
1:
797:10.1016/S0021-9258(18)52927-6
737:Pike, LJ (30 December 2005).
54:distances (<100 nm).
1210:10.1016/j.bbamem.2019.183091
1107:10.1016/j.bbamem.2019.183091
976:10.1016/j.bbalip.2015.01.011
756:10.1016/j.bbamcr.2005.05.005
387:10.1016/j.bbamem.2019.183091
324:Membrane-mediated anesthesia
78:to yield a 40-42 amino acid
716:10.1016/j.yexcr.2006.08.027
290:polyunsaturated fatty acids
1286:
704:Experimental Cell Research
437:10.1016/j.tibs.2019.04.001
927:10.1101/2020.05.09.086249
833:10.1101/2020.05.09.086249
487:10.3390/membranes13020250
198:Receptor Tyrosine Kinases
68:Amyloid precursor protein
63:Amyloid precursor protein
330:and inhaled anesthetics
239:Mechanisms of activation
1034:10.1073/pnas.1016184107
958:Hansen, SB (May 2015).
613:10.1073/pnas.1016184107
546:10.1073/pnas.2102191118
235:symptoms in children.
173:
28:Substrate presentation
24:
21:Substrate presentation
921:: 2020.05.09.086249.
870:Cell Chemical Biology
827:: 2020.05.09.086249.
650:Nature Communications
270:organelle trafficking
187:tumor necrosis factor
164:
111:cholesterol signaling
48:allosteric regulation
19:
1270:Biological processes
166:Enzyme translocation
133:". The substrate of
105:. For lipid rafts,
70:(APP) is cleaved by
1159:10.7554/eLife.89465
1025:2010PNAS..10722050L
670:10.1038/ncomms13873
662:2016NatCo...713873P
604:2010PNAS..10722050L
537:2021PNAS..11802191W
531:(33): e2102191118.
149:). PLD2 has a PIP2
139:phosphatidylcholine
88:Alzheimer's disease
174:
25:
710:(20): 3969–3980.
1277:
1254:
1253:
1251:
1239:
1233:
1232:
1222:
1212:
1188:
1182:
1181:
1171:
1161:
1136:
1130:
1129:
1119:
1109:
1085:
1079:
1078:
1076:
1063:
1057:
1056:
1046:
1036:
1004:
998:
997:
987:
955:
949:
948:
938:
910:
904:
903:
893:
861:
855:
854:
844:
816:
810:
809:
799:
775:
769:
768:
758:
734:
728:
727:
698:
692:
691:
681:
642:
636:
635:
625:
615:
583:
577:
576:
566:
548:
516:
510:
509:
499:
489:
465:
459:
458:
448:
416:
410:
409:
399:
389:
365:
310:Mechanosensation
206:Protein Kinase C
193:Kinase Signaling
117:Phospholipase D2
86:associated with
82:responsible for
1285:
1284:
1280:
1279:
1278:
1276:
1275:
1274:
1260:
1259:
1258:
1257:
1241:
1240:
1236:
1190:
1189:
1185:
1138:
1137:
1133:
1087:
1086:
1082:
1065:
1064:
1060:
1019:(51): 22050–4.
1006:
1005:
1001:
957:
956:
952:
912:
911:
907:
863:
862:
858:
818:
817:
813:
790:(12): 8669–74.
777:
776:
772:
736:
735:
731:
700:
699:
695:
644:
643:
639:
598:(51): 22050–4.
585:
584:
580:
518:
517:
513:
467:
466:
462:
418:
417:
413:
367:
366:
362:
357:
321:
312:
307:
305:Role in biology
298:
283:
278:
246:
241:
214:
204:
195:
179:
135:phospholipase D
119:
84:amyloid plaques
76:gamma secretase
65:
60:
12:
11:
5:
1283:
1281:
1273:
1272:
1262:
1261:
1256:
1255:
1249:10.1101/313973
1234:
1183:
1131:
1080:
1074:10.1101/758896
1058:
999:
950:
905:
876:(3): 233–234.
856:
811:
770:
729:
693:
637:
578:
511:
460:
431:(9): 795–806.
411:
359:
358:
356:
353:
320:
317:
311:
308:
306:
303:
297:
294:
282:
279:
277:
274:
245:
242:
240:
237:
213:
210:
194:
191:
178:
175:
170:palmitoylation
151:binding domain
118:
115:
107:palmitoylation
103:cell membranes
99:Hydrophobicity
64:
61:
59:
56:
13:
10:
9:
6:
4:
3:
2:
1282:
1271:
1268:
1267:
1265:
1250:
1245:
1238:
1235:
1230:
1226:
1221:
1216:
1211:
1206:
1203:(1): 183091.
1202:
1198:
1194:
1187:
1184:
1179:
1175:
1170:
1165:
1160:
1155:
1151:
1147:
1143:
1135:
1132:
1127:
1123:
1118:
1113:
1108:
1103:
1100:(1): 183091.
1099:
1095:
1091:
1084:
1081:
1075:
1070:
1062:
1059:
1054:
1050:
1045:
1040:
1035:
1030:
1026:
1022:
1018:
1014:
1010:
1003:
1000:
995:
991:
986:
981:
977:
973:
969:
965:
961:
954:
951:
946:
942:
937:
932:
928:
924:
920:
916:
909:
906:
901:
897:
892:
887:
883:
879:
875:
871:
867:
860:
857:
852:
848:
843:
838:
834:
830:
826:
822:
815:
812:
807:
803:
798:
793:
789:
785:
781:
774:
771:
766:
762:
757:
752:
749:(3): 260–73.
748:
744:
740:
733:
730:
725:
721:
717:
713:
709:
705:
697:
694:
689:
685:
680:
675:
671:
667:
663:
659:
655:
651:
648:
641:
638:
633:
629:
624:
619:
614:
609:
605:
601:
597:
593:
589:
582:
579:
574:
570:
565:
560:
556:
552:
547:
542:
538:
534:
530:
526:
522:
515:
512:
507:
503:
498:
493:
488:
483:
479:
475:
471:
464:
461:
456:
452:
447:
442:
438:
434:
430:
426:
422:
415:
412:
407:
403:
398:
393:
388:
383:
380:(1): 183091.
379:
375:
371:
364:
361:
354:
352:
349:
345:
344:diethyl ether
341:
337:
333:
329:
325:
318:
316:
309:
304:
302:
295:
293:
291:
287:
280:
275:
273:
271:
267:
262:
258:
256:
252:
244:Sequestration
243:
238:
236:
234:
230:
225:
223:
219:
211:
209:
207:
202:
199:
192:
190:
188:
184:
176:
171:
167:
163:
159:
156:
152:
148:
144:
140:
136:
132:
128:
127:palmitoylated
124:
116:
114:
112:
108:
104:
100:
96:
93:
89:
85:
81:
77:
73:
69:
62:
57:
55:
53:
49:
45:
41:
37:
33:
29:
22:
18:
1237:
1200:
1196:
1186:
1149:
1145:
1134:
1097:
1093:
1083:
1061:
1016:
1012:
1002:
970:(5): 620–8.
967:
963:
953:
918:
908:
873:
869:
859:
824:
814:
787:
783:
773:
746:
742:
732:
707:
703:
696:
653:
649:
640:
595:
591:
581:
528:
524:
514:
477:
473:
463:
428:
424:
414:
377:
373:
363:
322:
313:
299:
284:
263:
259:
247:
226:
215:
203:
196:
180:
177:Inflammation
165:
155:PIP2 domains
120:
97:
66:
27:
26:
20:
1152:: RP89465.
319:Anaesthesia
286:Cholesterol
281:Cholesterol
131:lipid rafts
480:(2): 250.
355:References
340:isoflurane
336:chloroform
276:Regulation
251:organelles
222:SARS-CoV-2
212:SARS-CoV-2
52:nanoscopic
656:: 13873.
555:0027-8424
474:Membranes
92:secretase
36:substrate
1264:Category
1229:31672538
1178:38407149
1169:10942622
1126:31672538
1053:21131568
994:25633344
945:32511366
900:36931249
891:10018748
851:32511366
765:15951036
724:17010968
688:27976674
632:21131568
573:34385305
506:36837753
455:31060927
406:31672538
328:propofol
58:Examples
1244:bioRxiv
1220:6907892
1117:6907892
1069:bioRxiv
1044:3009825
1021:Bibcode
985:4540326
936:7263494
919:bioRxiv
842:7263494
825:bioRxiv
806:8473310
679:5171650
658:Bibcode
623:3009825
600:Bibcode
564:8379952
533:Bibcode
497:9966874
446:6729126
397:6907892
255:vesicle
233:COVID19
80:peptide
32:protein
1246:
1227:
1217:
1176:
1166:
1124:
1114:
1071:
1051:
1041:
992:
982:
943:
933:
898:
888:
849:
839:
804:
763:
722:
686:
676:
630:
620:
571:
561:
553:
504:
494:
453:
443:
404:
394:
183:ADAM17
90:. The
44:ligand
40:enzyme
1146:eLife
332:xenon
296:PUFAs
218:Furin
1225:PMID
1201:1862
1174:PMID
1122:PMID
1098:1862
1049:PMID
990:PMID
968:1851
941:PMID
896:PMID
847:PMID
802:PMID
761:PMID
747:1746
720:PMID
684:PMID
628:PMID
569:PMID
551:ISSN
502:PMID
451:PMID
402:PMID
378:1862
288:and
253:and
229:ACE2
147:PIP2
123:PLD2
74:and
72:beta
1215:PMC
1205:doi
1164:PMC
1154:doi
1112:PMC
1102:doi
1039:PMC
1029:doi
1017:107
980:PMC
972:doi
931:PMC
923:doi
886:PMC
878:doi
837:PMC
829:doi
792:doi
788:268
751:doi
712:doi
708:312
674:PMC
666:doi
618:PMC
608:doi
596:107
559:PMC
541:doi
529:118
492:PMC
482:doi
441:PMC
433:doi
392:PMC
382:doi
268:or
137:is
1266::
1223:.
1213:.
1199:.
1195:.
1172:.
1162:.
1150:12
1148:.
1144:.
1120:.
1110:.
1096:.
1092:.
1047:.
1037:.
1027:.
1015:.
1011:.
988:.
978:.
966:.
962:.
939:.
929:.
917:.
894:.
884:.
874:30
872:.
868:.
845:.
835:.
823:.
800:.
786:.
782:.
759:.
745:.
741:.
718:.
706:.
682:.
672:.
664:.
652:.
626:.
616:.
606:.
594:.
590:.
567:.
557:.
549:.
539:.
527:.
523:.
500:.
490:.
478:13
476:.
472:.
449:.
439:.
429:44
427:.
423:.
400:.
390:.
376:.
372:.
342:,
338:,
334:,
113:.
1252:.
1231:.
1207::
1180:.
1156::
1128:.
1104::
1077:.
1055:.
1031::
1023::
996:.
974::
947:.
925::
902:.
880::
853:.
831::
808:.
794::
767:.
753::
726:.
714::
690:.
668::
660::
654:7
634:.
610::
602::
575:.
543::
535::
508:.
484::
457:.
435::
408:.
384::
227:(
216:(
181:(
145:(
121:(
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.