449:
426:
323:
348:
700:
5583:
5568:
5553:
5538:
455:
354:
5598:
1638:
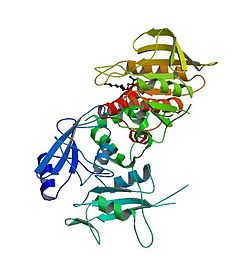
function as phospho-tyrosine binding domains and mediate the interaction of this PTP with its substrates. This PTP is widely expressed in most tissues and plays a regulatory role in various cell signaling events that are important for a diversity of cell functions, such as mitogenic activation, metabolic control, transcription regulation, and cell migration. Mutations in this gene are a cause of
7533:
1656:
1637:
PTPN11 is a member of the protein tyrosine phosphatase (PTP) family. PTPs are known to be signaling molecules that regulate a variety of cellular processes including cell growth, differentiation, mitotic cycle, and oncogenic transformation. This PTP contains two tandem Src homology-2 domains, which
44:
1812:
The mutations that cause
Leopard syndrome are restricted regions affecting the catalytic core of the enzyme producing catalytically impaired Shp2 variants. It is currently unclear how mutations that give rise to mutant variants of Shp2 with biochemically opposite characteristics result in similar
1803:
In the case of Noonan syndrome, mutations are broadly distributed throughout the coding region of the gene but all appear to result in hyper-activated, or unregulated mutant forms of the protein. Most of these mutations disrupt the binding interface between the N-SH2 domain and catalytic core
70:
3032:"The protein-tyrosine phosphatase SHP-2 binds platelet/endothelial cell adhesion molecule-1 (PECAM-1) and forms a distinct signaling complex during platelet aggregation. Evidence for a mechanistic link between PECAM-1- and integrin-mediated cellular signaling"
5288:
Ion A, Tartaglia M, Song X, Kalidas K, van der Burgt I, Shaw AC, Ming JE, Zampino G, Zackai EH, Dean JC, Somer M, Parenti G, Crosby AH, Patton MA, Gelb BD, Jeffery S (2002). "Absence of PTPN11 mutations in 28 cases of cardiofaciocutaneous (CFC) syndrome".
2553:
Bentires-Alj M, Paez JG, David FS, Keilhack H, Halmos B, Naoki K, Maris JM, Richardson A, Bardelli A, Sugarbaker DJ, Richards WG, Du J, Girard L, Minna JD, Loh ML, Fisher DE, Velculescu VE, Vogelstein B, Meyerson M, Sellers WR, Neel BG (December 2004).
3507:
Boudot C, Kadri Z, Petitfrère E, Lambert E, Chrétien S, Mayeux P, Haye B, Billat C (October 2002). "Phosphatidylinositol 3-kinase regulates glycosylphosphatidylinositol hydrolysis through PLC-gamma(2) activation in erythropoietin-stimulated cells".
4369:
Maegawa H, Ugi S, Adachi M, Hinoda Y, Kikkawa R, Yachi A, Shigeta Y, Kashiwagi A (March 1994). "Insulin receptor kinase phosphorylates protein tyrosine phosphatase containing Src homology 2 regions and modulates its PTPase activity in vitro".
1768:
in its N-terminus followed by a protein tyrosine phosphatase (PTP) domain. In the inactive state, the N-terminal SH2 domain binds the PTP domain and blocks access of potential substrates to the active site. Thus, Shp2 is auto-inhibited.
2991:"Recruitment and activation of SHP-1 protein-tyrosine phosphatase by human platelet endothelial cell adhesion molecule-1 (PECAM-1). Identification of immunoreceptor tyrosine-based inhibitory motif-like binding motifs and substrates"
5063:
Chin H, Saito T, Arai A, Yamamoto K, Kamiyama R, Miyasaka N, Miura O (October 1997). "Erythropoietin and IL-3 induce tyrosine phosphorylation of CrkL and its association with Shc, SHP-2, and Cbl in hematopoietic cells".
4659:
Maegawa H, Kashiwagi A, Fujita T, Ugi S, Hasegawa M, Obata T, Nishio Y, Kojima H, Hidaka H, Kikkawa R (November 1996). "SHPTP2 serves adapter protein linking between Janus kinase 2 and insulin receptor substrates".
2128:
into gastric epithelia. Once activated by SRC phosphorylation, CagA binds to SHP2, allosterically activating it. This leads to morphological changes, abnormal mitogenic signals and sustained activity can result in
2468:
Roberts AE, Araki T, Swanson KD, Montgomery KT, Schiripo TA, Joshi VA, Li L, Yassin Y, Tamburino AM, Neel BG, Kucherlapati RS (January 2007). "Germline gain-of-function mutations in SOS1 cause Noonan syndrome".
2901:"Platelet-endothelial cell adhesion molecule-1 (CD31), a scaffolding molecule for selected catenin family members whose binding is mediated by different tyrosine and serine/threonine phosphorylation"
3325:
Kurokawa K, Iwashita T, Murakami H, Hayashi H, Kawai K, Takahashi M (April 2001). "Identification of SNT/FRS2 docking site on RET receptor tyrosine kinase and its role for signal transduction".
5582:
5567:
5552:
5537:
5232:
Marron MB, Hughes DP, McCarthy MJ, Beaumont ER, Brindle NP (2000). "Tie-1 Receptor
Tyrosine Kinase Endodomain Interaction with SHP2: Potential Signalling Mechanisms and Roles in Angiogenesis".
2417:
Sobreira NL, Cirulli ET, Avramopoulos D, Wohler E, Oswald GL, Stevens EL, Ge D, Shianna KV, Smith JP, Maia JM, Gumbs CE, Pevsner J, Thomas G, Valle D, Hoover-Fong JE, Goldstein DB (June 2010).
2595:
Patel SS, Kuo FC, Gibson CJ, Steensma DP, Soiffer RJ, Alyea EP, Chen YA, Fathi AT, Graubert TA, Brunner AM, Wadleigh M, Stone RM, DeAngelo DJ, Nardi V, Hasserjian RP, Weinberg OK (May 2018).
3743:
Lehmann U, Schmitz J, Weissenbach M, Sobota RM, Hortner M, Friederichs K, Behrmann I, Tsiaris W, Sasaki A, Schneider-Mergener J, Yoshimura A, Neel BG, Heinrich PC, Schaper F (January 2003).
2244:
Jamieson CR, van der Burgt I, Brady AF, van Reen M, Elsawi MM, Hol F, Jeffery S, Patton MA, Mariman E (December 1994). "Mapping a gene for Noonan syndrome to the long arm of chromosome 12".
3163:"Association of SH2 domain protein tyrosine phosphatases with the epidermal growth factor receptor in human tumor cells. Phosphatidic acid activates receptor dephosphorylation by PTP1C"
5638:
3073:"The carboxyl-terminal region of biliary glycoprotein controls its tyrosine phosphorylation and association with protein-tyrosine phosphatases SHP-1 and SHP-2 in epithelial cells"
1772:
Upon binding to target phospho-tyrosyl residues, the N-terminal SH2 domain is released from the PTP domain, catalytically activating the enzyme by relieving this auto-inhibition.
3633:
Crouin C, Arnaud M, Gesbert F, Camonis J, Bertoglio J (April 2001). "A yeast two-hybrid study of human p97/Gab2 interactions with its SH2 domain-containing binding partners".
255:, BPTP3, CFC, JMML, METCDS, NS1, PTP-1D, PTP2C, SH-PTP2, SH-PTP3, SHP2, protein tyrosine phosphatase, non-receptor type 11, protein tyrosine phosphatase non-receptor type 11
462:
361:
5014:
Morra M, Lu J, Poy F, Martin M, Sayos J, Calpe S, Gullo C, Howie D, Rietdijk S, Thompson A, Coyle AJ, Denny C, Yaffe MB, Engel P, Eck MJ, Terhorst C (November 2001).
4893:"Activation of the SH2-containing phosphotyrosine phosphatase SH-PTP2 by its binding site, phosphotyrosine 1009, on the human platelet-derived growth factor receptor"
3424:"Protein kinase C-alpha and protein kinase C-epsilon are required for Grb2-associated binder-1 tyrosine phosphorylation in response to platelet-derived growth factor"
4620:"Tyrosine 425 within the activated erythropoietin receptor binds Syp, reduces the erythropoietin required for Syp tyrosine phosphorylation, and promotes mitogenesis"
4842:
Keilhack H, Müller M, Böhmer SA, Frank C, Weidner KM, Birchmeier W, Ligensa T, Berndt A, Kosmehl H, Günther B, Müller T, Birchmeier C, Böhmer FD (January 2001).
4107:"Fyn kinase-directed activation of SH2 domain-containing protein-tyrosine phosphatase SHP-2 by Gi protein-coupled receptors in Madin-Darby canine kidney cells"
5631:
1881:, resulting in regenerative hyperplasia and spontaneous development of tumors. Decreased PTPN11/Shp2 expression was detected in a subfraction of human
1293:
1274:
284:
4695:
Fournier N, Chalus L, Durand I, Garcia E, Pin JJ, Churakova T, Patel S, Zlot C, Gorman D, Zurawski S, Abrams J, Bates EE, Garrone P (August 2000).
6607:
4238:"Mutation of the SHP-2 binding site in growth hormone (GH) receptor prolongs GH-promoted tyrosyl phosphorylation of GH receptor, JAK2, and STAT5B"
6854:
5624:
1948:
4932:
Chauhan D, Pandey P, Hideshima T, Treon S, Raje N, Davies FE, Shima Y, Tai YT, Rosen S, Avraham S, Kharbanda S, Anderson KC (September 2000).
7553:
5517:
5249:
4497:"Localization of the insulin-like growth factor I receptor binding sites for the SH2 domain proteins p85, Syp, and GTPase activating protein"
3220:
2226:
2007:
4844:"Negative regulation of Ros receptor tyrosine kinase signaling. An epithelial function of the SH2 domain protein tyrosine phosphatase SHP-1"
2208:
6768:
2649:
Bard-Chapeau EA, Li S, Ding J, Zhang SS, Zhu HH, Princen F, Fang DD, Han T, Bailly-Maitre B, Poli V, Varki NM, Wang H, Feng GS (May 2011).
67:
6869:
6863:
6664:
6414:
1529:
4975:"Molecular dissection of the signaling and costimulatory functions of CD150 (SLAM): CD150/SAP binding and CD150-mediated costimulation"
2940:
Pumphrey NJ, Taylor V, Freeman S, Douglas MR, Bradfield PF, Young SP, Lord JM, Wakelam MJ, Bird IN, Salmon M, Buckley CD (April 1999).
448:
6712:
5326:
Hugues L, Cavé H, Philippe N, Pereira S, Fenaux P, Preudhomme C (2006). "Mutations of PTPN11 are rare in adult myeloid malignancies".
5145:"Prolactin induces SHP-2 association with Stat5, nuclear translocation, and binding to the beta-casein gene promoter in mammary cells"
4066:"Induced direct binding of the adapter protein Nck to the GTPase-activating protein-associated protein p62 by epidermal growth factor"
3784:
Anhuf D, Weissenbach M, Schmitz J, Sobota R, Hermanns HM, Radtke S, Linnemann S, Behrmann I, Heinrich PC, Schaper F (September 2000).
1536:
7252:
6823:
6612:
3786:"Signal transduction of IL-6, leukemia-inhibitory factor, and oncostatin M: structural receptor requirements for signal attenuation"
2556:"Activating mutations of the noonan syndrome-associated SHP2/PTPN11 gene in human solid tumors and adult acute myelogenous leukemia"
1747:
1861:
patients, although the prognostic significance of such associations has not been clarified. These data suggests that Shp2 may be a
425:
2346:
1942:
1822:
6930:
6818:
2195:
2174:
1891:
has been associated with gastric cancer, and this is thought to be mediated in part by the interaction of its virulence factor
4197:"Epidermal growth factor induces coupling of protein-tyrosine phosphatase 1D to GRB2 via the COOH-terminal SH3 domain of GRB2"
4148:"Flt3 signaling involves tyrosyl-phosphorylation of SHP-2 and SHIP and their association with Grb2 and Shc in Baf3/Flt3 cells"
2942:"Differential association of cytoplasmic signalling molecules SHP-1, SHP-2, SHIP and phospholipase C-gamma1 with PECAM-1/CD31"
2289:"Identification of a human Src homology 2-containing protein-tyrosine-phosphatase: a putative homolog of Drosophila corkscrew"
6739:
6649:
6602:
1681:
347:
322:
7408:
5267:
Carter-Su C, Rui L, Stofega MR (2000). "SH2-B and SIRP: JAK2 binding proteins that modulate the actions of growth hormone".
2852:"SHP-1 binds and negatively modulates the c-Kit receptor by interaction with tyrosine 569 in the c-Kit juxtamembrane domain"
2191:
7161:
7102:
5680:
3375:"Binding of Shp2 tyrosine phosphatase to FRS2 is essential for fibroblast growth factor-induced PC12 cell differentiation"
5434:"How do Shp2 mutations that oppositely influence its biochemical activity result in syndromes with overlapping symptoms?"
4697:"FDF03, a novel inhibitory receptor of the immunoglobulin superfamily, is expressed by human dendritic and myeloid cells"
2362:"Refinement of evolutionary medicine predictions based on clinical evidence for the manifestations of Mendelian diseases"
2170:
7206:
6773:
6763:
6358:
6323:
6305:
5900:
5664:
5651:
4934:"SHP2 mediates the protective effect of interleukin-6 against dexamethasone-induced apoptosis in multiple myeloma cells"
3594:"Gab2, a new pleckstrin homology domain-containing adapter protein, acts to uncouple signaling from ERK kinase to Elk-1"
1631:
264:
7166:
7058:
6683:
4279:"Grb10 identified as a potential regulator of growth hormone (GH) signaling by cloning of GH receptor target proteins"
3239:
Neel BG, Gu H, Pao L (June 2003). "The 'Shp'ing news: SH2 domain-containing tyrosine phosphatases in cell signaling".
3208:
1904:
461:
360:
1728:
7523:
6752:
6748:
6744:
6660:
6493:
5597:
5376:
Ogata T, Yoshida R (2006). "PTPN11 mutations and genotype-phenotype correlations in Noonan and LEOPARD syndromes".
1850:
1700:
454:
353:
7393:
4407:"Adapter function of protein-tyrosine phosphatase 1D in insulin receptor/insulin receptor substrate-1 interaction"
7509:
7496:
7483:
7470:
7457:
7444:
7431:
7014:
6960:
6920:
6879:
6783:
6622:
6590:
6476:
6440:
5510:
7403:
4891:
Lechleider RJ, Sugimoto S, Bennett AM, Kashishian AS, Cooper JA, Shoelson SE, Walsh CT, Neel BG (October 1993).
4579:"The COOH-terminal tyrosine phosphorylation sites on IRS-1 bind SHP-2 and negatively regulate insulin signaling"
4495:
Seely BL, Reichart DR, Staubs PA, Jhun BH, Hsu D, Maegawa H, Milarski KL, Saltiel AR, Olefsky JM (August 1995).
2419:"Whole-genome sequencing of a single proband together with linkage analysis identifies a Mendelian disease gene"
7357:
7300:
6668:
6517:
6431:
5655:
1882:
1873:. In aged mouse model, hepatocyte-specific deletion of PTPN11/Shp2 promotes inflammatory signaling through the
1666:
1338:
272:
5016:"Structural basis for the interaction of the free SH2 domain EAT-2 with SLAM receptors in hematopoietic cells"
1707:
7305:
6537:
6407:
4737:
3288:
1989:
1685:
1677:
1670:
1319:
5490:
4448:"Concerted activity of tyrosine phosphatase SHP-2 and focal adhesion kinase in regulation of cell motility"
7093:
6732:
5591:: CRYSTAL STRUCTURES OF PEPTIDE COMPLEXES OF THE AMINO-TERMINAL SH2 DOMAIN OF THE SYP TYROSINE PHOSPHATASE
5576:: CRYSTAL STRUCTURES OF PEPTIDE COMPLEXES OF THE AMINO-TERMINAL SH2 DOMAIN OF THE SYP TYROSINE PHOSPHATASE
5561:: CRYSTAL STRUCTURES OF PEPTIDE COMPLEXES OF THE AMINO-TERMINAL SH2 DOMAIN OF THE SYP TYROSINE PHOSPHATASE
5546:: CRYSTAL STRUCTURES OF PEPTIDE COMPLEXES OF THE AMINO-TERMINAL SH2 DOMAIN OF THE SYP TYROSINE PHOSPHATASE
4405:
Kharitonenkov A, Schnekenburger J, Chen Z, Knyazev P, Ali S, Zwick E, White M, Ullrich A (December 1995).
1858:
1834:
4015:"Direct binding of Shc, Grb2, SHP-2 and p40 to the murine granulocyte colony-stimulating factor receptor"
7326:
7245:
7028:
6925:
6717:
6678:
6542:
6464:
5616:
3873:"Molecular characterization of specific interactions between SHP-2 phosphatase and JAK tyrosine kinases"
3465:"Determination of Gab1 (Grb2-associated binder-1) interaction with insulin receptor-signaling molecules"
1870:
1714:
336:
7398:
5399:"Shp2-mediated molecular signaling in control of embryonic stem cell self-renewal and differentiation"
7211:
7046:
7041:
6974:
6634:
6459:
5503:
4538:"The insulin receptor substrate 1 associates with the SH2-containing phosphotyrosine phosphatase Syp"
3967:
3745:"SHP2 and SOCS3 contribute to Tyr-759-dependent attenuation of interleukin-6 signaling through gp130"
2373:
2300:
3161:
Tomic S, Greiser U, Lammers R, Kharitonenkov A, Imyanitov E, Ullrich A, Böhmer FD (September 1995).
699:
251:
7362:
7066:
7036:
6840:
6835:
6759:
6695:
6481:
5432:
Edouard T, Montagner A, Dance M, Conte F, Yart A, Parfait B, Tauber M, Salles JP, Raynal P (2007).
4793:"Structural and functional consequences of tyrosine phosphorylation in the LRP1 cytoplasmic domain"
4636:
4619:
4320:"Constitutively active SHP2 cooperates with HoxA10 overexpression to induce acute myeloid leukemia"
2124:
1887:
1696:
1508:
1487:
1475:
1445:
1420:
1416:
7295:
7199:
7051:
6532:
6522:
6400:
6161:
5471:
5314:
5209:
4318:
Wang H, Lindsey S, Konieczna I, Bei L, Horvath E, Huang W, Saberwal G, Eklund EA (January 2009).
4177:
3955:"Protein-tyrosine-phosphatase SHPTP2 couples platelet-derived growth factor receptor beta to Ras"
3827:"Transmembrane domain of gp130 contributes to intracellular signal transduction in hepatic cells"
3658:
3463:
Rocchi S, Tartare-Deckert S, Murdaca J, Holgado-Madruga M, Wong AJ, Van
Obberghen E (July 1998).
3350:
2971:
2811:"The ubiquitously expressed Syp phosphatase interacts with c-kit and Grb2 in hematopoietic cells"
2742:
2494:
2269:
2138:
1792:
296:
6194:
2760:
Tanaka Y, Tanaka N, Saeki Y, Tanaka K, Murakami M, Hirano T, Ishii N, Sugamura K (August 2008).
1512:
1483:
1479:
1449:
1424:
1412:
1408:
5184:
Hatakeyama M (September 2004). "Oncogenic mechanisms of the
Helicobacter pylori CagA protein".
7558:
7000:
6945:
6913:
6788:
6507:
5463:
5420:
5385:
5364:
5335:
5306:
5276:
5255:
5245:
5201:
5166:
5125:
5081:
5045:
4996:
4955:
4914:
4873:
4824:
4773:
4718:
4677:
4641:
4600:
4559:
4518:
4477:
4428:
4387:
4351:
4300:
4259:
4218:
4169:
4128:
4087:
4046:
3995:
3935:
3894:
3848:
3807:
3766:
3717:
3699:
3650:
3615:
3574:
3525:
3486:
3445:
3404:
3342:
3307:
3264:
3256:
3216:
3184:
3143:
3094:
3053:
3012:
2963:
2922:
2881:
2832:
2791:
2734:
2680:
2626:
2577:
2535:
2486:
2450:
2399:
2328:
2261:
1846:
244:
60:
6091:
6086:
6055:
5972:
2514:"PTPN11 (Shp2) mutations in LEOPARD syndrome have dominant negative, not activating, effects"
7341:
7336:
7310:
7238:
7076:
6656:
6561:
6556:
6512:
6287:
6277:
6267:
6262:
6141:
6136:
6131:
6116:
6096:
6076:
5913:
5869:
5788:
5453:
5445:
5410:
5356:
5298:
5237:
5193:
5156:
5115:
5104:"Cytosolic tyrosine dephosphorylation of STAT5. Potential role of SHP-2 in STAT5 regulation"
5073:
5035:
5027:
4986:
4945:
4904:
4863:
4855:
4814:
4804:
4763:
4753:
4708:
4669:
4631:
4590:
4549:
4508:
4467:
4459:
4418:
4379:
4341:
4331:
4290:
4249:
4208:
4159:
4118:
4077:
4036:
4026:
3985:
3975:
3925:
3884:
3838:
3797:
3756:
3707:
3689:
3642:
3605:
3564:
3556:
3517:
3476:
3435:
3394:
3386:
3334:
3297:
3248:
3174:
3133:
3125:
3084:
3043:
3002:
2953:
2912:
2871:
2863:
2822:
2781:
2773:
2724:
2714:
2670:
2662:
2616:
2608:
2567:
2525:
2478:
2440:
2430:
2389:
2381:
2347:"Entrez Gene: PTPN11 protein tyrosine phosphatase, non-receptor type 11 (Noonan syndrome 1)"
2318:
2308:
2253:
2119:
2001:
1995:
1977:
1821:
Patients with a subset of Noonan syndrome PTPN11 mutations also have a higher prevalence of
1785:
541:
472:
416:
371:
292:
6252:
6247:
6237:
6232:
6151:
5957:
7388:
7372:
7285:
7178:
6992:
6908:
6903:
6898:
6811:
6806:
6566:
6444:
6376:
6184:
2597:"High NPM1 mutant allele burden at diagnosis predicts unfavorable outcomes in de novo AML"
1781:
1639:
516:
3071:
Huber M, Izzi L, Grondin P, Houde C, Kunath T, Veillette A, Beauchemin N (January 1999).
2809:
Tauchi T, Feng GS, Marshall MS, Shen R, Mantel C, Pawson T, Broxmeyer HE (October 1994).
5458:
5433:
5347:
Tartaglia M, Gelb BD (2005). "Germ-line and somatic PTPN11 mutations in human disease".
3971:
2729:
2702:
2377:
2304:
7537:
7426:
7367:
7151:
7146:
7141:
6527:
6502:
6498:
6471:
6454:
6217:
4868:
4843:
4819:
4792:
4346:
4319:
3138:
3113:
2786:
2761:
2675:
2650:
2621:
2596:
2445:
2418:
2394:
2361:
2146:
2025:
2019:
1866:
1862:
5040:
5015:
4909:
4892:
4758:
4741:
4554:
4537:
4031:
4014:
3712:
3677:
3676:
Wolf I, Jenkins BJ, Liu Y, Seiffert M, Custodio JM, Young P, Rohrschneider LR (2002).
3646:
3569:
3544:
3521:
3399:
3374:
3252:
2958:
2941:
2876:
2851:
2827:
2810:
1208:
1203:
1198:
1193:
1188:
1183:
1178:
1173:
1168:
1163:
1158:
1153:
1148:
1143:
1138:
1133:
1128:
1123:
1118:
1113:
1108:
1103:
1098:
1093:
1088:
1083:
1078:
1073:
1068:
1063:
1058:
1053:
1048:
1043:
1038:
1033:
1028:
1023:
1018:
1013:
1008:
1003:
998:
993:
988:
983:
978:
973:
968:
963:
958:
953:
948:
943:
938:
933:
928:
923:
918:
913:
908:
903:
898:
893:
888:
872:
867:
862:
857:
852:
847:
831:
826:
821:
816:
811:
806:
801:
796:
791:
786:
781:
776:
771:
766:
761:
756:
751:
746:
741:
7547:
7331:
7290:
6892:
6801:
6551:
4472:
4447:
4041:
3990:
3954:
2719:
2323:
2288:
2070:
1838:
1826:
728:
5475:
5318:
4181:
3662:
3354:
2975:
2498:
1849:. Recently, a relatively high prevalence of PTPN11 mutations (24%) were detected by
1721:
7280:
7019:
6950:
6673:
6594:
6209:
5213:
2746:
2273:
2142:
534:
313:
3694:
2572:
2555:
276:
4446:
Mañes S, Mira E, Gómez-Mouton C, Zhao ZJ, Lacalle RA, Martínez-A C (April 1999).
2612:
2435:
300:
7504:
7439:
7275:
7194:
6965:
6859:
6722:
6688:
6626:
6392:
6179:
5241:
5236:. Advances in Experimental Medicine and Biology. Vol. 476. pp. 35–46.
4713:
4696:
3802:
3785:
3545:"PKB-mediated negative feedback tightly regulates mitogenic signalling via Gab2"
2231:
National Center for
Biotechnology Information, U.S. National Library of Medicine
2213:
National Center for
Biotechnology Information, U.S. National Library of Medicine
1842:
1655:
7532:
5360:
5031:
4742:"LAIR-1, a novel inhibitory receptor expressed on human mononuclear leukocytes"
3726:
and associates transiently with the SH2 domain-containing proteins p85 and SHP2
2385:
617:
7171:
5449:
5302:
4973:
Howie D, Simarro M, Sayos J, Guirado M, Sancho J, Terhorst C (February 2000).
2850:
Kozlowski M, Larose L, Lee F, Le DM, Rottapel R, Siminovitch KA (April 1998).
2666:
1765:
433:
330:
280:
4595:
4578:
4513:
4496:
4423:
4406:
4295:
4278:
4213:
4196:
4123:
4106:
3843:
3826:
3703:
3610:
3593:
3260:
3179:
3162:
3007:
2990:
2313:
2133:
of the host cell. Epidemiological studies have shown roles of cagA- positive
7478:
7452:
7084:
6883:
6796:
6423:
4991:
4974:
4618:
Tauchi T, Damen JE, Toyama K, Feng GS, Broxmeyer HE, Krystal G (June 1996).
3980:
3560:
3302:
3283:
3048:
3031:
2130:
1788:. At least 79 disease-causing mutations in this gene have been discovered.
1238:
677:
555:
500:
487:
399:
386:
288:
5467:
5424:
5389:
5368:
5339:
5310:
5280:
5259:
5205:
5170:
5161:
5144:
5129:
5077:
5049:
5000:
4959:
4950:
4933:
4877:
4828:
4809:
4722:
4673:
4481:
4383:
4355:
4336:
4263:
4254:
4237:
4173:
4132:
4082:
4065:
3939:
3930:
3913:
3889:
3872:
3811:
3770:
3761:
3744:
3721:
3678:"Gab3, a New DOS/Gab Family Member, Facilitates Macrophage Differentiation"
3654:
3619:
3578:
3529:
3481:
3464:
3449:
3440:
3423:
3346:
3338:
3311:
3268:
3147:
2967:
2926:
2917:
2900:
2795:
2738:
2684:
2630:
2581:
2539:
2530:
2513:
2490:
2454:
2403:
5415:
5398:
5120:
5103:
5085:
4918:
4859:
4777:
4681:
4645:
4604:
4563:
4522:
4463:
4432:
4391:
4304:
4222:
4091:
4050:
3999:
3898:
3852:
3490:
3408:
3390:
3188:
3098:
3089:
3072:
3057:
3016:
2885:
2867:
2836:
2332:
2265:
1576:
1571:
7129:
7124:
7119:
6940:
6575:
6427:
5647:
4577:
Myers MG, Mendez R, Shi P, Pierce JH, Rhoads R, White MF (October 1998).
4164:
4147:
2777:
2651:"Ptpn11/Shp2 acts as a tumor suppressor in hepatocellular carcinogenesis"
1918:
1878:
1865:. However, it has been reported that PTPN11/Shp2 can act as either tumor
1830:
1560:
1383:
1364:
3912:
Ganju RK, Brubaker SA, Chernock RD, Avraham S, Groopman JE (June 2000).
7136:
7114:
7109:
6727:
3129:
2762:"c-Cbl-dependent monoubiquitination and lysosomal degradation of gp130"
2703:"Helicobacter pylori CagA: a new paradigm for bacterial carcinogenesis"
2257:
1936:
1350:
1305:
4768:
4236:
Stofega MR, Herrington J, Billestrup N, Carter-Su C (September 2000).
2512:
Kontaridis MI, Swanson KD, David FS, Barford D, Neel BG (March 2006).
1804:
necessary for the enzyme to maintain its auto-inhibited conformation.
221:
217:
213:
209:
205:
201:
197:
193:
189:
185:
181:
177:
173:
169:
165:
161:
157:
153:
149:
145:
141:
137:
133:
129:
125:
121:
117:
113:
109:
105:
101:
97:
93:
89:
7491:
7261:
7156:
7089:
6982:
6580:
6488:
6346:
6341:
6336:
6282:
6272:
6146:
6126:
6121:
6111:
6106:
6101:
6081:
6045:
6040:
6024:
6019:
6014:
5967:
5962:
5884:
5879:
5874:
5864:
5859:
5854:
5849:
5783:
5748:
4277:
Moutoussamy S, Renaudie F, Lago F, Kelly PA, Finidori J (June 1998).
2105:
2099:
2075:
2043:
1620:
1544:
1260:
5197:
17:
2482:
7465:
7098:
6849:
6845:
6331:
6257:
6242:
6227:
6222:
6199:
6156:
6071:
6050:
5952:
5947:
5942:
5937:
5932:
5927:
5922:
5839:
5834:
5829:
5824:
5819:
5814:
5809:
5804:
5778:
5773:
5768:
5763:
5758:
5753:
5743:
5738:
5733:
5728:
5723:
5718:
5713:
5708:
5703:
5698:
5693:
5688:
3914:"Beta-chemokine receptor CCR5 signals through SHP1, SHP2, and Syk"
2093:
2081:
2064:
2058:
2031:
1924:
1874:
1223:
1219:
6830:
6707:
6700:
6644:
6639:
6381:
6371:
6366:
6313:
6189:
5998:
5993:
5988:
3959:
3953:
Bennett AM, Tang TL, Sugimoto S, Walsh CT, Neel BG (July 1994).
3114:"Phosphotyrosine interactome of the ErbB-receptor kinase family"
2087:
2049:
2037:
2013:
1983:
1971:
1965:
1959:
1953:
1930:
1912:
1892:
1854:
1780:
Missense mutations in the PTPN11 locus are associated with both
1761:
1627:
268:
43:
7234:
6396:
5620:
5499:
708:
5495:
5143:
Chughtai N, Schimchowitsch S, Lebrun JJ, Ali S (August 2002).
4105:
Tang H, Zhao ZJ, Huang XY, Landon EJ, Inagami T (April 1999).
2053:
1649:
4013:
Ward AC, Monkhouse JL, Hamilton JA, Csar XF (November 1998).
1825:(JMML). Activating Shp2 mutations have also been detected in
1764:, possesses a domain structure that consists of two tandem
777:
non-membrane spanning protein tyrosine phosphatase activity
7230:
1034:
regulation of type I interferon-mediated signaling pathway
3422:
Saito Y, Hojo Y, Tanimoto T, Abe J, Berk BC (June 2002).
3373:
Hadari YR, Kouhara H, Lax I, Schlessinger J (July 1998).
1209:
positive regulation of insulin receptor signaling pathway
1104:
negative regulation of cell adhesion mediated by integrin
1064:
platelet-derived growth factor receptor signaling pathway
2989:
Hua CT, Gamble JR, Vadas MA, Jackson DE (October 1998).
524:
3282:
Delahaye L, Rocchi S, Van
Obberghen E (February 2000).
1199:
positive regulation of tumor necrosis factor production
787:
phosphatidylinositol-4,5-bisphosphate 3-kinase activity
4536:
Kuhné MR, Pawson T, Lienhard GE, Feng GS (June 1993).
7521:
3030:
Jackson DE, Ward CM, Wang R, Newman PJ (March 1997).
1605:
Src homology region 2 domain-containing phosphatase-2
1159:
cellular response to epidermal growth factor stimulus
1144:
phosphatidylinositol-3-phosphate biosynthetic process
689:
3284:"Potential involvement of FRS2 in insulin signaling"
7417:
7381:
7350:
7319:
7268:
7187:
7075:
7027:
7013:
6991:
6973:
6959:
6939:
6878:
6782:
6621:
6589:
6439:
6357:
6322:
6304:
6208:
6170:
6064:
6033:
6007:
5981:
5912:
5897:
5797:
5679:
5672:
5663:
1501:
1468:
1438:
1401:
1134:
phosphatidylinositol phosphate biosynthetic process
1019:
fibroblast growth factor receptor signaling pathway
4791:Betts GN, van der Geer P, Komives EA (June 2008).
2899:Ilan N, Cheung L, Pinter E, Madri JA (July 2000).
2187:
2185:
2183:
2166:
2164:
2162:
929:epidermal growth factor receptor signaling pathway
6718:Fructose 6-P,2-kinase:fructose 2,6-bisphosphatase
3207:L.A. Lai, C. Zhao, E.E. Zhang, G.S. Feng (2004).
2644:
2642:
2640:
1589:Tyrosine-protein phosphatase non-receptor type 11
1189:positive regulation of interferon-beta production
1164:positive regulation of protein kinase B signaling
1109:regulation of protein-containing complex assembly
471:
370:
5491:GeneReviews/NCBI/NIH/UW entry on Noonan syndrome
3871:Yin T, Shen R, Feng GS, Yang YC (January 1997).
3202:
3200:
3198:
2287:Freeman RM, Plutzky J, Neel BG (December 1992).
909:regulation of cell adhesion mediated by integrin
5097:
5095:
3738:
3736:
3734:
3502:
3500:
2696:
2694:
1194:positive regulation of interleukin-6 production
934:negative regulation of growth hormone secretion
4637:10.1182/blood.V87.11.4495.bloodjournal87114495
4146:Zhang S, Mantel C, Broxmeyer HE (March 1999).
3866:
3864:
3862:
3368:
3366:
3364:
2192:GRCm38: Ensembl release 89: ENSMUSG00000043733
1054:homeostasis of number of cells within a tissue
7246:
6408:
5632:
5511:
1129:multicellular organismal reproductive process
8:
3592:Zhao C, Yu DH, Shen R, Feng GS (July 1999).
3234:
3232:
3211:. In Joaquín Ariño, Denis Alexander (eds.).
1760:This phosphatase, along with its paralogue,
1154:positive regulation of ERK1 and ERK2 cascade
5102:Yu CL, Jin YJ, Burakoff SJ (January 2000).
2171:GRCh38: Ensembl release 89: ENSG00000179295
1684:. Unsourced material may be challenged and
1139:neurotrophin TRK receptor signaling pathway
959:regulation of multicellular organism growth
7253:
7239:
7231:
7024:
6970:
6956:
6415:
6401:
6393:
5909:
5676:
5669:
5639:
5625:
5617:
5518:
5504:
5496:
4736:Meyaard L, Adema GJ, Chang C, Woollatt E,
1234:
899:positive regulation of signal transduction
724:
512:
411:
308:
78:
5457:
5414:
5160:
5119:
5039:
4990:
4949:
4908:
4867:
4818:
4808:
4767:
4757:
4712:
4635:
4594:
4553:
4512:
4471:
4422:
4345:
4335:
4294:
4253:
4212:
4163:
4122:
4081:
4040:
4030:
3989:
3979:
3929:
3888:
3842:
3801:
3760:
3711:
3693:
3609:
3568:
3480:
3439:
3398:
3301:
3178:
3137:
3088:
3047:
3006:
2957:
2916:
2875:
2826:
2785:
2728:
2718:
2674:
2620:
2571:
2529:
2444:
2434:
2393:
2360:Šimčíková D, Heneberg P (December 2019).
2322:
2312:
1748:Learn how and when to remove this message
1069:negative regulation of cortisol secretion
1004:positive regulation of mitotic cell cycle
949:regulation of protein export from nucleus
4740:, Lanier LL, Phillips JH (August 1997).
1184:cellular response to mechanical stimulus
1174:interleukin-6-mediated signaling pathway
1099:positive regulation of hormone secretion
1084:negative regulation of hormone secretion
904:negative regulation of insulin secretion
792:1-phosphatidylinositol-3-kinase activity
7528:
6608:Ubiquitin carboxy-terminal hydrolase L1
5533:
2158:
1776:Genetic diseases associated with PTPN11
4064:Tang J, Feng GS, Li W (October 1997).
1179:cellular response to cytokine stimulus
33:
7188:either deoxy- or ribo-
2008:Insulin-like growth factor 1 receptor
1204:positive regulation of glucose import
802:protein tyrosine phosphatase activity
476:
437:
432:
375:
334:
329:
7:
6769:Protein serine/threonine phosphatase
5349:European Journal of Medical Genetics
1682:adding citations to reliable sources
919:intestinal epithelial cell migration
6870:Cyclic nucleotide phosphodiesterase
6864:Clostridium perfringens alpha toxin
6665:Tartrate-resistant acid phosphatase
3209:"14 The Shp-2 tyrosine phosphatase"
3112:Schulze WX, Deng L, Mann M (2005).
1642:as well as acute myeloid leukemia.
1169:cytokine-mediated signaling pathway
1074:peptidyl-tyrosine dephosphorylation
1049:Bergmann glial cell differentiation
1044:integrin-mediated signaling pathway
747:phosphoprotein phosphatase activity
570:dorsal motor nucleus of vagus nerve
6713:Pyruvate dehydrogenase phosphatase
4195:Wong L, Johnson GR (August 1996).
3825:Kim H, Baumann H (December 1997).
3543:Lynch DK, Daly RJ (January 2002).
1498:
1465:
1435:
1398:
1374:
1355:
1329:
1310:
1284:
1265:
1039:hormone-mediated signaling pathway
914:atrioventricular canal development
822:insulin receptor substrate binding
694:
612:
550:
529:
25:
6613:4-hydroxybenzoyl-CoA thioesterase
1877:pathway and hepatic inflammation/
1823:juvenile myelomonocytic leukemias
1791:It has also been associated with
1623:that in humans is encoded by the
969:ephrin receptor signaling pathway
7531:
5596:
5581:
5566:
5551:
5536:
2720:10.1111/j.1349-7006.2005.00130.x
2701:Hatakeyama M, Higashi H (2005).
1943:Epidermal growth factor receptor
1654:
767:peptide hormone receptor binding
762:receptor tyrosine kinase binding
698:
460:
453:
447:
424:
359:
352:
346:
321:
42:
6931:N-acetylglucosamine-6-sulfatase
6819:Sphingomyelin phosphodiesterase
5378:Pediatric Endocrinology Reviews
1885:(HCC) specimens. The bacterium
1613:protein-tyrosine phosphatase 2C
1597:protein-tyrosine phosphatase 1D
979:DNA damage checkpoint signaling
827:protein tyrosine kinase binding
812:protein domain specific binding
807:phosphotyrosine residue binding
6740:Inositol-phosphate phosphatase
6603:Palmitoyl protein thioesterase
3682:Molecular and Cellular Biology
3241:Trends in Biochemical Sciences
3215:. Springer. pp. 275–299.
2114:H Pylori CagA virulence factor
1089:triglyceride metabolic process
797:cell adhesion molecule binding
709:More reference expression data
678:More reference expression data
1:
7103:RNA-induced silencing complex
5652:protein tyrosine phosphatases
5066:Biochem. Biophys. Res. Commun
4910:10.1016/S0021-9258(20)80562-6
4759:10.1016/S1074-7613(00)80530-0
4662:Biochem. Biophys. Res. Commun
4555:10.1016/S0021-9258(19)50220-4
4372:Biochem. Biophys. Res. Commun
4032:10.1016/S0167-4889(98)00120-7
3960:Proc. Natl. Acad. Sci. U.S.A.
3695:10.1128/MCB.22.1.231-244.2002
3647:10.1016/S0014-5793(01)02373-0
3522:10.1016/S0898-6568(02)00036-0
3253:10.1016/S0968-0004(03)00091-4
2959:10.1016/S0014-5793(99)00446-9
2828:10.1016/S0021-9258(17)31518-1
2573:10.1158/0008-5472.CAN-04-1923
1817:Cancer associated with PTPN11
954:multicellular organism growth
445:
344:
29:Protein-coding gene in humans
7554:Genes on human chromosome 12
7207:Serratia marcescens nuclease
6774:Dual-specificity phosphatase
6764:Protein tyrosine phosphatase
5606:: TYROSINE PHOSPHATASE SHP-2
2613:10.1182/blood-2018-01-828467
2436:10.1371/journal.pgen.1000991
2293:Proc. Natl. Acad. Sci. U.S.A
1632:protein tyrosine phosphatase
817:D1 dopamine receptor binding
632:Epithelium of choroid plexus
598:Epithelium of choroid plexus
6684:Fructose 1,6-bisphosphatase
5242:10.1007/978-1-4615-4221-6_3
4714:10.4049/jimmunol.165.3.1197
3803:10.4049/jimmunol.165.5.2535
1119:cerebellar cortex formation
974:abortive mitotic cell cycle
7575:
5361:10.1016/j.ejmg.2005.03.001
2386:10.1038/s41598-019-54976-4
1851:next-generation sequencing
1530:Chr 12: 112.42 – 112.51 Mb
873:protein-containing complex
644:retinal pigment epithelium
7409:Michaelis–Menten kinetics
6921:Galactosamine-6 sulfatase
6477:6-phosphogluconolactonase
5531:
5450:10.1007/s00018-007-6509-0
5303:10.1007/s00439-002-0803-6
2667:10.1016/j.ccr.2011.03.023
2227:"Mouse PubMed Reference:"
2209:"Human PubMed Reference:"
1903:PTPN11 has been shown to
1813:human genetic syndromes.
1575:
1570:
1566:
1559:
1543:
1537:Chr 5: 121.27 – 121.33 Mb
1524:
1505:
1472:
1461:
1442:
1405:
1394:
1381:
1377:
1362:
1358:
1349:
1336:
1332:
1317:
1313:
1304:
1291:
1287:
1272:
1268:
1259:
1244:
1237:
1233:
1217:
1094:hormone metabolic process
984:protein dephosphorylation
894:megakaryocyte development
727:
723:
706:
697:
688:
675:
640:medullary collecting duct
624:
615:
562:
553:
523:
515:
511:
494:
481:
444:
423:
414:
410:
393:
380:
343:
320:
311:
307:
262:
259:
249:
242:
237:
86:
81:
64:
59:
54:
50:
41:
36:
7301:Diffusion-limited enzyme
6669:Purple acid phosphatases
5032:10.1093/emboj/20.21.5840
4596:10.1074/jbc.273.41.26908
4514:10.1074/jbc.270.32.19151
4424:10.1074/jbc.270.49.29189
4296:10.1074/jbc.273.26.15906
4214:10.1074/jbc.271.35.20981
4124:10.1074/jbc.274.18.12401
3844:10.1074/jbc.272.49.30741
3611:10.1074/jbc.274.28.19649
3180:10.1074/jbc.270.36.21277
3008:10.1074/jbc.273.43.28332
2314:10.1073/pnas.89.23.11239
1883:hepatocellular carcinoma
999:microvillus organization
752:insulin receptor binding
602:external globus pallidus
594:inferior olivary nucleus
566:internal globus pallidus
4992:10.1182/blood.V99.3.957
3981:10.1073/pnas.91.15.7335
3303:10.1210/endo.141.2.7298
3049:10.1074/jbc.272.11.6986
1990:Growth hormone receptor
7094:Microprocessor complex
6733:Beta-propeller phytase
6171:Phosphatase and tensin
5798:Non receptor type PTPs
5269:Recent Prog. Horm. Res
5162:10.1074/jbc.M200156200
5078:10.1006/bbrc.1997.7480
4951:10.1074/jbc.M003428200
4810:10.1074/jbc.M709514200
4674:10.1006/bbrc.1996.1626
4384:10.1006/bbrc.1994.1297
4337:10.1074/jbc.M804704200
4255:10.1210/mend.14.9.0513
4083:10.1038/sj.onc.1201351
4019:Biochim. Biophys. Acta
3931:10.1074/jbc.M000689200
3890:10.1074/jbc.272.2.1032
3762:10.1074/jbc.M210552200
3482:10.1210/mend.12.7.0141
3441:10.1074/jbc.M200605200
3339:10.1038/sj.onc.1204290
2918:10.1074/jbc.M001857200
2531:10.1074/jbc.M513068200
2137:in the development of
2118:CagA is a protein and
1859:acute myeloid leukemia
1835:acute myeloid leukemia
1646:Structure and function
1079:ERBB signaling pathway
832:protein kinase binding
7394:Eadie–Hofstee diagram
7327:Allosteric regulation
7029:Endodeoxyribonuclease
6926:Iduronate-2-sulfatase
6679:Glucose 6-phosphatase
6465:Butyrylcholinesterase
5416:10.1038/sj.cr.7310140
5186:Nature Reviews Cancer
5121:10.1074/jbc.275.1.599
4860:10.1083/jcb.152.2.325
4701:Journal of Immunology
4464:10.1128/mcb.19.4.3125
4042:10536/DRO/DU:30096477
3790:Journal of Immunology
3561:10.1093/emboj/21.1.72
3391:10.1128/MCB.18.7.3966
3090:10.1074/jbc.274.1.335
2868:10.1128/MCB.18.4.2089
1059:inner ear development
1009:genitalia development
742:phospholipase binding
337:Chromosome 12 (human)
7404:Lineweaver–Burk plot
7212:Micrococcal nuclease
7047:Deoxyribonuclease IV
7042:Deoxyribonuclease II
6975:Exodeoxyribonuclease
6635:Alkaline phosphatase
6460:Acetylcholinesterase
4165:10.1002/jlb.65.3.372
3213:Protein phosphatases
2778:10.1128/MCB.01784-07
1678:improve this section
989:T cell costimulation
757:phosphatase activity
439:Chromosome 5 (mouse)
82:List of PDB id codes
55:Available structures
7067:UvrABC endonuclease
7037:Deoxyribonuclease I
6760:Protein phosphatase
6696:Protein phosphatase
6494:Bile salt-dependent
6482:PAF acetylhydrolase
5438:Cell. Mol. Life Sci
3972:1994PNAS...91.7335B
2378:2019NatSR...918577S
2305:1992PNAS...8911239F
2125:Helicobacter pylori
1888:Helicobacter pylori
1124:leukocyte migration
1014:platelet activation
944:glucose homeostasis
660:endocardial cushion
586:subthalamic nucleus
7363:Enzyme superfamily
7296:Enzyme promiscuity
7200:Mung bean nuclease
7059:Restriction enzyme
7052:Restriction enzyme
5681:Receptor type PTPs
3130:10.1038/msb4100012
2366:Scientific Reports
2258:10.1038/ng1294-357
2139:atrophic gastritis
1793:metachondromatosis
1339:ENSMUSG00000043733
1114:face morphogenesis
994:platelet formation
882:Biological process
841:Cellular component
782:hydrolase activity
735:Molecular function
301:PTPN11 - orthologs
7519:
7518:
7228:
7227:
7224:
7223:
7220:
7219:
7009:
7008:
7001:Oligonucleotidase
6946:deoxyribonuclease
6914:Steroid sulfatase
6789:Phosphodiesterase
6518:Hormone-sensitive
6390:
6389:
6300:
6299:
6296:
6295:
5914:MAPK phosphatases
5893:
5892:
5614:
5613:
5251:978-1-4613-6895-3
3222:978-3-540-20560-9
2607:(25): 2816–2825.
2147:gastric carcinoma
1847:colorectal cancer
1758:
1757:
1750:
1732:
1586:
1585:
1582:
1581:
1555:
1554:
1520:
1519:
1495:
1494:
1457:
1456:
1432:
1431:
1390:
1389:
1371:
1370:
1345:
1344:
1326:
1325:
1300:
1299:
1281:
1280:
1229:
1228:
1029:brain development
1024:heart development
889:dephosphorylation
719:
718:
715:
714:
684:
683:
671:
670:
609:
608:
507:
506:
406:
405:
233:
232:
229:
228:
65:Ortholog search:
16:(Redirected from
7566:
7536:
7535:
7527:
7399:Hanes–Woolf plot
7342:Enzyme activator
7337:Enzyme inhibitor
7311:Enzyme catalysis
7255:
7248:
7241:
7232:
7077:Endoribonuclease
7063:
7057:
7025:
6971:
6957:
6657:Acid phosphatase
6538:Monoacylglycerol
6448:ester hydrolases
6417:
6410:
6403:
6394:
6173:homologs (PTENs)
5910:
5677:
5670:
5641:
5634:
5627:
5618:
5600:
5585:
5570:
5555:
5540:
5520:
5513:
5506:
5497:
5479:
5461:
5428:
5418:
5397:Feng GS (2007).
5393:
5372:
5343:
5322:
5284:
5263:
5218:
5217:
5181:
5175:
5174:
5164:
5155:(34): 31107–14.
5140:
5134:
5133:
5123:
5099:
5090:
5089:
5060:
5054:
5053:
5043:
5011:
5005:
5004:
4994:
4970:
4964:
4963:
4953:
4944:(36): 27845–50.
4929:
4923:
4922:
4912:
4903:(29): 21478–81.
4888:
4882:
4881:
4871:
4839:
4833:
4832:
4822:
4812:
4803:(23): 15656–64.
4788:
4782:
4781:
4771:
4761:
4733:
4727:
4726:
4716:
4692:
4686:
4685:
4656:
4650:
4649:
4639:
4630:(11): 4495–501.
4615:
4609:
4608:
4598:
4589:(41): 26908–14.
4574:
4568:
4567:
4557:
4548:(16): 11479–81.
4533:
4527:
4526:
4516:
4492:
4486:
4485:
4475:
4443:
4437:
4436:
4426:
4417:(49): 29189–93.
4402:
4396:
4395:
4366:
4360:
4359:
4349:
4339:
4315:
4309:
4308:
4298:
4289:(26): 15906–12.
4274:
4268:
4267:
4257:
4233:
4227:
4226:
4216:
4192:
4186:
4185:
4167:
4143:
4137:
4136:
4126:
4102:
4096:
4095:
4085:
4061:
4055:
4054:
4044:
4034:
4010:
4004:
4003:
3993:
3983:
3950:
3944:
3943:
3933:
3909:
3903:
3902:
3892:
3868:
3857:
3856:
3846:
3822:
3816:
3815:
3805:
3781:
3775:
3774:
3764:
3740:
3729:
3728:
3715:
3697:
3673:
3667:
3666:
3630:
3624:
3623:
3613:
3604:(28): 19649–54.
3589:
3583:
3582:
3572:
3540:
3534:
3533:
3504:
3495:
3494:
3484:
3460:
3454:
3453:
3443:
3434:(26): 23216–22.
3419:
3413:
3412:
3402:
3370:
3359:
3358:
3322:
3316:
3315:
3305:
3279:
3273:
3272:
3236:
3227:
3226:
3204:
3193:
3192:
3182:
3173:(36): 21277–84.
3158:
3152:
3151:
3141:
3109:
3103:
3102:
3092:
3068:
3062:
3061:
3051:
3027:
3021:
3020:
3010:
3001:(43): 28332–40.
2986:
2980:
2979:
2961:
2937:
2931:
2930:
2920:
2911:(28): 21435–43.
2896:
2890:
2889:
2879:
2847:
2841:
2840:
2830:
2821:(40): 25206–11.
2806:
2800:
2799:
2789:
2757:
2751:
2750:
2732:
2722:
2698:
2689:
2688:
2678:
2646:
2635:
2634:
2624:
2592:
2586:
2585:
2575:
2550:
2544:
2543:
2533:
2509:
2503:
2502:
2465:
2459:
2458:
2448:
2438:
2414:
2408:
2407:
2397:
2357:
2351:
2350:
2343:
2337:
2336:
2326:
2316:
2299:(23): 11239–43.
2284:
2278:
2277:
2241:
2235:
2234:
2223:
2217:
2216:
2205:
2199:
2189:
2178:
2168:
2120:virulence factor
2002:Insulin receptor
1978:Glycoprotein 130
1808:Leopard syndrome
1786:Leopard syndrome
1753:
1746:
1742:
1739:
1733:
1731:
1690:
1658:
1650:
1595:) also known as
1568:
1567:
1539:
1532:
1515:
1499:
1490:
1466:
1462:RefSeq (protein)
1452:
1436:
1427:
1399:
1375:
1356:
1330:
1311:
1285:
1266:
1235:
964:lipid metabolism
725:
711:
702:
695:
680:
664:primitive streak
648:substantia nigra
620:
618:Top expressed in
613:
574:Brodmann area 23
558:
556:Top expressed in
551:
530:
513:
503:
490:
479:
464:
457:
451:
440:
428:
412:
402:
389:
378:
363:
356:
350:
339:
325:
309:
303:
254:
247:
224:
79:
73:
52:
51:
46:
34:
21:
7574:
7573:
7569:
7568:
7567:
7565:
7564:
7563:
7544:
7543:
7542:
7530:
7522:
7520:
7515:
7427:Oxidoreductases
7413:
7389:Enzyme kinetics
7377:
7373:List of enzymes
7346:
7315:
7286:Catalytic triad
7264:
7259:
7229:
7216:
7183:
7071:
7061:
7055:
7018:
7005:
6993:Exoribonuclease
6987:
6964:
6948:
6944:
6935:
6909:Arylsulfatase L
6904:Arylsulfatase B
6899:Arylsulfatase A
6874:
6787:
6778:
6617:
6585:
6447:
6435:
6421:
6391:
6386:
6353:
6318:
6292:
6204:
6172:
6166:
6060:
6029:
6003:
5977:
5905:
5902:
5899:
5889:
5793:
5659:
5645:
5615:
5610:
5607:
5601:
5592:
5586:
5577:
5571:
5562:
5556:
5547:
5541:
5527:
5524:
5487:
5482:
5444:(13): 1585–90.
5431:
5396:
5375:
5346:
5325:
5287:
5266:
5252:
5231:
5227:
5225:Further reading
5222:
5221:
5198:10.1038/nrc1433
5183:
5182:
5178:
5142:
5141:
5137:
5101:
5100:
5093:
5062:
5061:
5057:
5026:(21): 5840–52.
5013:
5012:
5008:
4972:
4971:
4967:
4931:
4930:
4926:
4890:
4889:
4885:
4841:
4840:
4836:
4790:
4789:
4785:
4735:
4734:
4730:
4707:(3): 1197–209.
4694:
4693:
4689:
4658:
4657:
4653:
4617:
4616:
4612:
4576:
4575:
4571:
4535:
4534:
4530:
4507:(32): 19151–7.
4494:
4493:
4489:
4452:Mol. Cell. Biol
4445:
4444:
4440:
4404:
4403:
4399:
4368:
4367:
4363:
4317:
4316:
4312:
4276:
4275:
4271:
4242:Mol. Endocrinol
4235:
4234:
4230:
4207:(35): 20981–4.
4194:
4193:
4189:
4152:J. Leukoc. Biol
4145:
4144:
4140:
4117:(18): 12401–7.
4104:
4103:
4099:
4076:(15): 1823–32.
4063:
4062:
4058:
4012:
4011:
4007:
3952:
3951:
3947:
3924:(23): 17263–8.
3911:
3910:
3906:
3870:
3869:
3860:
3837:(49): 30741–7.
3824:
3823:
3819:
3783:
3782:
3778:
3742:
3741:
3732:
3675:
3674:
3670:
3632:
3631:
3627:
3591:
3590:
3586:
3542:
3541:
3537:
3506:
3505:
3498:
3469:Mol. Endocrinol
3462:
3461:
3457:
3421:
3420:
3416:
3379:Mol. Cell. Biol
3372:
3371:
3362:
3333:(16): 1929–38.
3324:
3323:
3319:
3281:
3280:
3276:
3238:
3237:
3230:
3223:
3206:
3205:
3196:
3160:
3159:
3155:
3118:Mol. Syst. Biol
3111:
3110:
3106:
3070:
3069:
3065:
3042:(11): 6986–93.
3029:
3028:
3024:
2988:
2987:
2983:
2939:
2938:
2934:
2898:
2897:
2893:
2856:Mol. Cell. Biol
2849:
2848:
2844:
2808:
2807:
2803:
2772:(15): 4805–18.
2766:Mol. Cell. Biol
2759:
2758:
2754:
2713:(12): 835–843.
2700:
2699:
2692:
2648:
2647:
2638:
2594:
2593:
2589:
2566:(24): 8816–20.
2552:
2551:
2547:
2524:(10): 6785–92.
2511:
2510:
2506:
2467:
2466:
2462:
2429:(6): e1000991.
2416:
2415:
2411:
2359:
2358:
2354:
2345:
2344:
2340:
2286:
2285:
2281:
2243:
2242:
2238:
2225:
2224:
2220:
2207:
2206:
2202:
2190:
2181:
2169:
2160:
2155:
2116:
2111:
1901:
1853:in a cohort of
1819:
1810:
1801:
1799:Noonan syndrome
1782:Noonan syndrome
1778:
1754:
1743:
1737:
1734:
1691:
1689:
1675:
1659:
1648:
1640:Noonan syndrome
1577:View/Edit Mouse
1572:View/Edit Human
1535:
1528:
1525:Location (UCSC)
1511:
1507:
1486:
1482:
1478:
1474:
1448:
1444:
1423:
1419:
1415:
1411:
1407:
1320:ENSG00000179295
1213:
877:
836:
772:protein binding
707:
676:
667:
662:
658:
654:
650:
646:
642:
638:
634:
630:
628:renal corpuscle
616:
605:
600:
596:
592:
590:pars reticulata
588:
584:
582:parietal pleura
580:
578:visceral pleura
576:
572:
568:
554:
498:
485:
477:
467:
466:
465:
458:
438:
415:Gene location (
397:
384:
376:
366:
365:
364:
357:
335:
312:Gene location (
263:
250:
243:
88:
66:
30:
23:
22:
15:
12:
11:
5:
7572:
7570:
7562:
7561:
7556:
7546:
7545:
7541:
7540:
7517:
7516:
7514:
7513:
7500:
7487:
7474:
7461:
7448:
7435:
7421:
7419:
7415:
7414:
7412:
7411:
7406:
7401:
7396:
7391:
7385:
7383:
7379:
7378:
7376:
7375:
7370:
7365:
7360:
7354:
7352:
7351:Classification
7348:
7347:
7345:
7344:
7339:
7334:
7329:
7323:
7321:
7317:
7316:
7314:
7313:
7308:
7303:
7298:
7293:
7288:
7283:
7278:
7272:
7270:
7266:
7265:
7260:
7258:
7257:
7250:
7243:
7235:
7226:
7225:
7222:
7221:
7218:
7217:
7215:
7214:
7209:
7204:
7203:
7202:
7191:
7189:
7185:
7184:
7182:
7181:
7176:
7175:
7174:
7169:
7164:
7159:
7149:
7144:
7139:
7134:
7133:
7132:
7127:
7122:
7117:
7107:
7106:
7105:
7096:
7081:
7079:
7073:
7072:
7070:
7069:
7064:
7049:
7044:
7039:
7033:
7031:
7022:
7011:
7010:
7007:
7006:
7004:
7003:
6997:
6995:
6989:
6988:
6986:
6985:
6979:
6977:
6968:
6954:
6937:
6936:
6934:
6933:
6928:
6923:
6918:
6917:
6916:
6911:
6906:
6901:
6888:
6886:
6876:
6875:
6873:
6872:
6867:
6857:
6852:
6843:
6838:
6833:
6828:
6827:
6826:
6816:
6815:
6814:
6809:
6799:
6793:
6791:
6780:
6779:
6777:
6776:
6771:
6766:
6757:
6756:
6755:
6737:
6736:
6735:
6725:
6720:
6715:
6710:
6705:
6704:
6703:
6693:
6692:
6691:
6681:
6676:
6671:
6654:
6653:
6652:
6647:
6642:
6631:
6629:
6619:
6618:
6616:
6615:
6610:
6605:
6599:
6597:
6587:
6586:
6584:
6583:
6578:
6572:
6571:
6570:
6569:
6564:
6559:
6548:
6547:
6546:
6545:
6543:Diacylglycerol
6540:
6535:
6530:
6525:
6520:
6515:
6510:
6505:
6496:
6485:
6484:
6479:
6474:
6472:Pectinesterase
6469:
6468:
6467:
6462:
6455:Cholinesterase
6451:
6449:
6437:
6436:
6422:
6420:
6419:
6412:
6405:
6397:
6388:
6387:
6385:
6384:
6379:
6374:
6369:
6363:
6361:
6355:
6354:
6352:
6351:
6350:
6349:
6344:
6339:
6328:
6326:
6320:
6319:
6317:
6316:
6310:
6308:
6302:
6301:
6298:
6297:
6294:
6293:
6291:
6290:
6285:
6280:
6275:
6270:
6265:
6260:
6255:
6250:
6245:
6240:
6235:
6230:
6225:
6220:
6214:
6212:
6206:
6205:
6203:
6202:
6197:
6192:
6187:
6182:
6176:
6174:
6168:
6167:
6165:
6164:
6159:
6154:
6149:
6144:
6139:
6134:
6129:
6124:
6119:
6114:
6109:
6104:
6099:
6094:
6089:
6084:
6079:
6074:
6068:
6066:
6062:
6061:
6059:
6058:
6053:
6048:
6043:
6037:
6035:
6031:
6030:
6028:
6027:
6022:
6017:
6011:
6009:
6005:
6004:
6002:
6001:
5996:
5991:
5985:
5983:
5979:
5978:
5976:
5975:
5970:
5965:
5960:
5955:
5950:
5945:
5940:
5935:
5930:
5925:
5919:
5917:
5907:
5895:
5894:
5891:
5890:
5888:
5887:
5882:
5877:
5872:
5867:
5862:
5857:
5852:
5847:
5842:
5837:
5832:
5827:
5822:
5817:
5812:
5807:
5801:
5799:
5795:
5794:
5792:
5791:
5786:
5781:
5776:
5771:
5766:
5761:
5756:
5751:
5746:
5741:
5736:
5731:
5726:
5721:
5716:
5711:
5706:
5701:
5696:
5691:
5685:
5683:
5674:
5673:Classical PTPs
5667:
5661:
5660:
5646:
5644:
5643:
5636:
5629:
5621:
5612:
5611:
5609:
5608:
5602:
5595:
5593:
5587:
5580:
5578:
5572:
5565:
5563:
5557:
5550:
5548:
5542:
5535:
5532:
5529:
5528:
5525:
5523:
5522:
5515:
5508:
5500:
5494:
5493:
5486:
5485:External links
5483:
5481:
5480:
5429:
5394:
5373:
5344:
5323:
5297:(4–5): 421–7.
5285:
5264:
5250:
5228:
5226:
5223:
5220:
5219:
5176:
5135:
5114:(1): 599–604.
5091:
5055:
5006:
4965:
4924:
4883:
4834:
4783:
4728:
4687:
4651:
4610:
4569:
4528:
4487:
4458:(4): 3125–35.
4438:
4397:
4361:
4330:(4): 2549–67.
4310:
4269:
4248:(9): 1338–50.
4228:
4187:
4138:
4097:
4056:
4005:
3966:(15): 7335–9.
3945:
3904:
3858:
3817:
3796:(5): 2535–43.
3776:
3730:
3688:(1): 231–244.
3668:
3625:
3584:
3555:(1–2): 72–82.
3535:
3516:(10): 869–78.
3496:
3455:
3414:
3385:(7): 3966–73.
3360:
3317:
3274:
3247:(6): 284–293.
3228:
3221:
3194:
3153:
3104:
3063:
3022:
2981:
2952:(1–2): 77–83.
2932:
2891:
2862:(4): 2089–99.
2842:
2801:
2752:
2707:Cancer Science
2690:
2636:
2587:
2545:
2504:
2483:10.1038/ng1926
2460:
2409:
2352:
2338:
2279:
2236:
2218:
2200:
2179:
2157:
2156:
2154:
2151:
2135:H. pylori
2115:
2112:
2110:
2109:
2103:
2097:
2091:
2085:
2079:
2073:
2068:
2062:
2056:
2047:
2041:
2035:
2029:
2026:Janus kinase 2
2023:
2020:Janus kinase 1
2017:
2011:
2005:
1999:
1993:
1987:
1981:
1975:
1969:
1963:
1957:
1951:
1946:
1940:
1934:
1928:
1922:
1916:
1909:
1900:
1897:
1863:proto-oncogene
1818:
1815:
1809:
1806:
1800:
1797:
1777:
1774:
1756:
1755:
1662:
1660:
1653:
1647:
1644:
1630:. PTPN11 is a
1584:
1583:
1580:
1579:
1574:
1564:
1563:
1557:
1556:
1553:
1552:
1550:
1548:
1541:
1540:
1533:
1526:
1522:
1521:
1518:
1517:
1503:
1502:
1496:
1493:
1492:
1470:
1469:
1463:
1459:
1458:
1455:
1454:
1440:
1439:
1433:
1430:
1429:
1403:
1402:
1396:
1392:
1391:
1388:
1387:
1379:
1378:
1372:
1369:
1368:
1360:
1359:
1353:
1347:
1346:
1343:
1342:
1334:
1333:
1327:
1324:
1323:
1315:
1314:
1308:
1302:
1301:
1298:
1297:
1289:
1288:
1282:
1279:
1278:
1270:
1269:
1263:
1257:
1256:
1251:
1246:
1242:
1241:
1231:
1230:
1227:
1226:
1215:
1214:
1212:
1211:
1206:
1201:
1196:
1191:
1186:
1181:
1176:
1171:
1166:
1161:
1156:
1151:
1146:
1141:
1136:
1131:
1126:
1121:
1116:
1111:
1106:
1101:
1096:
1091:
1086:
1081:
1076:
1071:
1066:
1061:
1056:
1051:
1046:
1041:
1036:
1031:
1026:
1021:
1016:
1011:
1006:
1001:
996:
991:
986:
981:
976:
971:
966:
961:
956:
951:
946:
941:
936:
931:
926:
921:
916:
911:
906:
901:
896:
891:
885:
883:
879:
878:
876:
875:
870:
865:
860:
855:
850:
844:
842:
838:
837:
835:
834:
829:
824:
819:
814:
809:
804:
799:
794:
789:
784:
779:
774:
769:
764:
759:
754:
749:
744:
738:
736:
732:
731:
721:
720:
717:
716:
713:
712:
704:
703:
692:
686:
685:
682:
681:
673:
672:
669:
668:
666:
665:
661:
657:
653:
649:
645:
641:
637:
633:
629:
625:
622:
621:
610:
607:
606:
604:
603:
599:
595:
591:
587:
583:
579:
575:
571:
567:
563:
560:
559:
547:
546:
538:
527:
521:
520:
517:RNA expression
509:
508:
505:
504:
496:
492:
491:
483:
480:
475:
469:
468:
459:
452:
446:
442:
441:
436:
430:
429:
421:
420:
408:
407:
404:
403:
395:
391:
390:
382:
379:
374:
368:
367:
358:
351:
345:
341:
340:
333:
327:
326:
318:
317:
305:
304:
261:
257:
256:
248:
240:
239:
235:
234:
231:
230:
227:
226:
84:
83:
75:
74:
63:
57:
56:
48:
47:
39:
38:
28:
24:
14:
13:
10:
9:
6:
4:
3:
2:
7571:
7560:
7557:
7555:
7552:
7551:
7549:
7539:
7534:
7529:
7525:
7511:
7507:
7506:
7501:
7498:
7494:
7493:
7488:
7485:
7481:
7480:
7475:
7472:
7468:
7467:
7462:
7459:
7455:
7454:
7449:
7446:
7442:
7441:
7436:
7433:
7429:
7428:
7423:
7422:
7420:
7416:
7410:
7407:
7405:
7402:
7400:
7397:
7395:
7392:
7390:
7387:
7386:
7384:
7380:
7374:
7371:
7369:
7368:Enzyme family
7366:
7364:
7361:
7359:
7356:
7355:
7353:
7349:
7343:
7340:
7338:
7335:
7333:
7332:Cooperativity
7330:
7328:
7325:
7324:
7322:
7318:
7312:
7309:
7307:
7304:
7302:
7299:
7297:
7294:
7292:
7291:Oxyanion hole
7289:
7287:
7284:
7282:
7279:
7277:
7274:
7273:
7271:
7267:
7263:
7256:
7251:
7249:
7244:
7242:
7237:
7236:
7233:
7213:
7210:
7208:
7205:
7201:
7198:
7197:
7196:
7193:
7192:
7190:
7186:
7180:
7177:
7173:
7170:
7168:
7165:
7163:
7160:
7158:
7155:
7154:
7153:
7150:
7148:
7145:
7143:
7140:
7138:
7135:
7131:
7128:
7126:
7123:
7121:
7118:
7116:
7113:
7112:
7111:
7108:
7104:
7100:
7097:
7095:
7091:
7088:
7087:
7086:
7083:
7082:
7080:
7078:
7074:
7068:
7065:
7060:
7053:
7050:
7048:
7045:
7043:
7040:
7038:
7035:
7034:
7032:
7030:
7026:
7023:
7021:
7016:
7012:
7002:
6999:
6998:
6996:
6994:
6990:
6984:
6981:
6980:
6978:
6976:
6972:
6969:
6967:
6962:
6958:
6955:
6952:
6947:
6942:
6938:
6932:
6929:
6927:
6924:
6922:
6919:
6915:
6912:
6910:
6907:
6905:
6902:
6900:
6897:
6896:
6895:
6894:
6893:arylsulfatase
6890:
6889:
6887:
6885:
6881:
6877:
6871:
6868:
6865:
6861:
6858:
6856:
6853:
6851:
6847:
6844:
6842:
6839:
6837:
6834:
6832:
6829:
6825:
6822:
6821:
6820:
6817:
6813:
6810:
6808:
6805:
6804:
6803:
6802:Phospholipase
6800:
6798:
6795:
6794:
6792:
6790:
6785:
6781:
6775:
6772:
6770:
6767:
6765:
6761:
6758:
6754:
6750:
6746:
6743:
6742:
6741:
6738:
6734:
6731:
6730:
6729:
6726:
6724:
6721:
6719:
6716:
6714:
6711:
6709:
6706:
6702:
6699:
6698:
6697:
6694:
6690:
6687:
6686:
6685:
6682:
6680:
6677:
6675:
6672:
6670:
6666:
6662:
6658:
6655:
6651:
6648:
6646:
6643:
6641:
6638:
6637:
6636:
6633:
6632:
6630:
6628:
6624:
6620:
6614:
6611:
6609:
6606:
6604:
6601:
6600:
6598:
6596:
6592:
6588:
6582:
6579:
6577:
6574:
6573:
6568:
6565:
6563:
6560:
6558:
6555:
6554:
6553:
6552:Phospholipase
6550:
6549:
6544:
6541:
6539:
6536:
6534:
6531:
6529:
6526:
6524:
6521:
6519:
6516:
6514:
6511:
6509:
6506:
6504:
6500:
6497:
6495:
6492:
6491:
6490:
6487:
6486:
6483:
6480:
6478:
6475:
6473:
6470:
6466:
6463:
6461:
6458:
6457:
6456:
6453:
6452:
6450:
6446:
6442:
6438:
6433:
6429:
6425:
6418:
6413:
6411:
6406:
6404:
6399:
6398:
6395:
6383:
6380:
6378:
6375:
6373:
6370:
6368:
6365:
6364:
6362:
6360:
6356:
6348:
6345:
6343:
6340:
6338:
6335:
6334:
6333:
6330:
6329:
6327:
6325:
6321:
6315:
6312:
6311:
6309:
6307:
6303:
6289:
6286:
6284:
6281:
6279:
6276:
6274:
6271:
6269:
6266:
6264:
6261:
6259:
6256:
6254:
6251:
6249:
6246:
6244:
6241:
6239:
6236:
6234:
6231:
6229:
6226:
6224:
6221:
6219:
6216:
6215:
6213:
6211:
6210:Myotubularins
6207:
6201:
6198:
6196:
6193:
6191:
6188:
6186:
6183:
6181:
6178:
6177:
6175:
6169:
6163:
6160:
6158:
6155:
6153:
6150:
6148:
6145:
6143:
6140:
6138:
6135:
6133:
6130:
6128:
6125:
6123:
6120:
6118:
6115:
6113:
6110:
6108:
6105:
6103:
6100:
6098:
6095:
6093:
6090:
6088:
6085:
6083:
6080:
6078:
6075:
6073:
6070:
6069:
6067:
6065:Atypical DSPs
6063:
6057:
6054:
6052:
6049:
6047:
6044:
6042:
6039:
6038:
6036:
6032:
6026:
6023:
6021:
6018:
6016:
6013:
6012:
6010:
6006:
6000:
5997:
5995:
5992:
5990:
5987:
5986:
5984:
5980:
5974:
5971:
5969:
5966:
5964:
5961:
5959:
5956:
5954:
5951:
5949:
5946:
5944:
5941:
5939:
5936:
5934:
5931:
5929:
5926:
5924:
5921:
5920:
5918:
5915:
5911:
5908:
5904:
5901:dual specific
5896:
5886:
5883:
5881:
5878:
5876:
5873:
5871:
5868:
5866:
5863:
5861:
5858:
5856:
5853:
5851:
5848:
5846:
5843:
5841:
5838:
5836:
5833:
5831:
5828:
5826:
5823:
5821:
5818:
5816:
5813:
5811:
5808:
5806:
5803:
5802:
5800:
5796:
5790:
5787:
5785:
5782:
5780:
5777:
5775:
5772:
5770:
5767:
5765:
5762:
5760:
5757:
5755:
5752:
5750:
5747:
5745:
5742:
5740:
5737:
5735:
5732:
5730:
5727:
5725:
5722:
5720:
5717:
5715:
5712:
5710:
5707:
5705:
5702:
5700:
5697:
5695:
5692:
5690:
5687:
5686:
5684:
5682:
5678:
5675:
5671:
5668:
5666:
5662:
5657:
5653:
5649:
5642:
5637:
5635:
5630:
5628:
5623:
5622:
5619:
5605:
5599:
5594:
5590:
5584:
5579:
5575:
5569:
5564:
5560:
5554:
5549:
5545:
5539:
5534:
5530:
5521:
5516:
5514:
5509:
5507:
5502:
5501:
5498:
5492:
5489:
5488:
5484:
5477:
5473:
5469:
5465:
5460:
5455:
5451:
5447:
5443:
5439:
5435:
5430:
5426:
5422:
5417:
5412:
5408:
5404:
5400:
5395:
5391:
5387:
5384:(4): 669–74.
5383:
5379:
5374:
5370:
5366:
5362:
5358:
5354:
5350:
5345:
5341:
5337:
5333:
5329:
5328:Haematologica
5324:
5320:
5316:
5312:
5308:
5304:
5300:
5296:
5292:
5286:
5282:
5278:
5274:
5270:
5265:
5261:
5257:
5253:
5247:
5243:
5239:
5235:
5230:
5229:
5224:
5215:
5211:
5207:
5203:
5199:
5195:
5192:(9): 688–94.
5191:
5187:
5180:
5177:
5172:
5168:
5163:
5158:
5154:
5150:
5149:J. Biol. Chem
5146:
5139:
5136:
5131:
5127:
5122:
5117:
5113:
5109:
5108:J. Biol. Chem
5105:
5098:
5096:
5092:
5087:
5083:
5079:
5075:
5071:
5067:
5059:
5056:
5051:
5047:
5042:
5037:
5033:
5029:
5025:
5021:
5017:
5010:
5007:
5002:
4998:
4993:
4988:
4985:(3): 957–65.
4984:
4980:
4976:
4969:
4966:
4961:
4957:
4952:
4947:
4943:
4939:
4938:J. Biol. Chem
4935:
4928:
4925:
4920:
4916:
4911:
4906:
4902:
4898:
4897:J. Biol. Chem
4894:
4887:
4884:
4879:
4875:
4870:
4865:
4861:
4857:
4854:(2): 325–34.
4853:
4849:
4845:
4838:
4835:
4830:
4826:
4821:
4816:
4811:
4806:
4802:
4798:
4797:J. Biol. Chem
4794:
4787:
4784:
4779:
4775:
4770:
4765:
4760:
4755:
4752:(2): 283–90.
4751:
4747:
4743:
4739:
4738:Sutherland GR
4732:
4729:
4724:
4720:
4715:
4710:
4706:
4702:
4698:
4691:
4688:
4683:
4679:
4675:
4671:
4667:
4663:
4655:
4652:
4647:
4643:
4638:
4633:
4629:
4625:
4621:
4614:
4611:
4606:
4602:
4597:
4592:
4588:
4584:
4583:J. Biol. Chem
4580:
4573:
4570:
4565:
4561:
4556:
4551:
4547:
4543:
4542:J. Biol. Chem
4539:
4532:
4529:
4524:
4520:
4515:
4510:
4506:
4502:
4501:J. Biol. Chem
4498:
4491:
4488:
4483:
4479:
4474:
4469:
4465:
4461:
4457:
4453:
4449:
4442:
4439:
4434:
4430:
4425:
4420:
4416:
4412:
4411:J. Biol. Chem
4408:
4401:
4398:
4393:
4389:
4385:
4381:
4377:
4373:
4365:
4362:
4357:
4353:
4348:
4343:
4338:
4333:
4329:
4325:
4321:
4314:
4311:
4306:
4302:
4297:
4292:
4288:
4284:
4283:J. Biol. Chem
4280:
4273:
4270:
4265:
4261:
4256:
4251:
4247:
4243:
4239:
4232:
4229:
4224:
4220:
4215:
4210:
4206:
4202:
4201:J. Biol. Chem
4198:
4191:
4188:
4183:
4179:
4175:
4171:
4166:
4161:
4158:(3): 372–80.
4157:
4153:
4149:
4142:
4139:
4134:
4130:
4125:
4120:
4116:
4112:
4111:J. Biol. Chem
4108:
4101:
4098:
4093:
4089:
4084:
4079:
4075:
4071:
4067:
4060:
4057:
4052:
4048:
4043:
4038:
4033:
4028:
4024:
4020:
4016:
4009:
4006:
4001:
3997:
3992:
3987:
3982:
3977:
3973:
3969:
3965:
3962:
3961:
3956:
3949:
3946:
3941:
3937:
3932:
3927:
3923:
3919:
3918:J. Biol. Chem
3915:
3908:
3905:
3900:
3896:
3891:
3886:
3883:(2): 1032–7.
3882:
3878:
3877:J. Biol. Chem
3874:
3867:
3865:
3863:
3859:
3854:
3850:
3845:
3840:
3836:
3832:
3831:J. Biol. Chem
3828:
3821:
3818:
3813:
3809:
3804:
3799:
3795:
3791:
3787:
3780:
3777:
3772:
3768:
3763:
3758:
3755:(1): 661–71.
3754:
3750:
3749:J. Biol. Chem
3746:
3739:
3737:
3735:
3731:
3727:
3723:
3719:
3714:
3709:
3705:
3701:
3696:
3691:
3687:
3683:
3679:
3672:
3669:
3664:
3660:
3656:
3652:
3648:
3644:
3641:(3): 148–53.
3640:
3636:
3629:
3626:
3621:
3617:
3612:
3607:
3603:
3599:
3598:J. Biol. Chem
3595:
3588:
3585:
3580:
3576:
3571:
3566:
3562:
3558:
3554:
3550:
3546:
3539:
3536:
3531:
3527:
3523:
3519:
3515:
3511:
3503:
3501:
3497:
3492:
3488:
3483:
3478:
3475:(7): 914–23.
3474:
3470:
3466:
3459:
3456:
3451:
3447:
3442:
3437:
3433:
3429:
3428:J. Biol. Chem
3425:
3418:
3415:
3410:
3406:
3401:
3396:
3392:
3388:
3384:
3380:
3376:
3369:
3367:
3365:
3361:
3356:
3352:
3348:
3344:
3340:
3336:
3332:
3328:
3321:
3318:
3313:
3309:
3304:
3299:
3295:
3291:
3290:
3289:Endocrinology
3285:
3278:
3275:
3270:
3266:
3262:
3258:
3254:
3250:
3246:
3242:
3235:
3233:
3229:
3224:
3218:
3214:
3210:
3203:
3201:
3199:
3195:
3190:
3186:
3181:
3176:
3172:
3168:
3167:J. Biol. Chem
3164:
3157:
3154:
3149:
3145:
3140:
3135:
3131:
3127:
3124:(1): E1–E13.
3123:
3119:
3115:
3108:
3105:
3100:
3096:
3091:
3086:
3083:(1): 335–44.
3082:
3078:
3077:J. Biol. Chem
3074:
3067:
3064:
3059:
3055:
3050:
3045:
3041:
3037:
3036:J. Biol. Chem
3033:
3026:
3023:
3018:
3014:
3009:
3004:
3000:
2996:
2995:J. Biol. Chem
2992:
2985:
2982:
2977:
2973:
2969:
2965:
2960:
2955:
2951:
2947:
2943:
2936:
2933:
2928:
2924:
2919:
2914:
2910:
2906:
2905:J. Biol. Chem
2902:
2895:
2892:
2887:
2883:
2878:
2873:
2869:
2865:
2861:
2857:
2853:
2846:
2843:
2838:
2834:
2829:
2824:
2820:
2816:
2815:J. Biol. Chem
2812:
2805:
2802:
2797:
2793:
2788:
2783:
2779:
2775:
2771:
2767:
2763:
2756:
2753:
2748:
2744:
2740:
2736:
2731:
2726:
2721:
2716:
2712:
2708:
2704:
2697:
2695:
2691:
2686:
2682:
2677:
2672:
2668:
2664:
2661:(5): 629–39.
2660:
2656:
2652:
2645:
2643:
2641:
2637:
2632:
2628:
2623:
2618:
2614:
2610:
2606:
2602:
2598:
2591:
2588:
2583:
2579:
2574:
2569:
2565:
2561:
2557:
2549:
2546:
2541:
2537:
2532:
2527:
2523:
2519:
2518:J. Biol. Chem
2515:
2508:
2505:
2500:
2496:
2492:
2488:
2484:
2480:
2476:
2472:
2464:
2461:
2456:
2452:
2447:
2442:
2437:
2432:
2428:
2424:
2420:
2413:
2410:
2405:
2401:
2396:
2391:
2387:
2383:
2379:
2375:
2371:
2367:
2363:
2356:
2353:
2348:
2342:
2339:
2334:
2330:
2325:
2320:
2315:
2310:
2306:
2302:
2298:
2294:
2290:
2283:
2280:
2275:
2271:
2267:
2263:
2259:
2255:
2252:(4): 357–60.
2251:
2247:
2240:
2237:
2232:
2228:
2222:
2219:
2214:
2210:
2204:
2201:
2197:
2193:
2188:
2186:
2184:
2180:
2176:
2172:
2167:
2165:
2163:
2159:
2152:
2150:
2148:
2144:
2140:
2136:
2132:
2127:
2126:
2121:
2113:
2107:
2104:
2101:
2098:
2095:
2092:
2089:
2086:
2083:
2080:
2077:
2074:
2072:
2069:
2066:
2063:
2060:
2057:
2055:
2051:
2048:
2045:
2042:
2039:
2036:
2033:
2030:
2027:
2024:
2021:
2018:
2015:
2012:
2009:
2006:
2003:
2000:
1997:
1994:
1991:
1988:
1985:
1982:
1979:
1976:
1973:
1970:
1967:
1964:
1961:
1958:
1955:
1952:
1950:
1947:
1944:
1941:
1938:
1935:
1932:
1929:
1926:
1923:
1920:
1917:
1914:
1911:
1910:
1908:
1906:
1898:
1896:
1894:
1890:
1889:
1884:
1880:
1876:
1872:
1868:
1864:
1860:
1856:
1852:
1848:
1844:
1840:
1839:breast cancer
1836:
1832:
1828:
1827:neuroblastoma
1824:
1816:
1814:
1807:
1805:
1798:
1796:
1794:
1789:
1787:
1783:
1775:
1773:
1770:
1767:
1763:
1752:
1749:
1741:
1738:November 2023
1730:
1727:
1723:
1720:
1716:
1713:
1709:
1706:
1702:
1699: –
1698:
1694:
1693:Find sources:
1687:
1683:
1679:
1673:
1672:
1668:
1663:This section
1661:
1657:
1652:
1651:
1645:
1643:
1641:
1635:
1633:
1629:
1626:
1622:
1618:
1614:
1610:
1606:
1602:
1598:
1594:
1590:
1578:
1573:
1569:
1565:
1562:
1558:
1551:
1549:
1546:
1542:
1538:
1534:
1531:
1527:
1523:
1516:
1514:
1510:
1504:
1500:
1497:
1491:
1489:
1485:
1481:
1477:
1471:
1467:
1464:
1460:
1453:
1451:
1447:
1441:
1437:
1434:
1428:
1426:
1422:
1418:
1414:
1410:
1404:
1400:
1397:
1395:RefSeq (mRNA)
1393:
1386:
1385:
1380:
1376:
1373:
1367:
1366:
1361:
1357:
1354:
1352:
1348:
1341:
1340:
1335:
1331:
1328:
1322:
1321:
1316:
1312:
1309:
1307:
1303:
1296:
1295:
1290:
1286:
1283:
1277:
1276:
1271:
1267:
1264:
1262:
1258:
1255:
1252:
1250:
1247:
1243:
1240:
1236:
1232:
1225:
1221:
1216:
1210:
1207:
1205:
1202:
1200:
1197:
1195:
1192:
1190:
1187:
1185:
1182:
1180:
1177:
1175:
1172:
1170:
1167:
1165:
1162:
1160:
1157:
1155:
1152:
1150:
1149:axon guidance
1147:
1145:
1142:
1140:
1137:
1135:
1132:
1130:
1127:
1125:
1122:
1120:
1117:
1115:
1112:
1110:
1107:
1105:
1102:
1100:
1097:
1095:
1092:
1090:
1087:
1085:
1082:
1080:
1077:
1075:
1072:
1070:
1067:
1065:
1062:
1060:
1057:
1055:
1052:
1050:
1047:
1045:
1042:
1040:
1037:
1035:
1032:
1030:
1027:
1025:
1022:
1020:
1017:
1015:
1012:
1010:
1007:
1005:
1002:
1000:
997:
995:
992:
990:
987:
985:
982:
980:
977:
975:
972:
970:
967:
965:
962:
960:
957:
955:
952:
950:
947:
945:
942:
940:
937:
935:
932:
930:
927:
925:
922:
920:
917:
915:
912:
910:
907:
905:
902:
900:
897:
895:
892:
890:
887:
886:
884:
881:
880:
874:
871:
869:
866:
864:
861:
859:
858:mitochondrion
856:
854:
851:
849:
846:
845:
843:
840:
839:
833:
830:
828:
825:
823:
820:
818:
815:
813:
810:
808:
805:
803:
800:
798:
795:
793:
790:
788:
785:
783:
780:
778:
775:
773:
770:
768:
765:
763:
760:
758:
755:
753:
750:
748:
745:
743:
740:
739:
737:
734:
733:
730:
729:Gene ontology
726:
722:
710:
705:
701:
696:
693:
691:
687:
679:
674:
663:
659:
655:
651:
647:
643:
639:
635:
631:
627:
626:
623:
619:
614:
611:
601:
597:
593:
589:
585:
581:
577:
573:
569:
565:
564:
561:
557:
552:
549:
548:
545:
543:
539:
537:
536:
532:
531:
528:
526:
522:
518:
514:
510:
502:
497:
493:
489:
484:
474:
470:
463:
456:
450:
443:
435:
431:
427:
422:
418:
413:
409:
401:
396:
392:
388:
383:
373:
369:
362:
355:
349:
342:
338:
332:
328:
324:
319:
315:
310:
306:
302:
298:
294:
290:
286:
282:
278:
274:
270:
266:
258:
253:
246:
241:
236:
225:
223:
219:
215:
211:
207:
203:
199:
195:
191:
187:
183:
179:
175:
171:
167:
163:
159:
155:
151:
147:
143:
139:
135:
131:
127:
123:
119:
115:
111:
107:
103:
99:
95:
91:
85:
80:
77:
76:
72:
69:
62:
58:
53:
49:
45:
40:
35:
32:
27:
19:
7505:Translocases
7502:
7489:
7476:
7463:
7450:
7440:Transferases
7437:
7424:
7281:Binding site
7062:}}
7056:{{
7020:Endonuclease
6951:ribonuclease
6891:
6674:Nucleotidase
6595:Thioesterase
5903:phosphatases
5844:
5603:
5588:
5573:
5558:
5543:
5441:
5437:
5409:(1): 37–41.
5406:
5402:
5381:
5377:
5355:(2): 81–96.
5352:
5348:
5334:(6): 853–4.
5331:
5327:
5294:
5290:
5272:
5268:
5234:Angiogenesis
5233:
5189:
5185:
5179:
5152:
5148:
5138:
5111:
5107:
5072:(2): 412–7.
5069:
5065:
5058:
5023:
5019:
5009:
4982:
4978:
4968:
4941:
4937:
4927:
4900:
4896:
4886:
4851:
4848:J. Cell Biol
4847:
4837:
4800:
4796:
4786:
4749:
4745:
4731:
4704:
4700:
4690:
4668:(1): 122–7.
4665:
4661:
4654:
4627:
4623:
4613:
4586:
4582:
4572:
4545:
4541:
4531:
4504:
4500:
4490:
4455:
4451:
4441:
4414:
4410:
4400:
4378:(2): 780–5.
4375:
4371:
4364:
4327:
4323:
4313:
4286:
4282:
4272:
4245:
4241:
4231:
4204:
4200:
4190:
4155:
4151:
4141:
4114:
4110:
4100:
4073:
4069:
4059:
4022:
4018:
4008:
3963:
3958:
3948:
3921:
3917:
3907:
3880:
3876:
3834:
3830:
3820:
3793:
3789:
3779:
3752:
3748:
3725:
3685:
3681:
3671:
3638:
3634:
3628:
3601:
3597:
3587:
3552:
3548:
3538:
3513:
3510:Cell. Signal
3509:
3472:
3468:
3458:
3431:
3427:
3417:
3382:
3378:
3330:
3326:
3320:
3296:(2): 621–8.
3293:
3287:
3277:
3244:
3240:
3212:
3170:
3166:
3156:
3121:
3117:
3107:
3080:
3076:
3066:
3039:
3035:
3025:
2998:
2994:
2984:
2949:
2945:
2935:
2908:
2904:
2894:
2859:
2855:
2845:
2818:
2814:
2804:
2769:
2765:
2755:
2710:
2706:
2658:
2654:
2604:
2600:
2590:
2563:
2559:
2548:
2521:
2517:
2507:
2474:
2470:
2463:
2426:
2422:
2412:
2372:(1): 18577.
2369:
2365:
2355:
2341:
2296:
2292:
2282:
2249:
2245:
2239:
2230:
2221:
2212:
2203:
2145:disease and
2143:peptic ulcer
2134:
2123:
2122:inserted by
2117:
1902:
1899:Interactions
1886:
1820:
1811:
1802:
1790:
1779:
1771:
1759:
1744:
1735:
1725:
1718:
1711:
1704:
1692:
1676:Please help
1664:
1636:
1634:(PTP) Shp2.
1624:
1616:
1612:
1608:
1604:
1600:
1596:
1592:
1588:
1587:
1509:NP_001103462
1506:
1488:NP_001361554
1476:NP_001317366
1473:
1446:NM_001109992
1443:
1421:NM_001374625
1417:NM_001330437
1406:
1382:
1363:
1337:
1318:
1292:
1273:
1253:
1248:
939:axonogenesis
924:organ growth
652:ciliary body
540:
533:
499:121,329,460
486:121,268,596
398:112,509,918
385:112,418,351
260:External IDs
87:
31:
26:
7276:Active site
7195:Nuclease S1
6966:Exonuclease
6860:Lecithinase
6689:Calcineurin
6627:Phosphatase
6533:Lipoprotein
6523:Endothelial
5898:VH1-like or
5526:PDB gallery
5275:: 293–311.
4324:J Biol Chem
4025:(1): 70–6.
2655:Cancer Cell
2477:(1): 70–4.
1895:with SHP2.
1843:lung cancer
1766:SH2 domains
868:nucleoplasm
238:Identifiers
7548:Categories
7479:Isomerases
7453:Hydrolases
7320:Regulation
6508:Pancreatic
6445:Carboxylic
5982:Slingshots
5291:Hum. Genet
4769:2066/26173
2560:Cancer Res
2471:Nat. Genet
2423:PLOS Genet
2246:Nat. Genet
2198:, May 2017
2177:, May 2017
2153:References
1871:suppressor
1708:newspapers
544:(ortholog)
478:5|5 F
281:HomoloGene
7358:EC number
7085:RNase III
6943:(includes
6884:Sulfatase
6797:Autotaxin
6661:Prostatic
6513:Lysosomal
6428:esterases
6424:Hydrolase
6324:Class III
5658:3.1.3.48)
3704:0270-7306
3635:FEBS Lett
3261:0968-0004
2946:FEBS Lett
2131:apoptosis
1857:-mutated
1665:does not
1513:NP_035332
1484:NP_542168
1480:NP_002825
1450:NM_011202
1425:NM_018508
1413:NM_080601
1409:NM_002834
1239:Orthologs
848:cytoplasm
289:GeneCards
7559:EC 3.1.3
7382:Kinetics
7306:Cofactor
7269:Activity
7179:RNase T1
6941:Nuclease
6576:Cutinase
6359:Class IV
6306:Class II
5648:Esterase
5476:25934330
5468:17453145
5459:11136329
5425:17211446
5403:Cell Res
5390:16208280
5369:16053901
5340:15951301
5319:27085702
5311:12384786
5281:11036942
5260:10949653
5206:15343275
5171:12060651
5130:10617656
5050:11689425
5001:11806999
4960:10880513
4878:11266449
4829:18381291
4746:Immunity
4723:10903717
4482:10082579
4356:19022774
4264:10976913
4182:38211235
4174:10080542
4133:10212213
4070:Oncogene
3940:10747947
3812:10946280
3771:12403768
3722:11739737
3663:24499468
3655:11334882
3620:10391903
3579:11782427
3530:12135708
3450:11940581
3355:25346661
3347:11360177
3327:Oncogene
3312:10650943
3269:12826400
3148:16729043
2976:31471121
2968:10350061
2927:10801826
2796:18519587
2739:16367902
2730:11159386
2685:21575863
2631:29724895
2582:15604238
2540:16377799
2499:10222262
2491:17143285
2455:20577567
2404:31819097
2194:–
2173:–
1919:Cbl gene
1905:interact
1879:necrosis
1867:promoter
1831:melanoma
1697:"PTPN11"
1619:) is an
1561:Wikidata
1218:Sources:
377:12q24.13
7538:Biology
7492:Ligases
7262:Enzymes
7152:RNase E
7147:RNase Z
7142:RNase A
7137:RNase P
7110:RNase H
6728:Phytase
6528:Hepatic
6503:Lingual
6499:Gastric
6092:DUSP13B
6087:DUSP13A
6056:PTP9Q22
5973:MK-STYX
5665:Class I
5214:1218835
5086:9344843
4919:7691811
4869:2199605
4820:2414285
4778:9285412
4682:8912646
4646:8639815
4605:9756938
4564:8505282
4523:7642582
4433:7493946
4392:8135823
4347:2629090
4305:9632636
4223:8702859
4092:9362449
4051:9824671
4000:8041791
3968:Bibcode
3899:8995399
3853:9388212
3491:9658397
3409:9632781
3189:7673163
3139:1681463
3099:9867848
3058:9054388
3017:9774457
2886:9528781
2837:7523381
2787:2493370
2747:5721063
2676:3098128
2622:6265642
2446:2887469
2395:6901466
2374:Bibcode
2333:1280823
2301:Bibcode
2274:1582162
2266:7894486
2196:Ensembl
2175:Ensembl
1937:CEACAM1
1722:scholar
1686:removed
1671:sources
1351:UniProt
1306:Ensembl
1245:Species
1224:QuickGO
863:nucleus
853:cytosol
636:utricle
519:pattern
245:Aliases
7524:Portal
7466:Lyases
7090:Drosha
7015:3.1.21
6983:RecBCD
6961:3.1.11
6581:PETase
6489:Lipase
6347:CDC25C
6342:CDC25B
6337:CDC25A
6288:MTMR15
6283:MTMR14
6278:MTMR13
6273:MTMR12
6268:MTMR11
6263:MTMR10
6147:DUSP27
6142:DUSP26
6137:DUSP25
6132:DUSP24
6127:DUSP23
6122:DUSP22
6117:DUSP21
6112:DUSP19
6107:DUSP18
6102:DUSP15
6097:DUSP14
6082:DUSP12
6077:DUSP11
6046:CDC14B
6041:CDC14A
6034:CDC14s
6025:PTP4A3
6020:PTP4A2
6015:PTP4A1
5968:DUSP16
5963:DUSP10
5916:(MKPs)
5906:(DSPs)
5885:PTPN23
5880:PTPN22
5875:PTPN21
5870:PTPN20
5865:PTPN18
5860:PTPN14
5855:PTPN13
5850:PTPN12
5845:PTPN11
5789:PTPRZ2
5784:PTPRZ1
5749:PTPRN2
5474:
5466:
5456:
5423:
5388:
5367:
5338:
5317:
5309:
5279:
5258:
5248:
5212:
5204:
5169:
5128:
5084:
5048:
5041:125701
5038:
5020:EMBO J
4999:
4958:
4917:
4876:
4866:
4827:
4817:
4776:
4721:
4680:
4644:
4603:
4562:
4521:
4480:
4470:
4431:
4390:
4354:
4344:
4303:
4262:
4221:
4180:
4172:
4131:
4090:
4049:
3998:
3988:
3938:
3897:
3851:
3810:
3769:
3720:
3713:134230
3710:
3702:
3661:
3653:
3618:
3577:
3570:125816
3567:
3549:EMBO J
3528:
3489:
3448:
3407:
3400:108981
3397:
3353:
3345:
3310:
3267:
3259:
3219:
3187:
3146:
3136:
3097:
3056:
3015:
2974:
2966:
2925:
2884:
2877:121439
2874:
2835:
2794:
2784:
2745:
2737:
2727:
2683:
2673:
2629:
2619:
2580:
2538:
2497:
2489:
2453:
2443:
2402:
2392:
2331:
2321:
2272:
2264:
2106:STAT5B
2100:STAT5A
2076:SLAMF1
2044:PDGFRB
1996:HoxA10
1724:
1717:
1710:
1703:
1695:
1625:PTPN11
1621:enzyme
1617:PTP-2C
1611:), or
1601:PTP-1D
1593:PTPN11
1547:search
1545:PubMed
1384:P35235
1365:Q06124
1261:Entrez
690:BioGPS
293:PTPN11
269:176876
252:PTPN11
37:PTPN11
7418:Types
7099:Dicer
7054:;see
6880:3.1.6
6850:PDE4B
6846:PDE4A
6784:3.1.4
6753:IMPA3
6749:IMPA2
6745:IMPA1
6623:3.1.3
6591:3.1.2
6441:3.1.1
6332:Cdc25
6258:MTMR9
6253:MTMR8
6248:MTMR7
6243:MTMR6
6238:MTMR5
6233:MTMR4
6228:MTMR3
6223:MTMR2
6200:TENC1
6157:RNGTT
6152:EMP2A
6072:DUSP3
6051:CDKN3
5958:DUSP9
5953:DUSP8
5948:DUSP7
5943:DUSP6
5938:DUSP5
5933:DUSP4
5928:DUSP2
5923:DUSP1
5840:PTPN9
5835:PTPN7
5830:PTPN6
5825:PTPN5
5820:PTPN4
5815:PTPN3
5810:PTPN2
5805:PTPN1
5779:PTPRU
5774:PTPRT
5769:PTPRS
5764:PTPRR
5759:PTPRQ
5754:PTPRO
5744:PTPRN
5739:PTPRM
5734:PTPRK
5729:PTPRJ
5724:PTPRH
5719:PTPRG
5714:PTPRF
5709:PTPRE
5704:PTPRD
5699:PTPRC
5694:PTPRB
5689:PTPRA
5472:S2CID
5315:S2CID
5210:S2CID
4979:Blood
4624:Blood
4473:84106
4178:S2CID
3991:44394
3659:S2CID
3351:S2CID
2972:S2CID
2743:S2CID
2601:Blood
2495:S2CID
2324:50525
2270:S2CID
2102:, and
2094:STAT3
2082:SOCS3
2065:PTK2B
2059:PLCG2
2032:LAIR1
1925:CD117
1907:with
1875:STAT3
1729:JSTOR
1715:books
1609:SHP-2
1294:19247
1254:Mouse
1249:Human
1220:Amigo
542:Mouse
535:Human
482:Start
417:Mouse
381:Start
314:Human
277:99511
7510:list
7503:EC7
7497:list
7490:EC6
7484:list
7477:EC5
7471:list
7464:EC4
7458:list
7451:EC3
7445:list
7438:EC2
7432:list
7425:EC1
7017:-31:
6963:-16:
6949:and
6855:PDE5
6841:PDE3
6836:PDE2
6831:PDE1
6723:PTEN
6708:OCRL
6701:PP2A
6650:ALPP
6645:ALPL
6640:ALPI
6434:3.1)
6382:EYA4
6377:EYA3
6372:EYA2
6367:EYA1
6314:ACP1
6218:MTM1
6190:TPTE
6185:TPIP
6180:PTEN
6162:STYX
6008:PRLs
5999:SSH3
5994:SSH2
5989:SSH1
5604:2shp
5589:1ayd
5574:1ayc
5559:1ayb
5544:1aya
5464:PMID
5421:PMID
5386:PMID
5365:PMID
5336:PMID
5307:PMID
5277:PMID
5256:PMID
5246:ISBN
5202:PMID
5167:PMID
5126:PMID
5082:PMID
5046:PMID
4997:PMID
4956:PMID
4915:PMID
4874:PMID
4825:PMID
4774:PMID
4719:PMID
4678:PMID
4642:PMID
4601:PMID
4560:PMID
4519:PMID
4478:PMID
4429:PMID
4388:PMID
4352:PMID
4301:PMID
4260:PMID
4219:PMID
4170:PMID
4129:PMID
4088:PMID
4047:PMID
4023:1448
3996:PMID
3936:PMID
3895:PMID
3849:PMID
3808:PMID
3767:PMID
3718:PMID
3700:ISSN
3651:PMID
3616:PMID
3575:PMID
3526:PMID
3487:PMID
3446:PMID
3405:PMID
3343:PMID
3308:PMID
3265:PMID
3257:ISSN
3217:ISBN
3185:PMID
3144:PMID
3095:PMID
3054:PMID
3013:PMID
2964:PMID
2923:PMID
2882:PMID
2833:PMID
2792:PMID
2735:PMID
2681:PMID
2627:PMID
2578:PMID
2536:PMID
2487:PMID
2451:PMID
2400:PMID
2329:PMID
2262:PMID
2088:SOS1
2050:PI3K
2038:LRP1
2014:IRS1
1984:Grb2
1972:GAB3
1966:GAB2
1960:GAB1
1954:FRS2
1931:CD31
1913:CagA
1893:CagA
1855:NPM1
1784:and
1762:Shp1
1701:news
1669:any
1667:cite
1628:gene
1275:5781
656:iris
525:Bgee
473:Band
434:Chr.
372:Band
331:Chr.
285:2122
265:OMIM
222:5IBM
218:5I6V
214:5EHR
210:5EHP
206:5IBS
202:5DF6
198:4QSY
194:4RDD
190:4PVG
186:4OHL
182:4OHI
178:4OHH
174:4OHE
170:4OHD
166:4NWG
162:4NWF
158:4JMG
154:4H34
150:3ZM3
146:3ZM2
142:3ZM1
138:3ZM0
134:4JEG
130:4JE4
126:4H1O
122:4GWF
118:4DGX
114:4DGP
110:3TL0
106:3TKZ
102:3O5X
98:3MOW
94:3B7O
90:2SHP
71:RCSB
68:PDBe
18:Shp2
7172:4/5
6195:TNS
5454:PMC
5446:doi
5411:doi
5357:doi
5299:doi
5295:111
5238:doi
5194:doi
5157:doi
5153:277
5116:doi
5112:275
5074:doi
5070:239
5036:PMC
5028:doi
4987:doi
4946:doi
4942:275
4905:doi
4901:268
4864:PMC
4856:doi
4852:152
4815:PMC
4805:doi
4801:283
4764:hdl
4754:doi
4709:doi
4705:165
4670:doi
4666:228
4632:doi
4591:doi
4587:273
4550:doi
4546:268
4509:doi
4505:270
4468:PMC
4460:doi
4419:doi
4415:270
4380:doi
4376:199
4342:PMC
4332:doi
4328:284
4291:doi
4287:273
4250:doi
4209:doi
4205:271
4160:doi
4119:doi
4115:274
4078:doi
4037:hdl
4027:doi
3986:PMC
3976:doi
3926:doi
3922:275
3885:doi
3881:272
3839:doi
3835:272
3798:doi
3794:165
3757:doi
3753:278
3708:PMC
3690:doi
3643:doi
3639:495
3606:doi
3602:274
3565:PMC
3557:doi
3518:doi
3477:doi
3436:doi
3432:277
3395:PMC
3387:doi
3335:doi
3298:doi
3294:141
3249:doi
3175:doi
3171:270
3134:PMC
3126:doi
3085:doi
3081:274
3044:doi
3040:272
3003:doi
2999:273
2954:doi
2950:450
2913:doi
2909:275
2872:PMC
2864:doi
2823:doi
2819:269
2782:PMC
2774:doi
2725:PMC
2715:doi
2671:PMC
2663:doi
2617:PMC
2609:doi
2605:131
2568:doi
2526:doi
2522:281
2479:doi
2441:PMC
2431:doi
2390:PMC
2382:doi
2319:PMC
2309:doi
2254:doi
2071:Ras
2054:Akt
1949:Erk
1869:or
1680:by
1603:),
495:End
394:End
297:OMA
273:MGI
61:PDB
7550::
7130:2C
7125:2B
7120:2A
7101::
7092::
6882::
6762::
6751:,
6747:,
6663:)/
6625::
6593::
6562:A2
6557:A1
6443::
6432:EC
6426::
5656:EC
5650::
5470:.
5462:.
5452:.
5442:64
5440:.
5436:.
5419:.
5407:17
5405:.
5401:.
5380:.
5363:.
5353:48
5351:.
5332:90
5330:.
5313:.
5305:.
5293:.
5273:55
5271:.
5254:.
5244:.
5208:.
5200:.
5188:.
5165:.
5151:.
5147:.
5124:.
5110:.
5106:.
5094:^
5080:.
5068:.
5044:.
5034:.
5024:20
5022:.
5018:.
4995:.
4983:99
4981:.
4977:.
4954:.
4940:.
4936:.
4913:.
4899:.
4895:.
4872:.
4862:.
4850:.
4846:.
4823:.
4813:.
4799:.
4795:.
4772:.
4762:.
4748:.
4744:.
4717:.
4703:.
4699:.
4676:.
4664:.
4640:.
4628:87
4626:.
4622:.
4599:.
4585:.
4581:.
4558:.
4544:.
4540:.
4517:.
4503:.
4499:.
4476:.
4466:.
4456:19
4454:.
4450:.
4427:.
4413:.
4409:.
4386:.
4374:.
4350:.
4340:.
4326:.
4322:.
4299:.
4285:.
4281:.
4258:.
4246:14
4244:.
4240:.
4217:.
4203:.
4199:.
4176:.
4168:.
4156:65
4154:.
4150:.
4127:.
4113:.
4109:.
4086:.
4074:15
4072:.
4068:.
4045:.
4035:.
4021:.
4017:.
3994:.
3984:.
3974:.
3964:91
3957:.
3934:.
3920:.
3916:.
3893:.
3879:.
3875:.
3861:^
3847:.
3833:.
3829:.
3806:.
3792:.
3788:.
3765:.
3751:.
3747:.
3733:^
3724:.
3716:.
3706:.
3698:.
3686:22
3684:.
3680:.
3657:.
3649:.
3637:.
3614:.
3600:.
3596:.
3573:.
3563:.
3553:21
3551:.
3547:.
3524:.
3514:14
3512:.
3499:^
3485:.
3473:12
3471:.
3467:.
3444:.
3430:.
3426:.
3403:.
3393:.
3383:18
3381:.
3377:.
3363:^
3349:.
3341:.
3331:20
3329:.
3306:.
3292:.
3286:.
3263:.
3255:.
3245:28
3243:.
3231:^
3197:^
3183:.
3169:.
3165:.
3142:.
3132:.
3120:.
3116:.
3093:.
3079:.
3075:.
3052:.
3038:.
3034:.
3011:.
2997:.
2993:.
2970:.
2962:.
2948:.
2944:.
2921:.
2907:.
2903:.
2880:.
2870:.
2860:18
2858:.
2854:.
2831:.
2817:.
2813:.
2790:.
2780:.
2770:28
2768:.
2764:.
2741:.
2733:.
2723:.
2711:96
2709:.
2705:.
2693:^
2679:.
2669:.
2659:19
2657:.
2653:.
2639:^
2625:.
2615:.
2603:.
2599:.
2576:.
2564:64
2562:.
2558:.
2534:.
2520:.
2516:.
2493:.
2485:.
2475:39
2473:.
2449:.
2439:.
2425:.
2421:.
2398:.
2388:.
2380:.
2368:.
2364:.
2327:.
2317:.
2307:.
2297:89
2295:.
2291:.
2268:.
2260:.
2248:.
2229:.
2211:.
2182:^
2161:^
2149:.
2141:,
2052:→
1845:,
1841:,
1837:,
1833:,
1829:,
1795:.
1222:/
501:bp
488:bp
400:bp
387:bp
295:;
291::
287:;
283::
279:;
275::
271:;
267::
220:,
216:,
212:,
208:,
204:,
200:,
196:,
192:,
188:,
184:,
180:,
176:,
172:,
168:,
164:,
160:,
156:,
152:,
148:,
144:,
140:,
136:,
132:,
128:,
124:,
120:,
116:,
112:,
108:,
104:,
100:,
96:,
92:,
7526::
7512:)
7508:(
7499:)
7495:(
7486:)
7482:(
7473:)
7469:(
7460:)
7456:(
7447:)
7443:(
7434:)
7430:(
7254:e
7247:t
7240:v
7167:3
7162:2
7157:1
7115:1
6953:)
6866:)
6862:(
6848:/
6824:1
6812:D
6807:C
6786::
6667:/
6659:(
6567:B
6501:/
6430:(
6416:e
6409:t
6402:v
5654:(
5640:e
5633:t
5626:v
5519:e
5512:t
5505:v
5478:.
5448::
5427:.
5413::
5392:.
5382:2
5371:.
5359::
5342:.
5321:.
5301::
5283:.
5262:.
5240::
5216:.
5196::
5190:4
5173:.
5159::
5132:.
5118::
5088:.
5076::
5052:.
5030::
5003:.
4989::
4962:.
4948::
4921:.
4907::
4880:.
4858::
4831:.
4807::
4780:.
4766::
4756::
4750:7
4725:.
4711::
4684:.
4672::
4648:.
4634::
4607:.
4593::
4566:.
4552::
4525:.
4511::
4484:.
4462::
4435:.
4421::
4394:.
4382::
4358:.
4334::
4307:.
4293::
4266:.
4252::
4225:.
4211::
4184:.
4162::
4135:.
4121::
4094:.
4080::
4053:.
4039::
4029::
4002:.
3978::
3970::
3942:.
3928::
3901:.
3887::
3855:.
3841::
3814:.
3800::
3773:.
3759::
3692::
3665:.
3645::
3622:.
3608::
3581:.
3559::
3532:.
3520::
3493:.
3479::
3452:.
3438::
3411:.
3389::
3357:.
3337::
3314:.
3300::
3271:.
3251::
3225:.
3191:.
3177::
3150:.
3128::
3122:1
3101:.
3087::
3060:.
3046::
3019:.
3005::
2978:.
2956::
2929:.
2915::
2888:.
2866::
2839:.
2825::
2798:.
2776::
2749:.
2717::
2687:.
2665::
2633:.
2611::
2584:.
2570::
2542:.
2528::
2501:.
2481::
2457:.
2433::
2427:6
2406:.
2384::
2376::
2370:9
2349:.
2335:.
2311::
2303::
2276:.
2256::
2250:8
2233:.
2215:.
2108:.
2096:,
2090:,
2084:,
2078:,
2067:,
2061:,
2046:,
2040:,
2034:,
2028:,
2022:,
2016:,
2010:,
2004:,
1998:,
1992:,
1986:,
1980:,
1974:,
1968:,
1962:,
1956:,
1945:,
1939:,
1933:,
1927:,
1921:,
1915:,
1751:)
1745:(
1740:)
1736:(
1726:·
1719:·
1712:·
1705:·
1688:.
1674:.
1615:(
1607:(
1599:(
1591:(
419:)
316:)
299::
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.