99:
49:). Single-stranded DNA is produced during all aspects of DNA metabolism: replication, recombination, and repair. As well as stabilizing this single-stranded DNA, SSB proteins bind to and modulate the function of numerous proteins involved in all of these processes.
60:
subunits. Binding of single-stranded DNA to the tetramer can occur in different "modes", with SSB occupying different numbers of DNA bases depending on a number of factors, including salt concentration. For example, the
431:"Crystal structure of the homo-tetrameric DNA binding domain of Escherichia coli single-stranded DNA-binding protein determined by multiwavelength x-ray diffraction on the selenomethionyl protein at 2.9-A resolution"
75:
binding mode, in which about 35 nucleotides bind to only two of the SSB subunits, tends to form. Further work is required to elucidate the functions of the various binding modes
238:
65:
binding mode, in which approximately 65 nucleotides of DNA wrap around the SSB tetramer and contact all four of its subunits, is favoured at high salt concentrations
311:
333:"Intramolecular binding mode of the C-terminus of Escherichia coli single-stranded DNA binding protein determined by nuclear magnetic resonance spectroscopy"
534:
1098:
1061:
174:
1181:
1190:
756:
1243:
1291:
304:
42:
1103:
258:
1296:
913:
763:
527:
904:
572:
547:
507:
1033:
1009:
878:
710:
657:
639:
551:
1276:
1087:
497:
1281:
1248:
520:
501:
246:
650:
581:
1147:
1078:
1073:
842:
512:
242:
1120:
1093:
1044:
187:
850:
299:
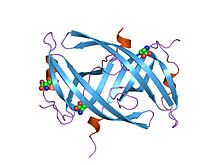
282:, repair, and recombination. It has a structure of three beta-strands to a single six-stranded
1286:
1056:
873:
462:
411:
362:
233:
1013:
862:
857:
749:
452:
442:
401:
393:
352:
344:
57:
29:
225:
1226:
1018:
892:
610:
560:
543:
279:
331:
Shishmarev D, Wang Y, Mason CE, Su XC, Oakley AJ, Graham B, et al. (February 2014).
866:
846:
357:
332:
271:
406:
381:
1270:
1068:
995:
605:
457:
430:
287:
179:
130:
1207:
397:
221:
143:
486:
199:
155:
1152:
435:
Proceedings of the
National Academy of Sciences of the United States of America
278:
are important in its function of maintaining DNA metabolism, more specifically
1238:
1212:
1000:
985:
283:
1176:
990:
980:
447:
366:
466:
415:
348:
183:
98:
1253:
1234:
806:
797:
595:
482:
275:
150:
67:
38:
817:
792:
787:
619:
167:
162:
77:
24:
253:
103:
Crystal structure of PriB- a primosomal DNA replication protein of
1167:
1162:
1157:
1140:
1135:
1130:
1125:
1113:
1108:
826:
821:
778:
773:
768:
1049:
973:
968:
963:
958:
953:
948:
943:
938:
933:
928:
923:
918:
810:
740:
735:
730:
725:
720:
715:
703:
696:
687:
682:
677:
672:
667:
662:
624:
600:
588:
478:
215:
194:
137:
125:
516:
46:
382:"The single-stranded DNA-binding protein of Escherichia coli"
429:
Raghunathan S, Ricard CS, Lohman TM, Waksman G (June 1997).
477:
This article incorporates text from the public domain
1225:
1199:
1029:
900:
891:
835:
635:
568:
559:
252:
232:
214:
209:
193:
173:
161:
149:
136:
124:
116:
111:
91:
312:Comparison of nucleic acid simulation software
528:
8:
897:
565:
535:
521:
513:
206:
97:
500:at the U.S. National Library of Medicine
456:
446:
405:
356:
71:. At lower salt concentrations, the (SSB)
323:
88:
56:SSB is composed of four identical 19
7:
498:Single-Stranded+DNA+Binding+Proteins
380:Meyer RR, Laine PS (December 1990).
14:
1191:Control of chromosome duplication
757:Autonomously replicating sequence
17:Single-strand DNA-binding protein
43:binds to single-stranded regions
305:Single-stranded binding protein
398:10.1128/MMBR.54.4.342-380.1990
1:
914:DNA polymerase III holoenzyme
764:Single-strand binding protein
210:Available protein structures:
1313:
1010:Prokaryotic DNA polymerase
711:Minichromosome maintenance
658:Origin recognition complex
476:
45:of deoxyribonucleic acid (
1088:Eukaryotic DNA polymerase
205:
96:
502:Medical Subject Headings
651:Pre-replication complex
582:Pre-replication complex
448:10.1073/pnas.94.13.6652
386:Microbiological Reviews
1292:DNA-binding substances
337:Nucleic Acids Research
1074:Replication protein A
843:Origin of replication
1297:DNA-binding proteins
1045:Replication factor C
349:10.1093/nar/gkt1238
300:DNA-binding protein
1264:
1263:
1221:
1220:
1057:Flap endonuclease
887:
886:
874:Okazaki fragments
441:(13): 6652–6657.
268:
267:
264:
263:
259:structure summary
1304:
1277:Protein families
1014:DNA polymerase I
898:
858:Replication fork
750:Licensing factor
566:
537:
530:
523:
514:
471:
470:
460:
450:
426:
420:
419:
409:
377:
371:
370:
360:
343:(4): 2750–2757.
328:
207:
105:Escherichia coli
101:
89:
30:Escherichia coli
1312:
1311:
1307:
1306:
1305:
1303:
1302:
1301:
1282:DNA replication
1267:
1266:
1265:
1260:
1217:
1195:
1035:
1031:
1025:
1019:Klenow fragment
902:
883:
867:leading strands
831:
641:
637:
631:
570:
555:
544:DNA replication
541:
494:
489:
475:
474:
428:
427:
423:
379:
378:
374:
330:
329:
325:
320:
296:
280:DNA replication
272:protein domains
107:
87:
74:
64:
12:
11:
5:
1310:
1308:
1300:
1299:
1294:
1289:
1284:
1279:
1269:
1268:
1262:
1261:
1259:
1258:
1257:
1256:
1251:
1246:
1231:
1229:
1223:
1222:
1219:
1218:
1216:
1215:
1210:
1203:
1201:
1197:
1196:
1194:
1193:
1187:
1186:
1185:
1184:
1173:
1172:
1171:
1170:
1165:
1160:
1155:
1145:
1144:
1143:
1138:
1133:
1128:
1118:
1117:
1116:
1111:
1106:
1101:
1091:
1084:
1083:
1082:
1081:
1071:
1066:
1065:
1064:
1054:
1053:
1052:
1041:
1039:
1027:
1026:
1024:
1023:
1022:
1021:
1006:
1005:
1004:
1003:
993:
988:
983:
978:
977:
976:
971:
966:
961:
956:
951:
946:
941:
936:
931:
926:
921:
910:
908:
895:
889:
888:
885:
884:
882:
881:
876:
871:
870:
869:
854:
853:
839:
837:
833:
832:
830:
829:
824:
814:
813:
803:
802:
801:
800:
795:
784:
783:
782:
781:
776:
771:
760:
759:
753:
752:
746:
745:
744:
743:
738:
733:
728:
723:
718:
707:
706:
700:
699:
693:
692:
691:
690:
685:
680:
675:
670:
665:
654:
653:
647:
645:
640:preparation in
633:
632:
630:
629:
628:
627:
616:
615:
614:
613:
608:
603:
592:
591:
585:
584:
578:
576:
563:
557:
556:
542:
540:
539:
532:
525:
517:
511:
510:
505:
493:
492:External links
490:
473:
472:
421:
392:(4): 342–380.
372:
322:
321:
319:
316:
315:
314:
309:
308:
307:
295:
292:
266:
265:
262:
261:
256:
250:
249:
236:
230:
229:
219:
212:
211:
203:
202:
197:
191:
190:
177:
171:
170:
165:
159:
158:
153:
147:
146:
141:
134:
133:
128:
122:
121:
118:
114:
113:
109:
108:
102:
94:
93:
86:
83:
72:
62:
13:
10:
9:
6:
4:
3:
2:
1309:
1298:
1295:
1293:
1290:
1288:
1285:
1283:
1280:
1278:
1275:
1274:
1272:
1255:
1252:
1250:
1247:
1245:
1242:
1241:
1240:
1236:
1233:
1232:
1230:
1228:
1224:
1214:
1211:
1209:
1205:
1204:
1202:
1198:
1192:
1189:
1188:
1183:
1180:
1179:
1178:
1175:
1174:
1169:
1166:
1164:
1161:
1159:
1156:
1154:
1151:
1150:
1149:
1146:
1142:
1139:
1137:
1134:
1132:
1129:
1127:
1124:
1123:
1122:
1119:
1115:
1112:
1110:
1107:
1105:
1102:
1100:
1097:
1096:
1095:
1092:
1089:
1086:
1085:
1080:
1077:
1076:
1075:
1072:
1070:
1069:Topoisomerase
1067:
1063:
1060:
1059:
1058:
1055:
1051:
1048:
1047:
1046:
1043:
1042:
1040:
1037:
1028:
1020:
1017:
1016:
1015:
1011:
1008:
1007:
1002:
999:
998:
997:
996:Topoisomerase
994:
992:
989:
987:
984:
982:
979:
975:
972:
970:
967:
965:
962:
960:
957:
955:
952:
950:
947:
945:
942:
940:
937:
935:
932:
930:
927:
925:
922:
920:
917:
916:
915:
912:
911:
909:
906:
899:
896:
894:
890:
880:
877:
875:
872:
868:
864:
861:
860:
859:
856:
855:
852:
848:
844:
841:
840:
838:
834:
828:
825:
823:
819:
816:
815:
812:
808:
805:
804:
799:
796:
794:
791:
790:
789:
786:
785:
780:
777:
775:
772:
770:
767:
766:
765:
762:
761:
758:
755:
754:
751:
748:
747:
742:
739:
737:
734:
732:
729:
727:
724:
722:
719:
717:
714:
713:
712:
709:
708:
705:
702:
701:
698:
695:
694:
689:
686:
684:
681:
679:
676:
674:
671:
669:
666:
664:
661:
660:
659:
656:
655:
652:
649:
648:
646:
643:
634:
626:
623:
622:
621:
618:
617:
612:
609:
607:
604:
602:
599:
598:
597:
594:
593:
590:
587:
586:
583:
580:
579:
577:
574:
567:
564:
562:
558:
553:
549:
545:
538:
533:
531:
526:
524:
519:
518:
515:
509:
506:
503:
499:
496:
495:
491:
488:
484:
480:
468:
464:
459:
454:
449:
444:
440:
436:
432:
425:
422:
417:
413:
408:
403:
399:
395:
391:
387:
383:
376:
373:
368:
364:
359:
354:
350:
346:
342:
338:
334:
327:
324:
317:
313:
310:
306:
303:
302:
301:
298:
297:
293:
291:
289:
288:protein dimer
285:
281:
277:
273:
260:
257:
255:
251:
248:
244:
240:
237:
235:
231:
227:
223:
220:
217:
213:
208:
204:
201:
198:
196:
192:
189:
185:
181:
178:
176:
172:
169:
166:
164:
160:
157:
154:
152:
148:
145:
142:
139:
135:
132:
129:
127:
123:
119:
115:
110:
106:
100:
95:
90:
85:Bacterial SSB
84:
82:
80:
79:
70:
69:
59:
55:
50:
48:
44:
40:
36:
32:
31:
26:
22:
18:
1208:Processivity
1034:synthesis in
438:
434:
424:
389:
385:
375:
340:
336:
326:
269:
104:
76:
66:
53:
51:
34:
28:
20:
16:
15:
1227:Termination
901:Prokaryotic
893:Replication
569:Prokaryotic
548:prokaryotic
546:(comparing
508:SSB in PFAM
112:Identifiers
1271:Categories
1239:Telomerase
1213:DNA ligase
1206:Movement:
1030:Eukaryotic
1001:DNA gyrase
986:DNA ligase
905:elongation
636:Eukaryotic
573:initiation
561:Initiation
552:eukaryotic
318:References
286:to form a
284:beta-sheet
222:structures
1177:DNA clamp
991:DNA clamp
981:Replisome
487:IPR000635
168:PDOC00602
156:IPR000424
27:found in
1287:Proteins
1235:Telomere
851:Replicon
807:Helicase
798:RNASEH2A
642:G1 phase
596:Helicase
483:InterPro
367:24288378
294:See also
276:bacteria
239:RCSB PDB
151:InterPro
68:in vitro
39:bacteria
1148:epsilon
1036:S phase
863:Lagging
818:Primase
793:RNASEH1
788:RNase H
620:Primase
467:9192620
416:2087220
358:3936761
163:PROSITE
131:PF00436
78:in vivo
54:E. coli
52:Active
41:, that
35:E. coli
25:protein
23:) is a
879:Primer
504:(MeSH)
465:
455:
414:
407:372786
404:
365:
355:
254:PDBsum
228:
218:
188:SUPFAM
144:CL0021
117:Symbol
1168:POLE4
1163:POLE3
1158:POLE2
1141:POLD4
1136:POLD3
1131:POLD2
1126:POLD1
1121:delta
1114:PRIM2
1109:PRIM1
1104:POLA2
1099:POLA1
1094:alpha
827:PRIM2
822:PRIM1
779:SSBP4
774:SSBP3
769:SSBP2
458:21213
200:3.A.7
184:SCOPe
175:SCOP2
61:(SSB)
1254:DKC1
1249:TERC
1244:TERT
1200:Both
1182:PCNA
1153:POLE
1079:RPA1
1062:FEN1
1050:RFC1
974:holE
969:holD
964:holC
959:holB
954:holA
949:dnaX
944:dnaT
939:dnaQ
934:dnaN
929:dnaH
924:dnaE
919:dnaC
865:and
836:Both
811:HFM1
741:MCM7
736:MCM6
731:MCM5
726:MCM4
721:MCM3
716:MCM2
704:Cdt1
697:Cdc6
688:ORC6
683:ORC5
678:ORC4
673:ORC3
668:ORC2
663:ORC1
625:dnaG
606:dnaB
601:dnaA
589:dnaC
481:and
479:Pfam
463:PMID
412:PMID
363:PMID
270:SSB
247:PDBj
243:PDBe
226:ECOD
216:Pfam
195:TCDB
180:1kaw
140:clan
138:Pfam
126:Pfam
847:Ori
550:to
453:PMC
443:doi
402:PMC
394:doi
353:PMC
345:doi
274:in
234:PDB
120:SSB
92:SSB
58:kDa
47:DNA
21:SSB
1273::
1237::
1012::
820::
809::
611:T7
485::
461:.
451:.
439:94
437:.
433:.
410:.
400:.
390:54
388:.
384:.
361:.
351:.
341:42
339:.
335:.
290:.
245:;
241:;
224:/
186:/
182:/
81:.
73:35
63:65
37:)
1090::
1038:)
1032:(
907:)
903:(
849:/
845:/
644:)
638:(
575:)
571:(
554:)
536:e
529:t
522:v
469:.
445::
418:.
396::
369:.
347::
33:(
19:(
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.