121:
146:
2105:
1083:(SNURF). Multiple transcription initiation sites have been identified and extensive alternative splicing occurs in the 5' untranslated region. Additional splice variants have been described but sequences for the complete transcripts have not been determined. The 5' UTR of this gene has been identified as an imprinting center. Alternative splicing or deletion caused by a translocation event in this paternally-expressed region is responsible for
152:
29:
2366:
1094:
of chromosome 15. After fluorescent-in-situ-hybridization has confirmed the presence of either SNRPN or UBE3A (a neighboring gene that is also imprinted), the methylation test (of SNRPN) can reveal whether the patient has uniparental disomy. SNRPN is maternally methylated (silenced). UBE3A appears
1082:
events. Although individual snRNPs are believed to recognize specific nucleic acid sequences through RNA-RNA base pairing, the specific role of this family member is unknown. The protein arises from a bicistronic transcript that also encodes a protein identified as the SNRPN upstream reading frame
1077:
The protein encoded by this gene is one polypeptide of a small nuclear ribonucleoprotein complex and belongs to the snRNP SMB/SMN family. The protein plays a role in pre-mRNA processing, possibly tissue-specific
1886:"The chromosome 15 imprinting centre (IC) region has undergone multiple duplication events and contains an upstream exon of SNRPN that is deleted in all Angelman syndrome patients with an IC microdeletion"
2104:
1314:
Ozçelik T, Leff S, Robinson W, et al. (1993). "Small nuclear ribonucleoprotein polypeptide N (SNRPN), an expressed gene in the Prader-Willi syndrome critical region".
1431:"Expression of the major human ribonucleoprotein (RNP) autoantigens in Escherichia coli and their use in an EIA for screening sera from patients with autoimmune diseases"
1573:
Esposito F, Fiore F, Cimino F, Russo T (1993). "Isolation and structural characterization of the rat gene encoding the brain specific snRNP-associated polypeptide "N"".
159:
1270:"Imprinted segments in the human genome: different DNA methylation patterns in the Prader-Willi/Angelman syndrome region as determined by the genomic sequencing method"
1746:
Suzuki Y, Yoshitomo-Nakagawa K, Maruyama K, et al. (1997). "Construction and characterization of a full length-enriched and a 5'-end-enriched cDNA library".
2219:
2214:
2194:
2174:
2015:
Albuquerque D, Manco L, González L, et al. (2017). "Polymorphisms in the SRNPN gene are associated with obesity susceptibility among
Spanish population".
2145:
1229:"Quantitative analysis of SNRPN(correction of SRNPN) gene methylation by pyrosequencing as a diagnostic test for Prader-Willi syndrome and Angelman syndrome"
573:
82:
554:
1637:
Dittrich B, Buiting K, Korn B, et al. (1996). "Imprint switching on human chromosome 15 may involve alternative transcripts of the SNRPN gene".
2257:
2407:
2426:
2084:
1151:
1133:
1775:
Fury MG, Zhang W, Christodoulopoulos I, Zieve GW (1998). "Multiple protein: protein interactions between the snRNP common core proteins".
1544:
Maruyama K, Sugano S (1994). "Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides".
1466:"Isolation of cDNA clones encoding the human Sm B/B' auto-immune antigen and specifically reacting with human anti-Sm auto-immune sera"
2204:
2199:
1003:
2138:
1213:
1120:
145:
120:
1804:
Yang T, Adamson TE, Resnick JL, et al. (1998). "A mouse model for Prader-Willi syndrome imprinting-centre mutations".
1116:
62:
2348:
2131:
158:
2400:
1171:"The gene encoding the small nuclear ribonucleoprotein-associated protein N is expressed at high levels in neurons"
1717:
Buiting K, Dittrich B, Endele S, Horsthemke B (1997). "Identification of novel exons 3' to the human SNRPN gene".
151:
2077:
70:
599:
2431:
2393:
1507:
Reed ML, Leff SE (1994). "Maternal imprinting of human SNRPN, a gene deleted in Prader-Willi syndrome".
1084:
134:
1396:"Primary structure of a human small nuclear ribonucleoprotein polypeptide as deduced by cDNA analysis"
2070:
1932:
1477:
1079:
49:
53:, HCERN3, PWCR, RT-LI, SM-D, SMN, SNRNP-N, SNURF-sm-N, Small nuclear ribonucleoprotein polypeptide N
2332:
2240:
2154:
986:
982:
978:
974:
964:
960:
952:
948:
933:
929:
925:
921:
917:
913:
909:
905:
901:
897:
893:
889:
885:
881:
877:
873:
869:
865:
861:
857:
807:
803:
782:
778:
774:
770:
766:
762:
758:
754:
730:
726:
722:
718:
714:
710:
706:
696:
692:
688:
684:
680:
2050:
1829:
1662:
1532:
1339:
1091:
94:
956:
847:
843:
839:
835:
831:
811:
750:
746:
742:
738:
734:
2322:
2158:
2042:
2003:
1960:
1907:
1872:
1821:
1792:
1763:
1734:
1705:
1654:
1625:
1590:
1561:
1524:
1495:
1452:
1417:
1382:
1331:
1291:
1250:
1192:
42:
2377:
2032:
2024:
1993:
1985:
1950:
1940:
1897:
1862:
1854:
1813:
1784:
1755:
1726:
1695:
1687:
1646:
1615:
1582:
1553:
1516:
1485:
1442:
1407:
1372:
1364:
1323:
1281:
1240:
1182:
238:
169:
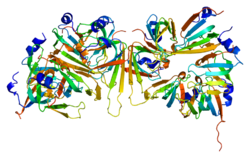
2113:: CRYSTAL STRUCTURE OF THE D3B SUBCOMPLEX OF THE HUMAN CORE SNRNP DOMAIN AT 2.0A RESOLUTION
90:
2123:
213:
1974:"Concerted regulation and molecular evolution of the duplicated SNRPB'/B and SNRPN loci"
1936:
1676:"Breakage in the SNRPN locus in a balanced 46,XY,t(15;19) Prader-Willi syndrome patient"
1481:
1867:
1842:
1700:
1675:
1620:
1603:
28:
1998:
1973:
1759:
1412:
1395:
1377:
1352:
1187:
1170:
488:
483:
478:
462:
457:
452:
447:
442:
437:
432:
427:
422:
417:
412:
407:
391:
386:
2420:
1955:
1920:
1557:
1490:
1465:
373:
1833:
1666:
1536:
1343:
2054:
1604:"Gene structure, DNA methylation, and imprinted expression of the human SNRPN gene"
231:
111:
74:
1921:"An imprinted, mammalian bicistronic transcript encodes two independent proteins"
1447:
1430:
1353:"A comparison of snRNP-associated Sm-autoantigens: human N, rat N and human B/B'"
1245:
1228:
98:
1268:
Zeschnigk M, Schmitz B, Dittrich B, Buiting K, Horsthemke B, Doerfler W (1997).
1156:
National Center for
Biotechnology Information, U.S. National Library of Medicine
1138:
National Center for
Biotechnology Information, U.S. National Library of Medicine
2296:
128:
78:
1989:
1945:
1368:
518:
331:
252:
197:
184:
86:
2046:
2007:
1964:
1788:
1730:
1586:
1254:
1911:
1902:
1885:
1876:
1825:
1796:
1767:
1738:
1709:
1691:
1658:
1629:
1594:
1565:
1528:
1499:
1456:
1421:
1386:
1335:
1295:
1286:
1269:
1196:
1048:
1043:
2312:
1032:
661:
642:
2373:
2265:
2247:
2236:
2037:
1650:
1520:
1327:
1064:
628:
585:
2365:
1817:
2280:
2275:
2270:
2209:
2189:
2179:
2028:
1016:
540:
1858:
2224:
2184:
2166:
1214:"Entrez Gene: SNRPN small nuclear ribonucleoprotein polypeptide N"
1843:"Prader-Willi syndrome is caused by disruption of the SNRPN gene"
503:
499:
1071:
66:
2127:
2066:
2062:
1972:
Gray TA, Smithwick MJ, Schaldach MA, et al. (2000).
1351:
Schmauss C, McAllister G, Ohosone Y, et al. (1989).
1841:
Kuslich CD, Kobori JA, Mohapatra G, et al. (1999).
1464:
Sharpe NG, Williams DG, Howarth DN, et al. (1989).
221:
2381:
1394:
Rokeach LA, Jannatipour M, Haselby JA, Hoch SO (1989).
1884:
Färber C, Dittrich B, Buiting K, Horsthemke B (1999).
343:
1061:
Small nuclear ribonucleoprotein-associated protein N
2341:
2321:
2305:
2289:
2256:
2165:
1674:Sun Y, Nicholls RD, Butler MG, et al. (1996).
947:
830:
796:
679:
1227:White HE, Durston VJ, Harvey JF, Cross NC (2006).
1112:
1110:
1108:
1602:Glenn CC, Saitoh S, Jong MT, et al. (1996).
1429:Renz M, Heim C, Bräunling O, et al. (1989).
168:
16:Protein-coding gene in the species Homo sapiens
2401:
2139:
2078:
1169:Schmauss C, Brines ML, Lerner MR (May 1992).
8:
1117:GRCh38: Ensembl release 89: ENSG00000128739
2408:
2394:
2146:
2132:
2124:
2085:
2071:
2063:
944:
827:
676:
514:
369:
209:
106:
2036:
1997:
1954:
1944:
1901:
1866:
1699:
1619:
1489:
1446:
1411:
1376:
1285:
1244:
1186:
1208:
1206:
1095:to be paternally methylated (silenced).
1087:due to parental imprint switch failure.
2100:
1919:Gray TA, Saitoh S, Nicholls RD (1999).
1104:
438:small nuclear ribonucleoprotein complex
18:
173:
132:
127:
7:
2362:
2360:
1090:SNRPN-methylation is used to detect
2380:. You can help Knowledge (XXG) by
793:
652:
633:
609:
590:
564:
545:
348:
309:
247:
226:
14:
1067:that in humans is encoded by the
2364:
2103:
157:
150:
144:
119:
27:
484:mRNA splicing, via spliceosome
332:More reference expression data
291:right hemisphere of cerebellum
1:
1760:10.1016/S0378-1119(97)00411-3
1575:Biochem. Biophys. Res. Commun
1413:10.1016/S0021-9258(18)83693-6
1188:10.1016/S0021-9258(18)42475-1
142:
2427:Genes on human chromosome 15
1925:Proc. Natl. Acad. Sci. U.S.A
1558:10.1016/0378-1119(94)90802-8
1491:10.1016/0014-5793(89)80801-4
1246:10.1373/clinchem.2005.065086
423:U4/U6 x U5 tri-snRNP complex
418:catalytic step 2 spliceosome
2349:Signal recognition particle
2448:
2359:
1448:10.1093/clinchem/35.9.1861
2098:
1152:"Mouse PubMed Reference:"
1134:"Human PubMed Reference:"
1047:
1042:
1038:
1031:
1015:
998:
971:
854:
823:
800:
703:
672:
659:
655:
640:
636:
627:
616:
612:
597:
593:
584:
571:
567:
552:
548:
539:
524:
517:
513:
497:
372:
368:
356:
351:
342:
329:
316:
312:
259:
250:
220:
212:
208:
191:
178:
141:
118:
109:
105:
60:
57:
47:
40:
35:
26:
21:
1004:Chr 15: 24.82 – 24.98 Mb
2323:Vault ribonucleoprotein
1946:10.1073/pnas.96.10.5616
2376:-related article is a
1990:10.1093/nar/27.23.4577
1789:10.1006/excr.1997.3750
1731:10.1006/geno.1996.4571
1587:10.1006/bbrc.1993.2047
443:U2-type prespliceosome
263:superior frontal gyrus
1369:10.1093/nar/17.4.1733
1085:Prader-Willi syndrome
283:cerebellar hemisphere
271:primary visual cortex
135:Chromosome 15 (human)
1080:alternative splicing
428:spliceosomal complex
2333:Major vault protein
2290:Other transcription
2241:U1 spliceosomal RNA
2155:RNA-binding protein
1937:1999PNAS...96.5616G
1903:10.1093/hmg/8.2.337
1692:10.1093/hmg/5.4.517
1482:1989FEBSL.250..585S
1287:10.1093/hmg/6.3.387
489:response to hormone
2159:Ribonucleoproteins
1651:10.1038/ng1096-163
1521:10.1038/ng0294-163
1328:10.1038/ng1292-265
1092:uniparental disomy
472:Biological process
401:Cellular component
380:Molecular function
287:right frontal lobe
2389:
2388:
2357:
2356:
2121:
2120:
1978:Nucleic Acids Res
1847:Am. J. Hum. Genet
1818:10.1038/ng0598-25
1608:Am. J. Hum. Genet
1357:Nucleic Acids Res
1058:
1057:
1054:
1053:
1027:
1026:
994:
993:
941:
940:
819:
818:
790:
789:
668:
667:
649:
648:
623:
622:
606:
605:
580:
579:
561:
560:
509:
508:
364:
363:
360:
359:
338:
337:
325:
324:
306:
305:
279:cerebellar cortex
267:prefrontal cortex
204:
203:
99:SNRPN - orthologs
2439:
2410:
2403:
2396:
2368:
2361:
2148:
2141:
2134:
2125:
2107:
2087:
2080:
2073:
2064:
2058:
2040:
2029:10.1002/jgm.2956
2011:
2001:
1968:
1958:
1948:
1915:
1905:
1880:
1870:
1837:
1800:
1771:
1742:
1713:
1703:
1670:
1633:
1623:
1598:
1569:
1540:
1503:
1493:
1460:
1450:
1425:
1415:
1390:
1380:
1347:
1300:
1299:
1289:
1265:
1259:
1258:
1248:
1224:
1218:
1217:
1210:
1201:
1200:
1190:
1166:
1160:
1159:
1148:
1142:
1141:
1130:
1124:
1114:
1040:
1039:
1011:
1006:
989:
945:
936:
828:
824:RefSeq (protein)
814:
794:
785:
677:
653:
634:
610:
591:
565:
546:
515:
370:
349:
334:
310:
255:
253:Top expressed in
248:
227:
210:
200:
187:
176:
161:
154:
148:
137:
123:
107:
101:
52:
45:
31:
19:
2447:
2446:
2442:
2441:
2440:
2438:
2437:
2436:
2417:
2416:
2415:
2414:
2358:
2353:
2337:
2324:
2317:
2301:
2285:
2252:
2161:
2152:
2122:
2117:
2114:
2108:
2094:
2091:
2061:
2014:
1984:(23): 4577–84.
1971:
1931:(10): 5616–21.
1918:
1890:Hum. Mol. Genet
1883:
1840:
1803:
1774:
1754:(1–2): 149–56.
1745:
1716:
1680:Hum. Mol. Genet
1673:
1636:
1601:
1572:
1543:
1506:
1463:
1428:
1393:
1350:
1313:
1309:
1307:Further reading
1304:
1303:
1274:Hum. Mol. Genet
1267:
1266:
1262:
1226:
1225:
1221:
1212:
1211:
1204:
1168:
1167:
1163:
1150:
1149:
1145:
1132:
1131:
1127:
1115:
1106:
1101:
1049:View/Edit Mouse
1044:View/Edit Human
1009:
1002:
999:Location (UCSC)
985:
981:
977:
973:
967:
963:
959:
955:
951:
932:
928:
924:
920:
916:
912:
908:
904:
900:
896:
892:
888:
884:
880:
876:
872:
868:
864:
860:
856:
850:
846:
842:
838:
834:
810:
806:
802:
781:
777:
773:
769:
765:
761:
757:
753:
749:
745:
741:
737:
733:
729:
725:
721:
717:
713:
709:
705:
699:
695:
691:
687:
683:
600:ENSG00000128739
493:
467:
396:
387:protein binding
330:
321:
302:
297:
295:Brodmann area 9
293:
289:
285:
281:
277:
273:
269:
265:
251:
195:
182:
174:
164:
163:
162:
155:
133:
110:Gene location (
61:
48:
41:
17:
12:
11:
5:
2445:
2443:
2435:
2434:
2429:
2419:
2418:
2413:
2412:
2405:
2398:
2390:
2387:
2386:
2369:
2355:
2354:
2352:
2351:
2345:
2343:
2339:
2338:
2336:
2335:
2329:
2327:
2319:
2318:
2316:
2315:
2309:
2307:
2303:
2302:
2300:
2299:
2293:
2291:
2287:
2286:
2284:
2283:
2278:
2273:
2268:
2262:
2260:
2254:
2253:
2251:
2250:
2244:
2243:
2233:
2232:
2227:
2222:
2217:
2212:
2207:
2202:
2197:
2192:
2187:
2182:
2177:
2171:
2169:
2163:
2162:
2153:
2151:
2150:
2143:
2136:
2128:
2119:
2118:
2116:
2115:
2109:
2102:
2099:
2096:
2095:
2092:
2090:
2089:
2082:
2075:
2067:
2060:
2059:
2012:
1969:
1916:
1881:
1859:10.1086/302177
1838:
1801:
1772:
1743:
1714:
1671:
1634:
1599:
1570:
1552:(1–2): 171–4.
1541:
1504:
1461:
1426:
1406:(9): 5024–30.
1391:
1363:(4): 1733–43.
1348:
1310:
1308:
1305:
1302:
1301:
1260:
1239:(6): 1005–13.
1219:
1202:
1181:(12): 8521–9.
1161:
1143:
1125:
1103:
1102:
1100:
1097:
1056:
1055:
1052:
1051:
1046:
1036:
1035:
1029:
1028:
1025:
1024:
1022:
1020:
1013:
1012:
1007:
1000:
996:
995:
992:
991:
969:
968:
942:
939:
938:
852:
851:
825:
821:
820:
817:
816:
798:
797:
791:
788:
787:
701:
700:
674:
670:
669:
666:
665:
657:
656:
650:
647:
646:
638:
637:
631:
625:
624:
621:
620:
614:
613:
607:
604:
603:
595:
594:
588:
582:
581:
578:
577:
569:
568:
562:
559:
558:
550:
549:
543:
537:
536:
531:
526:
522:
521:
511:
510:
507:
506:
495:
494:
492:
491:
486:
481:
475:
473:
469:
468:
466:
465:
460:
455:
450:
445:
440:
435:
430:
425:
420:
415:
410:
404:
402:
398:
397:
395:
394:
389:
383:
381:
377:
376:
366:
365:
362:
361:
358:
357:
354:
353:
346:
340:
339:
336:
335:
327:
326:
323:
322:
320:
317:
314:
313:
307:
304:
303:
301:
300:
296:
292:
288:
284:
280:
276:
272:
268:
264:
260:
257:
256:
244:
243:
235:
224:
218:
217:
214:RNA expression
206:
205:
202:
201:
193:
189:
188:
180:
177:
172:
166:
165:
156:
149:
143:
139:
138:
131:
125:
124:
116:
115:
103:
102:
59:
55:
54:
46:
38:
37:
33:
32:
24:
23:
15:
13:
10:
9:
6:
4:
3:
2:
2444:
2433:
2432:Protein stubs
2430:
2428:
2425:
2424:
2422:
2411:
2406:
2404:
2399:
2397:
2392:
2391:
2385:
2383:
2379:
2375:
2370:
2367:
2363:
2350:
2347:
2346:
2344:
2340:
2334:
2331:
2330:
2328:
2326:
2320:
2314:
2311:
2310:
2308:
2304:
2298:
2295:
2294:
2292:
2288:
2282:
2279:
2277:
2274:
2272:
2269:
2267:
2264:
2263:
2261:
2259:
2255:
2249:
2246:
2245:
2242:
2238:
2235:
2234:
2231:
2228:
2226:
2223:
2221:
2218:
2216:
2213:
2211:
2208:
2206:
2203:
2201:
2198:
2196:
2193:
2191:
2188:
2186:
2183:
2181:
2178:
2176:
2173:
2172:
2170:
2168:
2164:
2160:
2156:
2149:
2144:
2142:
2137:
2135:
2130:
2129:
2126:
2112:
2106:
2101:
2097:
2088:
2083:
2081:
2076:
2074:
2069:
2068:
2065:
2056:
2052:
2048:
2044:
2039:
2034:
2030:
2026:
2022:
2018:
2013:
2009:
2005:
2000:
1995:
1991:
1987:
1983:
1979:
1975:
1970:
1966:
1962:
1957:
1952:
1947:
1942:
1938:
1934:
1930:
1926:
1922:
1917:
1913:
1909:
1904:
1899:
1896:(2): 337–43.
1895:
1891:
1887:
1882:
1878:
1874:
1869:
1864:
1860:
1856:
1852:
1848:
1844:
1839:
1835:
1831:
1827:
1823:
1819:
1815:
1811:
1807:
1802:
1798:
1794:
1790:
1786:
1782:
1778:
1777:Exp. Cell Res
1773:
1769:
1765:
1761:
1757:
1753:
1749:
1744:
1740:
1736:
1732:
1728:
1724:
1720:
1715:
1711:
1707:
1702:
1697:
1693:
1689:
1686:(4): 517–24.
1685:
1681:
1677:
1672:
1668:
1664:
1660:
1656:
1652:
1648:
1645:(2): 163–70.
1644:
1640:
1635:
1631:
1627:
1622:
1617:
1614:(2): 335–46.
1613:
1609:
1605:
1600:
1596:
1592:
1588:
1584:
1581:(1): 317–26.
1580:
1576:
1571:
1567:
1563:
1559:
1555:
1551:
1547:
1542:
1538:
1534:
1530:
1526:
1522:
1518:
1514:
1510:
1505:
1501:
1497:
1492:
1487:
1483:
1479:
1476:(2): 585–90.
1475:
1471:
1467:
1462:
1458:
1454:
1449:
1444:
1441:(9): 1861–3.
1440:
1436:
1432:
1427:
1423:
1419:
1414:
1409:
1405:
1401:
1400:J. Biol. Chem
1397:
1392:
1388:
1384:
1379:
1374:
1370:
1366:
1362:
1358:
1354:
1349:
1345:
1341:
1337:
1333:
1329:
1325:
1321:
1317:
1312:
1311:
1306:
1297:
1293:
1288:
1283:
1280:(3): 387–95.
1279:
1275:
1271:
1264:
1261:
1256:
1252:
1247:
1242:
1238:
1234:
1230:
1223:
1220:
1215:
1209:
1207:
1203:
1198:
1194:
1189:
1184:
1180:
1176:
1172:
1165:
1162:
1157:
1153:
1147:
1144:
1139:
1135:
1129:
1126:
1122:
1118:
1113:
1111:
1109:
1105:
1098:
1096:
1093:
1088:
1086:
1081:
1075:
1073:
1070:
1066:
1062:
1050:
1045:
1041:
1037:
1034:
1030:
1023:
1021:
1018:
1014:
1008:
1005:
1001:
997:
990:
988:
984:
980:
976:
970:
966:
962:
958:
954:
950:
946:
943:
937:
935:
931:
927:
923:
919:
915:
911:
907:
903:
899:
895:
891:
887:
883:
879:
875:
871:
867:
863:
859:
853:
849:
845:
841:
837:
833:
829:
826:
822:
815:
813:
809:
805:
799:
795:
792:
786:
784:
780:
776:
772:
768:
764:
760:
756:
752:
748:
744:
740:
736:
732:
728:
724:
720:
716:
712:
708:
702:
698:
694:
690:
686:
682:
678:
675:
673:RefSeq (mRNA)
671:
664:
663:
658:
654:
651:
645:
644:
639:
635:
632:
630:
626:
619:
615:
611:
608:
602:
601:
596:
592:
589:
587:
583:
576:
575:
570:
566:
563:
557:
556:
551:
547:
544:
542:
538:
535:
532:
530:
527:
523:
520:
516:
512:
505:
501:
496:
490:
487:
485:
482:
480:
477:
476:
474:
471:
470:
464:
461:
459:
456:
454:
451:
449:
446:
444:
441:
439:
436:
434:
431:
429:
426:
424:
421:
419:
416:
414:
411:
409:
406:
405:
403:
400:
399:
393:
390:
388:
385:
384:
382:
379:
378:
375:
374:Gene ontology
371:
367:
355:
350:
347:
345:
341:
333:
328:
318:
315:
311:
308:
298:
294:
290:
286:
282:
278:
274:
270:
266:
262:
261:
258:
254:
249:
246:
245:
242:
240:
236:
234:
233:
229:
228:
225:
223:
219:
215:
211:
207:
199:
194:
190:
186:
181:
171:
167:
160:
153:
147:
140:
136:
130:
126:
122:
117:
113:
108:
104:
100:
96:
92:
88:
84:
80:
76:
72:
68:
64:
56:
51:
44:
39:
34:
30:
25:
20:
2382:expanding it
2371:
2229:
2110:
2023:(5): e2956.
2020:
2016:
1981:
1977:
1928:
1924:
1893:
1889:
1850:
1846:
1812:(1): 25–31.
1809:
1805:
1780:
1776:
1751:
1747:
1725:(1): 132–7.
1722:
1718:
1683:
1679:
1642:
1638:
1611:
1607:
1578:
1574:
1549:
1545:
1515:(2): 163–7.
1512:
1508:
1473:
1469:
1438:
1434:
1403:
1399:
1360:
1356:
1322:(4): 265–9.
1319:
1315:
1277:
1273:
1263:
1236:
1232:
1222:
1178:
1174:
1164:
1155:
1146:
1137:
1128:
1089:
1076:
1068:
1060:
1059:
987:NP_001336624
983:NP_001336623
979:NP_001336622
975:NP_001336621
972:
965:NP_001336620
961:NP_001336619
953:NP_001076431
949:NP_001076430
934:NP_001365186
930:NP_001365185
926:NP_001365184
922:NP_001365183
918:NP_001365182
914:NP_001365181
910:NP_001365180
906:NP_001365178
902:NP_001336394
898:NP_001336393
894:NP_001336392
890:NP_001336391
886:NP_001336390
882:NP_001336389
878:NP_001336388
874:NP_001336387
870:NP_001336386
866:NP_001336385
862:NP_001336384
858:NP_001336383
855:
808:NM_001082962
804:NM_001082961
801:
783:NM_001378257
779:NM_001378256
775:NM_001378255
771:NM_001378254
767:NM_001378253
763:NM_001378252
759:NM_001378251
755:NM_001378249
731:NM_001349465
727:NM_001349464
723:NM_001349463
719:NM_001349462
715:NM_001349461
711:NM_001349460
707:NM_001349459
704:
697:NM_001349458
693:NM_001349457
689:NM_001349456
685:NM_001349455
681:NM_001349454
660:
641:
617:
598:
572:
553:
533:
528:
479:RNA splicing
299:hypothalamus
237:
230:
58:External IDs
2306:Translation
2093:PDB gallery
2038:10230/60864
2017:J. Gene Med
1853:(1): 70–6.
1783:(1): 63–9.
1175:J Biol Chem
453:nucleoplasm
392:RNA binding
196:24,978,723
183:24,823,637
36:Identifiers
2421:Categories
2297:Telomerase
1806:Nat. Genet
1639:Nat. Genet
1509:Nat. Genet
1435:Clin. Chem
1316:Nat. Genet
1233:Clin. Chem
1123:, May 2017
1099:References
275:cerebellum
241:(ortholog)
79:HomoloGene
2325:particles
1470:FEBS Lett
957:NP_038698
848:NP_073719
844:NP_073718
840:NP_073717
836:NP_073716
832:NP_003088
812:NM_013670
751:NM_022808
747:NM_022807
743:NM_022806
739:NM_022805
735:NM_003097
519:Orthologs
408:cytoplasm
87:GeneCards
2313:Ribosome
2248:SNRNP200
2047:28387446
2008:10556313
1965:10318933
1834:20124015
1719:Genomics
1667:20943659
1537:39792921
1344:32755927
1255:16574761
1119:–
1033:Wikidata
498:Sources:
463:U2 snRNP
458:U1 snRNP
433:U4 snRNP
413:U5 snRNP
2374:protein
2055:1860814
1933:Bibcode
1912:9931342
1877:9915945
1868:1377704
1826:9590284
1797:9417867
1768:9373149
1739:9070929
1710:8845846
1701:6057871
1659:8841186
1630:8571960
1621:1914536
1595:8363612
1566:8125298
1529:7512861
1500:2753153
1478:Bibcode
1457:2528429
1422:2522449
1387:2522186
1336:1303277
1296:9147641
1197:1533223
1121:Ensembl
1065:protein
629:UniProt
586:Ensembl
525:Species
504:QuickGO
448:nucleus
216:pattern
175:15q11.2
43:Aliases
2053:
2045:
2006:
1999:148745
1996:
1963:
1953:
1910:
1875:
1865:
1832:
1824:
1795:
1766:
1737:
1708:
1698:
1665:
1657:
1628:
1618:
1593:
1564:
1535:
1527:
1498:
1455:
1420:
1385:
1378:331831
1375:
1342:
1334:
1294:
1253:
1195:
1019:search
1017:PubMed
662:P63163
643:P63162
541:Entrez
344:BioGPS
67:182279
2372:This
2342:Other
2258:hnRNP
2167:snRNP
2051:S2CID
1956:21909
1830:S2CID
1663:S2CID
1533:S2CID
1340:S2CID
1069:SNRPN
1063:is a
574:20646
534:Mouse
529:Human
500:Amigo
239:Mouse
232:Human
179:Start
112:Human
91:SNRPN
83:68297
75:98347
50:SNRPN
22:SNRPN
2378:stub
2111:1d3b
2043:PMID
2004:PMID
1961:PMID
1908:PMID
1873:PMID
1822:PMID
1793:PMID
1764:PMID
1748:Gene
1735:PMID
1706:PMID
1655:PMID
1626:PMID
1591:PMID
1562:PMID
1546:Gene
1525:PMID
1496:PMID
1453:PMID
1418:PMID
1383:PMID
1332:PMID
1292:PMID
1251:PMID
1193:PMID
1072:gene
555:6638
222:Bgee
170:Band
129:Chr.
63:OMIM
2237:70K
2033:hdl
2025:doi
1994:PMC
1986:doi
1951:PMC
1941:doi
1898:doi
1863:PMC
1855:doi
1814:doi
1785:doi
1781:237
1756:doi
1752:200
1727:doi
1696:PMC
1688:doi
1647:doi
1616:PMC
1583:doi
1579:195
1554:doi
1550:138
1517:doi
1486:doi
1474:250
1443:doi
1408:doi
1404:264
1373:PMC
1365:doi
1324:doi
1282:doi
1241:doi
1183:doi
1179:267
1010:n/a
618:n/a
352:n/a
319:n/a
192:End
95:OMA
71:MGI
2423::
2266:A0
2210:D3
2205:D2
2200:D1
2190:B2
2180:A1
2157::
2049:.
2041:.
2031:.
2021:19
2019:.
2002:.
1992:.
1982:27
1980:.
1976:.
1959:.
1949:.
1939:.
1929:96
1927:.
1923:.
1906:.
1892:.
1888:.
1871:.
1861:.
1851:64
1849:.
1845:.
1828:.
1820:.
1810:19
1808:.
1791:.
1779:.
1762:.
1750:.
1733:.
1723:40
1721:.
1704:.
1694:.
1682:.
1678:.
1661:.
1653:.
1643:14
1641:.
1624:.
1612:58
1610:.
1606:.
1589:.
1577:.
1560:.
1548:.
1531:.
1523:.
1511:.
1494:.
1484:.
1472:.
1468:.
1451:.
1439:35
1437:.
1433:.
1416:.
1402:.
1398:.
1381:.
1371:.
1361:17
1359:.
1355:.
1338:.
1330:.
1318:.
1290:.
1276:.
1272:.
1249:.
1237:52
1235:.
1231:.
1205:^
1191:.
1177:.
1173:.
1154:.
1136:.
1107:^
1074:.
502:/
198:bp
185:bp
93:;
89::
85:;
81::
77:;
73::
69:;
65::
2409:e
2402:t
2395:v
2384:.
2281:R
2276:L
2271:C
2239:/
2230:N
2225:G
2220:F
2215:E
2195:C
2185:B
2175:A
2147:e
2140:t
2133:v
2086:e
2079:t
2072:v
2057:.
2035::
2027::
2010:.
1988::
1967:.
1943::
1935::
1914:.
1900::
1894:8
1879:.
1857::
1836:.
1816::
1799:.
1787::
1770:.
1758::
1741:.
1729::
1712:.
1690::
1684:5
1669:.
1649::
1632:.
1597:.
1585::
1568:.
1556::
1539:.
1519::
1513:6
1502:.
1488::
1480::
1459:.
1445::
1424:.
1410::
1389:.
1367::
1346:.
1326::
1320:2
1298:.
1284::
1278:6
1257:.
1243::
1216:.
1199:.
1185::
1158:.
1140:.
114:)
97::
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.