213:
144:
285:
inducer/repressor system which provides temporal control of gene expression. To control gene expression spatially inkjet printers are under development for printing ligands on gel culture. Other popular method involves use of light to control gene expression in spatiotemporal fashion. Since light can
268:
tissues. Techniques that require fixation of tissue can only generate a single temporal time point per individual organism. However, using live animals instead of fixed tissue can be crucial in dynamically understanding expression patterns over an individual's lifespan. Either way, variation between
235:
to the mRNA of the gene, is added to the tissue. This probe is then chemically tagged so that it can be visualized later. This technique enables visualization specifically of mRNA-producing cells without any of the artifacts associated with immunohistochemistry. However, it is notoriously difficult,
184:
with specific affinity for the protein associated with the gene of interest. This distribution of this antibody can then be visualized by a technique such as fluorescent labeling. Immunohistochemistry has the advantages of being methodologically feasible and relatively inexpensive. Its disadvantages
216:
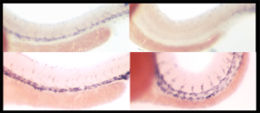
In situ hybridizations of genes expressed in arteries (top) and veins (bottom) in zebrafish. Blue staining indicates presence of the gene mRNAs. Panels on the left are normal animals, while animals on the right are mutated in the Notch gene. Fish lacking Notch have fewer arteries and more veins at
256:
proximal to the insertion point, the reporter gene will be expressed in particular tissues at particular points in development. While enhancer-trap derived expression patterns do not necessarily reflect the actual patterns of expression of specific genes, they reveal the variety of spatiotemporal
111:
What causes spatial and temporal differences in the expression of a single gene? Because current expression patterns depend strictly on previous expression patterns, there is a regressive problem of explaining what caused the first differences in gene expression. The process by which uniform gene
54:
which is expressed in all cells at all times in life. Some, on the other hand, are extraordinarily intricate and difficult to predict and model, with expression fluctuating wildly from minute to minute or from cell to cell. Spatiotemporal variation plays a key role in generating the diversity of
160:
downstream of its promoter. In this configuration, the promoter gene will cause the reporter gene to be expressed only where and when the gene of interest is expressed. The expression distribution of the reporter gene can be determined by visualizing it. For example, the reporter gene
20:
59:
found in developed organisms; since the identity of a cell is specified by the collection of genes actively expressed within that cell, if gene expression was uniform spatially and temporally, there could be at most one kind of cell.
641:
Govan, J. M; Deiters (2012) "Activation and
Deactivation of Antisense and RNA Interference Function with Light A. From Nucleic Acids Sequences to Molecular Medicine (eds V. A. Erdmann and J. Barciszewski)", Springer, Heidelberg,
251:
screening reveals the diversity of spatiotemporal gene expression patterns possible in an organism. In this technique, DNA that encodes a reporter gene is randomly inserted into the genome. Depending on the gene
595:
Tang, XinJing; Maegawa, Shingo; Weinberg, Eric S.; Dmochowski, Ivan J. (2007). "Regulating Gene
Expression in Zebrafish Embryos Using Light-Activated, Negatively Charged Peptide Nucleic Acids".
298:
demonstrating the control of gene expression spatiotemporally. Recently light based control has been shown at DNA level using transgene based system or caged triplex forming oligos
193:
results. Furthermore, since immunohistochemistry visualizes the protein generated by the gene, if the protein product diffuses between cells, or has a particularly short or long
286:
also be controlled easily in space, time and degree, several methods of controlling gene expression at DNA and RNA level have been developed and are under study. For example,
629:
Alexander Heckel, Günter Mayer, "The
Chemical Biology of Nucleic Acids", Chapter 13. Light-Responsive Nucleic Acids for the Spatiotemporal Control of Biological Processes.
81:
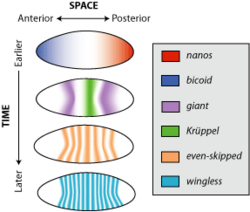
is expressed across almost the entire embryo in alternating stripes three cells separated. This pattern is lost by the time the organism develops into a larva, but
756:
501:
Jain, Piyush K.; Shah, Samit; Friedman, Simon H. (2011). "Patterning of Gene
Expression Using New Photolabile Groups Applied to Light Activated RNAi".
374:
Ando, Hideki; Furuta, Toshiaki; Tsien, Roger Y.; Okamoto, Hitoshi (2001). "Photo-mediated gene activation using caged RNA/DNA in zebrafish embryos".
290:
can be controlled using light and also patterning of gene expression has been performed in cell monolayer and in zebrafish embryos using caged
765:
232:
147:
The gamma-crystalline promoter drives expression of the green fluorescent protein reporter gene exclusively in the eye of an adult frog.
706:"Regulation of Transcription through Light-Activation and Light-Deactivation of Triplex-Forming Oligonucleotides in Mammalian Cells"
561:
787:
544:
Shestopalov, Ilya A; Chen, James K (2011). "Spatiotemporal
Control of Embryonic Gene Expression Using Caged Morpholinos".
176:
If the promoter of the gene of interest is unknown, there are several ways to identify its spatiotemporal distribution.
806:
98:
50:. Gene activation patterns vary widely in complexity. Some are straightforward and static, such as the pattern of
162:
811:
816:
73:
202:
90:
630:
221:
68:
47:
328:
295:
265:
177:
417:
Shah, Samit; Rangarajan, Subhashree; Friedman, Simon H. (2005). "Light‐Activated RNA Interference".
686:
526:
399:
253:
189:
identification of expression. Poor penetrance of the antibody into the target tissue can lead to
704:
Govan, Jeane M.; Uprety, Rajendra; Hemphill, James; Lively, Mark O.; Deiters, Alexander (2012).
735:
678:
612:
577:
557:
518:
483:
434:
391:
356:
113:
43:
725:
717:
668:
604:
567:
549:
510:
473:
465:
426:
383:
346:
336:
287:
212:
791:
278:
35:
19:
205:
the protein, this can lead to distorted interpretation of which cells are expressing the
785:
Expression patterns during
Drosophila embryogenesis as inferred by in situ hybridization
332:
260:
Reporter genes can be visualized in living organisms, but both immunohistochemistry and
774:
730:
705:
572:
553:
478:
453:
351:
316:
190:
186:
166:
800:
530:
248:
157:
133:
101:
of regulatory interactions consisting of the effects of many different genes such as
86:
690:
766:
Browse spatiotemporal gene expression patterns organized by human chromosome number
403:
237:
228:
170:
23:
Gene expression patterns are regulated both spatially and temporally in embryos of
779:
657:"Spatiotemporal control of gene expression by a light-switchable transgene system"
643:
341:
137:
784:
291:
156:
One way to identify the expression pattern of a particular gene is to place a
269:
individuals can confound the interpretation of temporal expression patterns.
194:
56:
739:
682:
616:
581:
522:
487:
438:
430:
395:
360:
317:"A Modified Consumer Inkjet for Spatiotemporal Control of Gene Expression"
143:
71:
family of genes. In the early embryonic development of the model organism
181:
673:
656:
469:
281:
spatially, temporally and in different degrees. One method is by using
721:
608:
514:
631:
http://onlinelibrary.wiley.com/doi/10.1002/9780470664001.ch13/summary
282:
165:
can be visualized by stimulating it with blue light and then using a
129:
112:
expression becomes spatially and temporally differential is known as
387:
211:
142:
770:
206:
198:
89:, patches of tissue that will develop into the adult wings. The
39:
454:"Light-dependent RNA interference with nucleobase-caged siRNAs"
780:
Search for mammalian genes with particular expression patterns
241:
136:(mRNA) for these genes in the poles of the egg before it is
85:
is still expressed in a variety of tissues such as the wing
548:. Methods in Cell Biology. Vol. 104. pp. 151–72.
227:
is an alternate method in which a "probe," a synthetic
185:include non-specificity of the antibody leading to
277:Several methods are being pursued for controlling
546:The Zebrafish: Genetics, Genomics and Informatics
273:Methods to control spatiotemporal gene expression
644:https://doi.org/10.1007%2F978-3-642-27426-8_11
315:Cohen, DJ; Morfino, RC; Maharbiz, MM (2009).
8:
257:patterns that are accessible to evolution.
729:
672:
571:
477:
350:
340:
597:Journal of the American Chemical Society
503:Journal of the American Chemical Society
116:. For example, in the case of embryonic
46:of an organism at specific times during
18:
452:Mikat, Vera; Heckel, Alexander (2007).
419:Angewandte Chemie International Edition
307:
244:corresponding to the gene of interest.
7:
128:are asymmetrically expressed in the
264:hybridization must be performed in
152:Identifying spatiotemporal patterns
97:gene expression is determined by a
655:Wang, X; Chen, X; Yang, Y (2012).
554:10.1016/B978-0-12-374814-0.00009-4
14:
217:this point in developmental time.
132:because maternal cells deposit
771:Spatiotemporal gene expression
236:and requires knowledge of the
32:Spatiotemporal gene expression
1:
342:10.1371/journal.pone.0007086
833:
761:expression in fruit flies
163:green fluorescent protein
120:development, the genes
74:Drosophila melanogaster
25:Drosophila melanogaster
431:10.1002/anie.200461458
218:
180:involves preparing an
148:
91:spatiotemporal pattern
28:
215:
146:
22:
296:peptide nucleic acid
178:Immunohistochemistry
603:(36): 11000–11001.
333:2009PLoSO...4.7086C
16:Activation of genes
807:Molecular genetics
790:2008-08-07 at the
757:FlyBase report of
674:10.1038/nmeth.1892
470:10.1261/rna.753407
219:
149:
63:Consider the gene
29:
722:10.1021/cb300161r
609:10.1021/ja073723s
515:10.1021/ja107226e
464:(12): 2341–2347.
114:symmetry breaking
824:
744:
743:
733:
716:(7): 1247–1256.
701:
695:
694:
676:
652:
646:
639:
633:
627:
621:
620:
592:
586:
585:
575:
541:
535:
534:
498:
492:
491:
481:
449:
443:
442:
425:(9): 1328–1332.
414:
408:
407:
371:
365:
364:
354:
344:
312:
288:RNA interference
247:A method called
231:with a sequence
201:that is used to
197:relative to the
169:to record green
77:, or fruit fly,
67:a member of the
42:within specific
832:
831:
827:
826:
825:
823:
822:
821:
812:Gene expression
797:
796:
792:Wayback Machine
753:
748:
747:
703:
702:
698:
654:
653:
649:
640:
636:
628:
624:
594:
593:
589:
564:
543:
542:
538:
500:
499:
495:
451:
450:
446:
416:
415:
411:
376:Nature Genetics
373:
372:
368:
314:
313:
309:
304:
279:gene expression
275:
154:
17:
12:
11:
5:
830:
828:
820:
819:
817:Space and time
814:
809:
799:
798:
795:
794:
782:
777:
775:Genevestigator
768:
763:
752:
751:External links
749:
746:
745:
710:ACS Chem. Biol
696:
647:
634:
622:
587:
562:
536:
509:(3): 440–446.
493:
444:
409:
382:(4): 317–325.
366:
306:
305:
303:
300:
274:
271:
191:false negative
187:false positive
167:digital camera
153:
150:
87:imaginal discs
15:
13:
10:
9:
6:
4:
3:
2:
829:
818:
815:
813:
810:
808:
805:
804:
802:
793:
789:
786:
783:
781:
778:
776:
772:
769:
767:
764:
762:
760:
755:
754:
750:
741:
737:
732:
727:
723:
719:
715:
711:
707:
700:
697:
692:
688:
684:
680:
675:
670:
666:
662:
658:
651:
648:
645:
638:
635:
632:
626:
623:
618:
614:
610:
606:
602:
598:
591:
588:
583:
579:
574:
569:
565:
563:9780123748140
559:
555:
551:
547:
540:
537:
532:
528:
524:
520:
516:
512:
508:
504:
497:
494:
489:
485:
480:
475:
471:
467:
463:
459:
455:
448:
445:
440:
436:
432:
428:
424:
420:
413:
410:
405:
401:
397:
393:
389:
388:10.1038/ng583
385:
381:
377:
370:
367:
362:
358:
353:
348:
343:
338:
334:
330:
326:
322:
318:
311:
308:
301:
299:
297:
293:
289:
284:
280:
272:
270:
267:
263:
258:
255:
250:
249:enhancer-trap
245:
243:
239:
234:
233:complementary
230:
226:
225:hybridization
224:
214:
210:
208:
204:
200:
196:
192:
188:
183:
179:
174:
172:
168:
164:
159:
158:reporter gene
151:
145:
141:
139:
135:
134:messenger RNA
131:
127:
123:
119:
115:
109:
108:
104:
100:
96:
92:
88:
84:
80:
76:
75:
70:
66:
61:
58:
53:
49:
45:
41:
37:
33:
26:
21:
758:
713:
709:
699:
667:(3): 266–9.
664:
660:
650:
637:
625:
600:
596:
590:
545:
539:
506:
502:
496:
461:
457:
447:
422:
418:
412:
379:
375:
369:
327:(9): e7086.
324:
320:
310:
276:
261:
259:
246:
229:nucleic acid
222:
220:
175:
155:
125:
121:
117:
110:
106:
103:even-skipped
102:
94:
82:
78:
72:
64:
62:
51:
31:
30:
24:
661:Nat Methods
171:fluorescent
48:development
801:Categories
302:References
292:morpholino
173:emission.
118:Drosophila
57:cell types
36:activation
531:207058522
254:promoters
203:translate
195:half-life
65:wingless,
788:Archived
759:wingless
740:22540192
691:26529717
683:22327833
617:17711280
582:21924162
523:21162570
488:17951332
439:15643658
396:11479592
361:19763256
321:PLOS ONE
238:sequence
182:antibody
107:Krüppel.
95:wingless
83:wingless
79:wingless
52:tubulin,
731:3401312
573:4408312
479:2080613
404:6773535
352:2739290
329:Bibcode
262:in situ
223:In situ
99:network
44:tissues
34:is the
738:
728:
689:
681:
615:
580:
570:
560:
529:
521:
486:
476:
437:
402:
394:
359:
349:
283:operon
130:oocyte
126:bicoid
687:S2CID
527:S2CID
400:S2CID
266:fixed
122:nanos
40:genes
736:PMID
679:PMID
613:PMID
578:PMID
558:ISBN
519:PMID
484:PMID
435:PMID
392:PMID
357:PMID
207:mRNA
199:mRNA
138:laid
124:and
105:and
773:in
726:PMC
718:doi
669:doi
605:doi
601:129
568:PMC
550:doi
511:doi
507:133
474:PMC
466:doi
458:RNA
427:doi
384:doi
347:PMC
337:doi
294:or
242:DNA
240:of
93:of
69:wnt
38:of
803::
734:.
724:.
712:.
708:.
685:.
677:.
663:.
659:.
611:.
599:.
576:.
566:.
556:.
525:.
517:.
505:.
482:.
472:.
462:13
460:.
456:.
433:.
423:44
421:.
398:.
390:.
380:28
378:.
355:.
345:.
335:.
323:.
319:.
209:.
140:.
742:.
720::
714:7
693:.
671::
665:9
619:.
607::
584:.
552::
533:.
513::
490:.
468::
441:.
429::
406:.
386::
363:.
339::
331::
325:4
27:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.