398:
457:
33:
2287:
1604:
253:
507:
types. The XPC gene is responsible for a protein which recognizes DNA during the early portion of the NER pathway. This gene can have polymorphisms at Intron 9 and SNPs in Exon 15 which have been correlated with cancer risk as well. Research has shown that a biallelic poly (AT) insertion/deletion polymorphism in Intron 9 of XPC is associated with increased risk for skin, breast and prostate cancers, especially in North Indian populations.
215:
3529:
231:) can also recognize some types of damage caused by UV light. Additionally, XPA performs a function in damage recognition that is as yet poorly defined. Upon identification of a damaged site, subsequent repair proteins are then recruited to the damaged DNA to verify presence of DNA damage, excise the damaged DNA surrounding the lesion then fill in the repair patch.
3065:
520:. In a study relapse rates of high-risk stage II and III colorectal cancers, XPD (ERCC2) polymorphism 2251A>C was significantly correlated with early relapse after chemotherapeutic treatment. Studies have indicated that the effects of polymorphic NER genes is additive, with greater frequency of variants, greater cancer risk presents.
260:
At any given time, most of the genome in an organism is not undergoing transcription; there is a difference in NER efficiency between transcriptionally silent and transcriptionally active regions of the genome. For many types of lesions, NER repairs the transcribed strands of transcriptionally active
2203:
Qiao Y, Spitz MR, Guo Z, Hadeyati M, Grossman L, Kraemer KH, Wei Q (November 2002). "Rapid assessment of repair of ultraviolet DNA damage with a modified host-cell reactivation assay using a luciferase reporter gene and correlation with polymorphisms of DNA repair genes in normal human lymphocytes".
351:
are the last two proteins associated with the main NER repair complex. These two proteins are present prior to TFIIH binding since they are involved with verifying DNA damage. They may also protect single-stranded DNA. After verification, the 5' side incision is made and DNA repair begins before the
200:
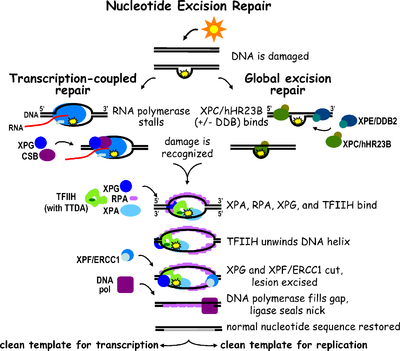
Eukaryotic nucleotide excision repair can be divided into two subpathways: global genomic NER (GG-NER) and transcription coupled NER (TC-NER). Three different sets of proteins are involved in recognizing DNA damage for each subpathway. After damage recognition, the three subpathways converge for the
515:
The study of a hereditary cancer, xeroderma pigmentosum has helped identify several genes which encode proteins in the NER pathway, two of which are XPC and XPD. XP is caused by a homozygous deficiency in UV DNA damage repair (GG-NER) which increases the patients' risk of skin cancer by 1000-fold.
506:
genes. XPD, also known as ERCC2, serves to open DNA around the site of damage during NER, in addition to other transcriptional activities. Studies have shown that polymorphisms at Exon 10 (G>A)(Asp312Asn) and Exon 23 (A>T)(Lys751Gln) are linked with genetic predisposition to several cancer
431:
4 nucleotides downstream of the DNA damage, and the UvrC cleaves a phosphodiester bond 8 nucleotides upstream of the DNA damage and created 12 nucleotide excised segment. DNA helicase II (sometimes called UvrD) then comes in and removes the excised segment by actively breaking the hydrogen bonds
2328:
Vermeij WP, Dollé ME, Reiling E, Jaarsma D, Payan-Gomez C, Bombardieri CR, Wu H, Roks AJ, Botter SM, van der Eerden BC, Youssef SA, Kuiper RV, Nagarajah B, van
Oostrom CT, Brandt RM, Barnhoorn S, Imholz S, Pennings JL, de Bruin A, Gyenis Á, Pothof J, Vijg J, van Steeg H, Hoeijmakers JH (2016).
343:
heterodimeric protein cuts on the 5' side. The dual incision leads to the removal of a ssDNA with a single strand gap of 25~30 nucleotides. The small, excised, damage-containing DNA (sedDNA) oligonucleotides are initially released from the duplex in complex with TFIIH but then dissociate in an
447:
that uses ATP hydrolysis to translocate on dsDNA upstream of the transcription bubble and forward translocate RNA polymerase, thus initiating dissociation of the RNA Polymerase ternary elongation complex. TRCF also recruits the Uvr(A)BC nucleotide excision repair machinery by direct physical
222:
Global genomic NER repairs damage in both transcribed and untranscribed DNA strands in active and inactive genes throughout the genome. This process is not dependent on transcription. This pathway employs several "damage sensing" proteins including the DNA-damage binding (DDB) and XPC-Rad23B
718:
As reviewed by
Gorbunova et al., studies of NER in different cells and tissues from young and old individuals frequently have shown a decrease in NER capacity with increasing age. This decline may be due to reduced constitutive levels of proteins employed in the NER pathway.
269:
stalls at a lesion in DNA: the blocked RNA polymerase serves as a damage recognition signal, which replaces the need for the distortion recognition properties of the XPC-RAD23B and DDB complexes. CS proteins (CSA and CSB) bind some types of DNA damage instead of XPC-Rad23B.
110:. NER can be divided into two subpathways: global genomic NER (GG-NER or GGR) and transcription coupled NER (TC-NER or TCR). The two subpathways differ in how they recognize DNA damage but they share the same process for lesion incision, repair, and ligation.
264:
TC-NER and GG-NER differ only in the initial steps of DNA damage recognition. The principal difference between TC-NER and GG-NER is that TC-NER does not require XPC or DDB proteins for distortion recognition in mammalian cells. Instead TC-NER initiates when
710:(CSB) mutations. Mutations in the CSA gene account for about 20% of CS cases. Individuals with CSA and CSB are characterised by severe postnatal growth and mental retardation and accelerated aging leading to premature death at the age of 12 to 16 years.
489:
cases found modest association between NER specific SNP polymorphisms and lung cancer risk. The results indicate that some inherited polymorphic variations in NER genes may result in predisposition to lung cancer, and potentially other cancer states.
480:
capacity resulting in an increase likelihood of cancer development. While the functional impact of all polymorphisms has not been characterized, some polymorphisms in DNA repair genes or their regulatory sequences do induce
2479:
Andressoo JO, Mitchell JR, de Wit J, Hoogstraten D, Volker M, Toussaint W, Speksnijder E, Beems RB, van Steeg H, Jans J, de Zeeuw CI, Jaspers NG, Raams A, Lehmann AR, Vermeulen W, Hoeijmakers JH, van der Horst GT (2006).
432:
between the complementary bases. The resultant gap is then filled in using DNA polymerase I and DNA ligase. The basic excision process is very similar in higher cells, but these cells usually involve many more proteins –
2634:
Barnhoorn S, Uittenboogaard LM, Jaarsma D, Vermeij WP, Tresini M, Weymaere M, Menoni H, Brandt RM, de Waard MC, Botter SM, Sarker AH, Jaspers NG, van der Horst GT, Cooper PK, Hoeijmakers JH, van der Pluijm I (2014).
281:
TC-NER initiates when RNA polymerase stalls at a lesion in DNA, whereupon protein complexes help move the polymerase backwards. Mutations in TC-NER machinery are responsible for multiple genetic disorders including:
2300:
Spitz MR, Wu X, Wang Y, Wang LE, Shete S, Amos CI, Guo Z, Lei L, Mohrenweiser H, Wei Q (February 2001). "Modulation of nucleotide excision repair capacity by XPD polymorphisms in lung cancer patients".
397:
476:(SNPs) and nonsynonymous coding SNPs (nsSNPs) are present at very low levels (>1%) in the human population. If located in NER genes or regulatory sequences, such mutations can negatively affect
669:
with pronounced degenerative phenotypes in both liver and brain. These mutant mice develop a multi-system premature aging degenerative phenotype that appears to strengthen the link between
344:
ATP-dependent manner and become bound to replication protein A (RPA). Inhibition of gap filling DNA synthesis and ligation results in an accumulation of RPA-bound sedDNAs in the cell.
2110:
Blankenburg S, König IR, Moessner R, Laspe P, Thoms KM, Krueger U, Khan SG, Westphal G, Berking C, Volkenandt M, Reich K, Neumann C, Ziegler A, Kraemer KH, Emmert S (June 2005).
98:
and 6,4-photoproducts. Recognition of the damage leads to removal of a short single-stranded DNA segment that contains the lesion. The undamaged single-stranded DNA remains and
419:
II (sometimes also known as UvrD in this complex). First, a UvrA-UvrB complex scans the DNA, with the UvrA subunit recognizing distortions in the helix, caused for example by
516:
In heterozygous patients, the risk of cancer is sporadic but can be predicted based on analytical assessment of polymorphisms in XP related DNA repair genes purified from
2244:"ERCC2 2251A>C genetic polymorphism was highly correlated with early relapse in high-risk stage II and stage III colorectal cancer patients: a preliminary study"
616:(XPD) mutant human and mouse show evidence of defective repair of oxidative DNA damages that may underlie the segmental progeroid (premature aging) symptoms (see
423:. When the complex recognizes such a distortion, the UvrA subunit leaves and an UvrC protein comes in and binds to the UvrB monomer and, hence, forms a new UvrBC
323:
act as a 5'-3' and 3'-5' helicase, respectively — they help unwind DNA and generate a junction between the double-stranded and single-stranded DNA around the
2432:"XPB and XPD helicases in TFIIH orchestrate DNA duplex opening and damage verification to coordinate repair with transcription and cell cycle via CAK kinase"
3462:
2950:"TFIIH-mediated nucleotide excision repair and initiation of mRNA transcription in an optimized cell-free DNA repair and RNA transcription assay"
2531:
Oh KS, Khan SG, Jaspers NG, Raams A, Ueda T, Lehmann A, Friedmann PS, Emmert S, Gratchev A, Lachlan K, Lucassan A, Baker CC, Kraemer KH (2006).
145:, there are 9 major proteins involved in NER. Deficiencies in certain proteins leads to disease; protein names are associated with the disease.
3436:
2061:"Genetic variation in nucleotide excision repair pathway genes influence prostate and bladder cancer susceptibility in North Indian population"
2938:
1626:
368:(PCNA) onto the DNA strand. This allows DNA polymerases implicated in repair (δ, ε and/or κ) to copy the undamaged strand via translocation.
3097:
2482:"An Xpd mouse model for the combined xeroderma pigmentosum/Cockayne syndrome exhibiting both cancer predisposition and segmental progeria"
3069:
3152:
468:
Though historical studies have shown inconsistent results, genetic variation or mutation to nucleotide excision repair genes can impact
103:
585:
mutant mice show features of accelerated aging, and have a limited lifespan. Accelerated aging in the mutant involves numerous organs.
3405:
3336:
365:
456:
3420:
2005:
Wang M, Gu D, Zhang Z, Zhou J, Zhang Z (2009). "XPD polymorphisms, cigarette smoking, and bladder cancer risk: a meta-analysis".
670:
464:. Unrepaired damage or malfunctioning proteins associated with excision repair could lead to unregulated cell growth and cancer.
2151:
Shore RE, Zeleniuch-Jacquotte A, Currie D, Mohrenweiser H, Afanasyeva Y, Koenig KL, Arslan AA, Toniolo P, Wirgin I (May 2008).
32:
1747:
Truglio JJ, Croteau DL, Van Houten B, Kisker C (February 2006). "Prokaryotic nucleotide excision repair: the UvrABC system".
473:
1923:
Sakoda LC, Loomis MM, Doherty JA, Julianto L, Barnett MJ, Neuhouser ML, Thornquist MD, Weiss NS, Goodman GE, Chen C (2012).
113:
The importance of NER is evidenced by the severe human diseases that result from in-born genetic mutations of NER proteins.
2737:"Cockayne syndrome protein A is a transcription factor of RNA polymerase I and stimulates ribosomal biogenesis and growth"
243:
Xeroderma pigmentosum (XP): severe photosensitivity, high cancer rates in areas of the body exposed to the sun (e.g. skin)
2533:"Phenotypic heterogeneity in the XPB DNA helicase gene (ERCC3): xeroderma pigmentosum without and with Cockayne syndrome"
3395:
3302:
52:
2112:"Assessment of 3 xeroderma pigmentosum group C gene polymorphisms and risk of cutaneous melanoma: a case-control study"
3324:
3141:
678:
617:
605:
440:
86:
Nucleotide excision repair (NER) is a particularly important excision mechanism that removes DNA damage induced by
2786:"Genome Instability in Development and Aging: Insights from Nucleotide Excision Repair in Humans, Mice, and Worms"
1382:
608:, retinal degeneration, white matter hypomethylation, central nervous system calcification, reduced stature, and
532:
in genes employed in NER cause features of premature aging. These genes and their corresponding proteins include
498:
Two important genes in the NER pathway for which polymorphism has shown functional and phenotypic impact are the
308:
193:, and others also participate in nucleotide excision repair. A more complete list of proteins involved in NER is
3319:
3241:
3090:
739:
63:. Three excision repair pathways exist to repair single stranded DNA damage: Nucleotide excision repair (NER),
36:
Diagram of both the TC-NER and GG-NER pathways. The two pathways differ only in initial DNA damage recognition.
1831:
Kwok PY, Gu Z (December 1999). "Single nucleotide polymorphism libraries: why and how are we building them?".
796:
2286:
1603:
401:
A schematic representation of models for the nucleotide excision repair pathway controlled by Uvr proteins.
3431:
3341:
3136:
3197:
650:
628:
593:
182:
166:
114:
1698:"Molecular mechanisms of the whole DNA repair system: a comparison of bacterial and eukaryotic systems"
2735:
Koch S, Garcia
Gonzalez O, Assfalg R, Schelling A, Schäfer P, Scharffetter-Kochanek K, Iben S (2014).
2637:"Cell-autonomous progeroid changes in conditional mouse models for repair endonuclease XPG deficiency"
1966:"The XPD variant alleles are associated with increased aromatic DNA adduct level and lung cancer risk"
1647:"True lies: the double life of the nucleotide excision repair factors in transcription and DNA repair"
3122:
2342:
2014:
1879:
1533:
361:
324:
76:
72:
64:
3553:
3532:
3219:
3083:
1539:
597:
428:
412:
392:
373:
286:
68:
2562:
2182:
2038:
1784:"Involvement of nucleotide excision and mismatch repair mechanisms in double strand break repair"
252:
3415:
3044:
3015:
2979:
2934:
2907:
2866:
2817:
2766:
2717:
2668:
2611:
2554:
2513:
2461:
2412:
2368:
2310:
2275:
2221:
2174:
2133:
2092:
2030:
1987:
1946:
1905:
1848:
1813:
1764:
1729:
1678:
1622:
1592:
654:
632:
601:
529:
424:
292:
118:
87:
1104:
Transcription elongation factor; involved in transcription coupling and chromatin remodelling
3036:
3005:
2969:
2961:
2897:
2856:
2848:
2807:
2797:
2756:
2748:
2707:
2699:
2658:
2648:
2601:
2593:
2544:
2503:
2493:
2451:
2443:
2402:
2358:
2350:
2265:
2255:
2213:
2164:
2123:
2082:
2072:
2022:
1977:
1936:
1895:
1887:
1840:
1803:
1795:
1756:
1719:
1709:
1668:
1658:
1582:
1572:
1420:
1417:
1414:
1305:
1272:
1071:
Involved in incision on 5' side of damage; stabilizes TFIIH; structure specific endonuclease
420:
407:
315:
are first recruited to the site of DNA damage (XPG stabilizes TFIIH). The TFIIH subunits of
17:
2688:"Elements That Regulate the DNA Damage Response of Proteins Defective in Cockayne Syndrome"
2331:"Restricted diet delays accelerated ageing and genomic stress in DNA-repair-deficient mice"
1239:
1206:
1140:
1107:
1074:
1041:
1008:
975:
942:
847:
352:
3' side incision. This helps reduce exposed single stranded DNA during the repair process.
3131:
1453:
1371:
1338:
1173:
909:
878:
818:
785:
1925:"Germ line variation in nucleotide excision repair genes and lung cancer risk in smokers"
1519:
1486:
604:(XPCS). TTD and CS both display features of premature aging. These features may include
2346:
2018:
1883:
239:
Mutations in GG-NER machinery are responsible for multiple genetic disorders including:
2861:
2836:
2812:
2785:
2761:
2736:
2712:
2687:
2663:
2636:
2606:
2581:
2456:
2431:
2363:
2330:
2270:
2243:
2087:
2060:
1941:
1924:
1900:
1867:
1808:
1783:
1724:
1697:
1673:
1646:
1587:
1560:
266:
99:
3010:
2993:
2974:
2949:
2217:
1844:
261:
genes faster than it repairs nontranscribed strands and transcriptionally silent DNA.
214:
3547:
3443:
3264:
1868:"Impact of DNA polymorphisms in key DNA base excision repair proteins on cancer risk"
162:
95:
2566:
2042:
3410:
3379:
3329:
3312:
3259:
3212:
2886:"Mechanisms and implications of the age-associated decrease in DNA repair capacity"
2186:
1118:
666:
574:
482:
416:
369:
328:
289:(TTD): some individuals are photosensitive, ichthyosis, mental/physical retardation
1964:
Hou SM, Fält S, Angelini S, Yang K, Nyberg F, Lambert B, Hemminki K (April 2002).
106:. Final ligation to complete NER and form a double stranded DNA is carried out by
3040:
2653:
2597:
2447:
1577:
2992:
Frit P, Kwon K, Coin F, Auriol J, Dubaele S, Salles B, Egly JM (December 2002).
733:
642:(XPF) in humans results in a variety of conditions including accelerated aging.
486:
227:-Rad23B complex is responsible for distortion recognition, while DDB1 and DDB2 (
142:
2391:"Nucleotide excision repair disorders and the balance between cancer and aging"
1799:
223:
complexes that constantly scan the genome and recognize helix distortions: the
3346:
3167:
3114:
3106:
2929:
Ellenberger T, Friedberg EC, Walker GS, Wolfram S, Wood RJ, Schultz R (2006).
2703:
2498:
2481:
2026:
1982:
1965:
1497:
517:
503:
477:
461:
380:
228:
224:
158:
154:
138:
134:
130:
107:
91:
44:
3027:
Mellon I (September 2005). "Transcription-coupled repair: a complex affair".
2965:
1891:
2582:"Physiological consequences of defects in ERCC1-XPF DNA repair endonuclease"
2128:
2111:
2077:
751:
80:
56:
3048:
3019:
2911:
2870:
2821:
2770:
2721:
2672:
2615:
2558:
2517:
2465:
2416:
2372:
2314:
2279:
2260:
2225:
2178:
2137:
2096:
2034:
1991:
1950:
1909:
1852:
1817:
1768:
1733:
1682:
1596:
415:
enzyme complex, which consists of four Uvr proteins: UvrA, UvrB, UvrC, and
177:
represent proteins linked to
Cockayne syndrome. Additionally, the proteins
3064:
2983:
2902:
2885:
1714:
1663:
1038:
Involved in incision on 3' side of damage; structure specific endonuclease
653:(XP) alone, or in combination with the severe neurodevelopmental disorder
3427:
2852:
2407:
2390:
763:
688:
685:
662:
609:
600:(TTD) or a combination of XP and TTD (XPTTD), or a combination of XP and
296:
2802:
2354:
661:(XPG) mutant mouse model presents features of premature aging including
3374:
3126:
2549:
2532:
1005:
ATPase and helicase activity; transcription factor II H (TFIIH) subunit
972:
ATPase and helicase activity; transcription factor II H (TFIIH) subunit
828:
706:(CSA) mutations generally give rise to a more moderate form of CS than
657:(CS) or the infantile lethal cerebro-oculo-facio-skeletal syndrome. An
60:
2884:
Goukassian D, Gad F, Yaar M, Eller MS, Nehal US, Gilchrest BA (2000).
2508:
2169:
2153:"Polymorphisms in XPC and ERCC2 genes, smoking and breast cancer risk"
2152:
1760:
273:
Other repair mechanisms are possible but less accurate and efficient.
3499:
3254:
3207:
3202:
2752:
1283:
1250:
674:
469:
444:
190:
186:
3514:
3509:
3504:
3494:
3487:
3482:
3477:
3472:
3467:
3457:
3452:
3307:
3271:
3192:
3187:
3182:
3172:
1217:
1184:
1085:
1052:
1019:
953:
920:
568:
562:
556:
542:
537:
499:
396:
377:
340:
336:
332:
316:
312:
213:
178:
174:
170:
79:. Similarly, the MMR pathway only targets mismatched Watson-Crick
939:
Involved in incision on 3' side of damage; forms complex with XPF
612:(loss of subcutaneous fat tissue). XPCS and TTD fibroblasts from
3400:
3356:
3351:
3286:
3281:
3276:
3249:
3229:
3224:
3177:
1431:
1349:
1316:
1151:
889:
858:
692:
3079:
311:(TFIIH) is the key enzyme involved in dual excision. TFIIH and
3162:
3157:
1464:
986:
551:
348:
320:
150:
146:
75:
in DNA, it can correct only damaged bases that are removed by
48:
3075:
2242:
Huang MY, Fang WY, Lee SC, Cheng TL, Wang JY, Lin SR (2008).
1137:
Ubiquitin ligase complex; interacts with CSB and p44 of TFIIH
1929:
International
Journal of Molecular Epidemiology and Genetics
1617:
Carroll SB; Wessler SR; Griffiths AJFl; Lewontin RC (2008).
485:
changes and are involved in cancer development. A study of
405:
The process of nucleotide excision repair is controlled in
256:
Schematic depicts binding of proteins involved with TC-NER.
218:
Schematic depicts binding of proteins involved with GG-NER.
1561:"DNA repair: dynamic defenders against cancer and aging"
649:(XPG) gene can cause either the cancer-prone condition
439:
TC-NER also exists in bacteria, and is mediated by the
1236:
Interacts with XPD and XPB subunits of TFIIH helicases
2054:
2052:
2007:
Journal of
Toxicology and Environmental Health Part A
1450:
Damage recognition; interacts with XPA, CSA, and CSB
51:
damage occurs constantly because of chemicals (e.g.
3388:
3367:
3295:
3240:
3113:
1696:Morita R, Nakane S, Shimada A, et al. (2010).
2389:Andressoo JO, Hoeijmakers JH, Mitchell JR (2006).
2994:"Transcriptional activators stimulate DNA repair"
1411:Involved in incision, forms complex around lesion
592:(XPD) gene can lead to various syndromes, either
327:. In addition to stabilizing TFIIH, XPG also has
2835:Gorbunova V, Seluanov A, Mao Z, Hine C (2007).
2629:
2627:
2625:
2580:Gregg SQ, Robinson AR, Niedernhofer LJ (2011).
2384:
2382:
460:DNA excision pathways work in tandem to repair
1866:Karahalil B, Bohr V, Wilson D (October 2012).
1640:
1638:
1621:. New York: W.H. Freeman and Co. p. 534.
201:steps of dual incision, repair, and ligation.
129:Nucleotide excision repair is more complex in
3091:
2988:Article on the relation between TFIIH and NER
2237:
2235:
121:are two examples of NER associated diseases.
8:
2198:
2196:
295:(CS): photosensitivity, mental retardation,
102:uses it as a template to synthesize a short
875:Damage recognition; forms complex with DDB2
71:(MMR). While the BER pathway can recognize
3098:
3084:
3076:
1302:Damage recognition, forms complex with XPC
1269:Damage recognition; forms complex with XPC
844:Damage recognition; forms complex with XPC
726:
494:NER dysfunction result of DNA polymorphism
3009:
2973:
2901:
2860:
2811:
2801:
2760:
2711:
2662:
2652:
2605:
2548:
2507:
2497:
2455:
2406:
2362:
2269:
2259:
2168:
2127:
2086:
2076:
1981:
1940:
1899:
1807:
1723:
1713:
1672:
1662:
1586:
1576:
194:
2948:Satoh MS, Hanawalt PC (September 1996).
455:
251:
31:
1551:
2059:Mittal RD, Mandal RK (January 2012).
1782:Zhang Y, Rohde LH, Wu H (June 2009).
645:In humans, mutational defects in the
248:Transcription coupled repair (TC-NER)
90:(UV). UV DNA damage results in bulky
7:
2837:"Changes in DNA repair during aging"
472:risk by affecting repair efficacy.
331:activity; it cuts DNA damage on the
1872:Human & Experimental Toxicology
684:Cockayne syndrome (CS) arises from
627:(XPB) gene can lead, in humans, to
448:interaction with the UvrA subunit.
3406:Proliferating Cell Nuclear Antigen
3337:Microhomology-mediated end joining
1645:Le May N, Egly JM, Coin F (2010).
782:CDK Activator Kinase (CAK) subunit
366:Proliferating Cell Nuclear Antigen
25:
137:, which express enzymes like the
3528:
3527:
3421:Meiotic recombination checkpoint
3063:
2784:Edifizi D, Schumacher B (2015).
2285:
2065:Indian Journal of Human Genetics
1619:Introduction to genetic analysis
1602:
1559:Fuss JO, Cooper PK (June 2006).
906:Damage recognition; recruits XPC
797:Cyclin Dependent Kinase (CDK) 7)
347:Replication protein A (RPA) and
2933:. Washington, D.C.: ASM Press.
2157:International Journal of Cancer
474:Single-nucleotide polymorphisms
1:
3011:10.1016/S1097-2765(02)00732-3
2218:10.1016/S0027-5107(02)00219-1
1845:10.1016/S1357-4310(99)01601-9
3303:Transcription-coupled repair
3041:10.1016/j.mrfmmm.2005.03.016
2654:10.1371/journal.pgen.1004686
2598:10.1016/j.dnarep.2011.04.026
2448:10.1016/j.dnarep.2011.04.028
1578:10.1371/journal.pbio.0040203
387:In prokaryotes: Uvr proteins
299:-like features, microcephaly
18:Transcription-coupled repair
2686:Iyama T, Wilson DM (2016).
2430:Fuss JO, Tainer JA (2011).
383:the nicks to complete NER.
210:Global genomic NER (GG-NER)
94:— these adducts are mostly
3570:
3325:Non-homologous end joining
3149:Nucleotide excision repair
3142:Poly ADP ribose polymerase
3070:Nucleotide excision repair
2931:DNA repair and mutagenesis
1800:10.2174/138920209788488544
679:DNA damage theory of aging
618:DNA damage theory of aging
511:Impact on cancer prognosis
390:
277:TC-NER associated diseases
235:GG-NER associated diseases
73:specific non-bulky lesions
41:Nucleotide excision repair
3523:
2704:10.1016/j.jmb.2015.11.020
2499:10.1016/j.ccr.2006.05.027
2027:10.1080/15287390902841029
1383:Transcription factor II H
714:Decline in NER with aging
631:(XP) or XP combined with
309:Transcription factor II H
3320:Homology directed repair
3242:Homologous recombination
1892:10.1177/0960327112444476
1833:Molecular Medicine Today
1702:Journal of Nucleic Acids
1651:Journal of Nucleic Acids
443:protein. TRCF is an SF2
378:Ligase-III-XRCC1 complex
2078:10.4103/0971-6866.96648
1983:10.1093/carcin/23.4.599
3342:Postreplication repair
3137:Uracil-DNA glycosylase
2966:10.1093/nar/24.18.3576
2954:Nucleic Acids Research
2261:10.1186/1471-2407-8-50
1368:Subunit of RFA complex
1335:Subunit of RFA complex
1203:Stabilizes CAK complex
606:sensorineural deafness
465:
402:
360:Replication factor C (
257:
219:
141:. In humans and other
104:complementary sequence
37:
2903:10.1096/fj.14.10.1325
2129:10.1093/carcin/bgi055
651:xeroderma pigmentosum
629:xeroderma pigmentosum
594:xeroderma pigmentosum
581:DNA repair-deficient
459:
436:is a simple example.
400:
391:Further information:
255:
217:
167:хeroderma pigmentosum
115:Xeroderma pigmentosum
77:specific glycosylases
35:
3448:core protein complex
3123:Base excision repair
3072:at Wikimedia Commons
2408:10.4161/cc.5.24.3565
1534:Base excision repair
730:Human Gene (Protein)
723:NER associated genes
528:In humans and mice,
325:transcription bubble
65:base excision repair
53:intercalating agents
27:DNA repair mechanism
3220:DNA mismatch repair
2803:10.3390/biom5031855
2355:10.1038/nature19329
2347:2016Natur.537..427V
2019:2009JTEHA..72..698W
1884:2012HETox..31..981K
1715:10.4061/2010/179594
1664:10.4061/2010/616342
598:trichothiodystrophy
429:phosphodiester bond
413:UvrABC endonuclease
393:UvrABC endonuclease
374:Flap endonuclease 1
356:Repair and ligation
287:Trichothiodystrophy
119:Cockayne's syndrome
69:DNA mismatch repair
2853:10.1093/nar/gkm756
2586:DNA Repair (Amst.)
2550:10.1002/humu.20392
2436:DNA Repair (Amst.)
2013:(11–12): 698–705.
1516:Damage recognition
1483:Damage recognition
466:
403:
258:
220:
205:Damage recognition
38:
3541:
3540:
3416:Adaptive response
3068:Media related to
2960:(18): 3576–3582.
2940:978-1-55581-319-2
2841:Nucleic Acids Res
2341:(7620): 427–431.
2206:Mutation Research
2170:10.1002/ijc.23361
1761:10.1021/cr040471u
1628:978-0-7167-6887-6
1525:
1524:
691:in either of two
655:Cockayne syndrome
633:Cockayne syndrome
623:Mutations in the
602:Cockayne syndrome
588:Mutations in the
530:germline mutation
427:. UvrB cleaves a
421:pyrimidine dimers
293:Cockayne syndrome
143:placental animals
88:ultraviolet light
16:(Redirected from
3561:
3531:
3530:
3100:
3093:
3086:
3077:
3067:
3052:
3035:(1–2): 155–161.
3023:
3013:
3004:(6): 1391–1401.
2987:
2977:
2944:
2916:
2915:
2905:
2881:
2875:
2874:
2864:
2832:
2826:
2825:
2815:
2805:
2781:
2775:
2774:
2764:
2753:10.4161/cc.29018
2732:
2726:
2725:
2715:
2683:
2677:
2676:
2666:
2656:
2647:(10): e1004686.
2631:
2620:
2619:
2609:
2577:
2571:
2570:
2552:
2543:(11): 1092–103.
2528:
2522:
2521:
2511:
2501:
2476:
2470:
2469:
2459:
2427:
2421:
2420:
2410:
2386:
2377:
2376:
2366:
2325:
2319:
2318:
2309:(4): 1354–1357.
2297:
2291:
2290:
2289:
2283:
2273:
2263:
2239:
2230:
2229:
2212:(1–2): 165–174.
2200:
2191:
2190:
2172:
2163:(9): 2101–2105.
2148:
2142:
2141:
2131:
2122:(6): 1085–1090.
2107:
2101:
2100:
2090:
2080:
2056:
2047:
2046:
2002:
1996:
1995:
1985:
1961:
1955:
1954:
1944:
1920:
1914:
1913:
1903:
1878:(10): 981–1005.
1863:
1857:
1856:
1828:
1822:
1821:
1811:
1788:Current Genomics
1779:
1773:
1772:
1749:Chemical Reviews
1744:
1738:
1737:
1727:
1717:
1693:
1687:
1686:
1676:
1666:
1642:
1633:
1632:
1614:
1608:
1607:
1606:
1600:
1590:
1580:
1556:
727:
408:Escherichia coli
165:all derive from
21:
3569:
3568:
3564:
3563:
3562:
3560:
3559:
3558:
3544:
3543:
3542:
3537:
3519:
3389:Other/ungrouped
3384:
3363:
3291:
3236:
3132:DNA glycosylase
3115:Excision repair
3109:
3104:
3060:
3055:
3026:
2991:
2947:
2941:
2928:
2924:
2922:Further reading
2919:
2896:(10): 1325–34.
2883:
2882:
2878:
2847:(22): 7466–74.
2834:
2833:
2829:
2783:
2782:
2778:
2747:(13): 2029–37.
2734:
2733:
2729:
2685:
2684:
2680:
2633:
2632:
2623:
2579:
2578:
2574:
2530:
2529:
2525:
2478:
2477:
2473:
2429:
2428:
2424:
2388:
2387:
2380:
2327:
2326:
2322:
2303:Cancer Research
2299:
2298:
2294:
2284:
2241:
2240:
2233:
2202:
2201:
2194:
2150:
2149:
2145:
2109:
2108:
2104:
2058:
2057:
2050:
2004:
2003:
1999:
1963:
1962:
1958:
1922:
1921:
1917:
1865:
1864:
1860:
1839:(12): 538–543.
1830:
1829:
1825:
1781:
1780:
1776:
1746:
1745:
1741:
1695:
1694:
1690:
1644:
1643:
1636:
1629:
1616:
1615:
1611:
1601:
1558:
1557:
1553:
1549:
1540:Mismatch repair
1530:
748:Function in NER
725:
716:
526:
513:
496:
454:
395:
389:
358:
335:side while the
306:
279:
250:
237:
212:
207:
127:
28:
23:
22:
15:
12:
11:
5:
3567:
3565:
3557:
3556:
3546:
3545:
3539:
3538:
3536:
3535:
3524:
3521:
3520:
3518:
3517:
3512:
3507:
3502:
3497:
3492:
3491:
3490:
3485:
3480:
3475:
3470:
3465:
3460:
3455:
3440:
3439:
3434:
3424:
3423:
3418:
3413:
3408:
3403:
3398:
3392:
3390:
3386:
3385:
3383:
3382:
3377:
3371:
3369:
3365:
3364:
3362:
3361:
3360:
3359:
3354:
3344:
3339:
3334:
3333:
3332:
3322:
3317:
3316:
3315:
3310:
3299:
3297:
3296:Other pathways
3293:
3292:
3290:
3289:
3284:
3279:
3274:
3269:
3268:
3267:
3257:
3252:
3246:
3244:
3238:
3237:
3235:
3234:
3233:
3232:
3227:
3217:
3216:
3215:
3210:
3205:
3200:
3195:
3190:
3185:
3180:
3175:
3170:
3165:
3160:
3146:
3145:
3144:
3139:
3134:
3119:
3117:
3111:
3110:
3105:
3103:
3102:
3095:
3088:
3080:
3074:
3073:
3059:
3058:External links
3056:
3054:
3053:
3024:
2989:
2945:
2939:
2925:
2923:
2920:
2918:
2917:
2876:
2827:
2796:(3): 1855–69.
2776:
2727:
2678:
2621:
2572:
2523:
2471:
2442:(7): 697–713.
2422:
2401:(24): 2886–8.
2378:
2320:
2292:
2231:
2192:
2143:
2116:Carcinogenesis
2102:
2048:
1997:
1976:(4): 599–603.
1970:Carcinogenesis
1956:
1915:
1858:
1823:
1794:(4): 250–258.
1774:
1755:(2): 233–252.
1739:
1688:
1634:
1627:
1609:
1550:
1548:
1545:
1544:
1543:
1537:
1529:
1526:
1523:
1522:
1517:
1514:
1511:
1506:
1501:
1490:
1489:
1484:
1481:
1478:
1473:
1468:
1457:
1456:
1451:
1448:
1445:
1440:
1435:
1424:
1423:
1412:
1409:
1406:
1395:
1386:
1375:
1374:
1369:
1366:
1363:
1358:
1353:
1342:
1341:
1336:
1333:
1330:
1325:
1320:
1309:
1308:
1303:
1300:
1297:
1292:
1287:
1276:
1275:
1270:
1267:
1264:
1259:
1254:
1243:
1242:
1237:
1234:
1231:
1226:
1221:
1210:
1209:
1204:
1201:
1198:
1193:
1188:
1177:
1176:
1171:
1170:Final ligation
1168:
1165:
1160:
1155:
1144:
1143:
1138:
1135:
1132:
1127:
1122:
1111:
1110:
1105:
1102:
1099:
1094:
1089:
1078:
1077:
1072:
1069:
1066:
1061:
1056:
1045:
1044:
1039:
1036:
1033:
1028:
1023:
1012:
1011:
1006:
1003:
1000:
995:
990:
979:
978:
973:
970:
967:
962:
957:
946:
945:
940:
937:
934:
929:
924:
913:
912:
907:
904:
901:
898:
893:
882:
881:
876:
873:
870:
867:
862:
851:
850:
845:
842:
839:
836:
831:
822:
821:
816:
813:
810:
805:
800:
789:
788:
783:
780:
777:
772:
767:
756:
755:
749:
746:
743:
737:
731:
724:
721:
715:
712:
638:Deficiency of
525:
522:
512:
509:
495:
492:
453:
450:
388:
385:
357:
354:
305:
302:
301:
300:
290:
278:
275:
267:RNA polymerase
249:
246:
245:
244:
236:
233:
211:
208:
206:
203:
126:
123:
100:DNA polymerase
96:thymine dimers
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
3566:
3555:
3552:
3551:
3549:
3534:
3526:
3525:
3522:
3516:
3513:
3511:
3508:
3506:
3503:
3501:
3498:
3496:
3493:
3489:
3486:
3484:
3481:
3479:
3476:
3474:
3471:
3469:
3466:
3464:
3461:
3459:
3456:
3454:
3451:
3450:
3449:
3445:
3444:FANC proteins
3442:
3441:
3438:
3435:
3433:
3429:
3426:
3425:
3422:
3419:
3417:
3414:
3412:
3409:
3407:
3404:
3402:
3399:
3397:
3394:
3393:
3391:
3387:
3381:
3378:
3376:
3373:
3372:
3370:
3366:
3358:
3355:
3353:
3350:
3349:
3348:
3345:
3343:
3340:
3338:
3335:
3331:
3328:
3327:
3326:
3323:
3321:
3318:
3314:
3311:
3309:
3306:
3305:
3304:
3301:
3300:
3298:
3294:
3288:
3285:
3283:
3280:
3278:
3275:
3273:
3270:
3266:
3265:RecQ helicase
3263:
3262:
3261:
3258:
3256:
3253:
3251:
3248:
3247:
3245:
3243:
3239:
3231:
3228:
3226:
3223:
3222:
3221:
3218:
3214:
3211:
3209:
3206:
3204:
3201:
3199:
3196:
3194:
3191:
3189:
3186:
3184:
3181:
3179:
3176:
3174:
3171:
3169:
3166:
3164:
3161:
3159:
3156:
3155:
3154:
3150:
3147:
3143:
3140:
3138:
3135:
3133:
3130:
3129:
3128:
3124:
3121:
3120:
3118:
3116:
3112:
3108:
3101:
3096:
3094:
3089:
3087:
3082:
3081:
3078:
3071:
3066:
3062:
3061:
3057:
3050:
3046:
3042:
3038:
3034:
3030:
3025:
3021:
3017:
3012:
3007:
3003:
2999:
2995:
2990:
2985:
2981:
2976:
2971:
2967:
2963:
2959:
2955:
2951:
2946:
2942:
2936:
2932:
2927:
2926:
2921:
2913:
2909:
2904:
2899:
2895:
2891:
2887:
2880:
2877:
2872:
2868:
2863:
2858:
2854:
2850:
2846:
2842:
2838:
2831:
2828:
2823:
2819:
2814:
2809:
2804:
2799:
2795:
2791:
2787:
2780:
2777:
2772:
2768:
2763:
2758:
2754:
2750:
2746:
2742:
2738:
2731:
2728:
2723:
2719:
2714:
2709:
2705:
2701:
2697:
2693:
2689:
2682:
2679:
2674:
2670:
2665:
2660:
2655:
2650:
2646:
2642:
2638:
2630:
2628:
2626:
2622:
2617:
2613:
2608:
2603:
2599:
2595:
2592:(7): 781–91.
2591:
2587:
2583:
2576:
2573:
2568:
2564:
2560:
2556:
2551:
2546:
2542:
2538:
2534:
2527:
2524:
2519:
2515:
2510:
2505:
2500:
2495:
2492:(2): 121–32.
2491:
2487:
2483:
2475:
2472:
2467:
2463:
2458:
2453:
2449:
2445:
2441:
2437:
2433:
2426:
2423:
2418:
2414:
2409:
2404:
2400:
2396:
2392:
2385:
2383:
2379:
2374:
2370:
2365:
2360:
2356:
2352:
2348:
2344:
2340:
2336:
2332:
2324:
2321:
2316:
2312:
2308:
2304:
2296:
2293:
2288:
2281:
2277:
2272:
2267:
2262:
2257:
2253:
2249:
2245:
2238:
2236:
2232:
2227:
2223:
2219:
2215:
2211:
2207:
2199:
2197:
2193:
2188:
2184:
2180:
2176:
2171:
2166:
2162:
2158:
2154:
2147:
2144:
2139:
2135:
2130:
2125:
2121:
2117:
2113:
2106:
2103:
2098:
2094:
2089:
2084:
2079:
2074:
2070:
2066:
2062:
2055:
2053:
2049:
2044:
2040:
2036:
2032:
2028:
2024:
2020:
2016:
2012:
2008:
2001:
1998:
1993:
1989:
1984:
1979:
1975:
1971:
1967:
1960:
1957:
1952:
1948:
1943:
1938:
1934:
1930:
1926:
1919:
1916:
1911:
1907:
1902:
1897:
1893:
1889:
1885:
1881:
1877:
1873:
1869:
1862:
1859:
1854:
1850:
1846:
1842:
1838:
1834:
1827:
1824:
1819:
1815:
1810:
1805:
1801:
1797:
1793:
1789:
1785:
1778:
1775:
1770:
1766:
1762:
1758:
1754:
1750:
1743:
1740:
1735:
1731:
1726:
1721:
1716:
1711:
1707:
1703:
1699:
1692:
1689:
1684:
1680:
1675:
1670:
1665:
1660:
1656:
1652:
1648:
1641:
1639:
1635:
1630:
1624:
1620:
1613:
1610:
1605:
1598:
1594:
1589:
1584:
1579:
1574:
1570:
1566:
1562:
1555:
1552:
1546:
1541:
1538:
1535:
1532:
1531:
1527:
1521:
1518:
1515:
1512:
1510:
1507:
1505:
1502:
1499:
1495:
1492:
1491:
1488:
1485:
1482:
1479:
1477:
1474:
1472:
1469:
1466:
1462:
1459:
1458:
1455:
1452:
1449:
1446:
1444:
1441:
1439:
1436:
1433:
1429:
1426:
1425:
1422:
1419:
1416:
1413:
1410:
1407:
1405:
1402:
1399:
1396:
1394:
1390:
1387:
1384:
1380:
1377:
1376:
1373:
1370:
1367:
1364:
1362:
1359:
1357:
1354:
1351:
1347:
1344:
1343:
1340:
1337:
1334:
1331:
1329:
1326:
1324:
1321:
1318:
1314:
1311:
1310:
1307:
1304:
1301:
1298:
1296:
1293:
1291:
1288:
1285:
1281:
1278:
1277:
1274:
1271:
1268:
1265:
1263:
1260:
1258:
1255:
1252:
1248:
1245:
1244:
1241:
1238:
1235:
1232:
1230:
1227:
1225:
1222:
1219:
1215:
1212:
1211:
1208:
1205:
1202:
1199:
1197:
1194:
1192:
1189:
1186:
1182:
1179:
1178:
1175:
1172:
1169:
1166:
1164:
1161:
1159:
1156:
1153:
1149:
1146:
1145:
1142:
1139:
1136:
1133:
1131:
1128:
1126:
1123:
1120:
1116:
1113:
1112:
1109:
1106:
1103:
1100:
1098:
1095:
1093:
1090:
1087:
1083:
1080:
1079:
1076:
1073:
1070:
1067:
1065:
1062:
1060:
1057:
1054:
1050:
1047:
1046:
1043:
1040:
1037:
1034:
1032:
1029:
1027:
1024:
1021:
1017:
1014:
1013:
1010:
1007:
1004:
1001:
999:
996:
994:
991:
988:
984:
981:
980:
977:
974:
971:
968:
966:
963:
961:
958:
955:
951:
948:
947:
944:
941:
938:
935:
933:
930:
928:
925:
922:
918:
915:
914:
911:
908:
905:
902:
899:
897:
894:
891:
887:
884:
883:
880:
877:
874:
871:
868:
866:
863:
860:
856:
853:
852:
849:
846:
843:
840:
837:
835:
832:
830:
827:
824:
823:
820:
817:
814:
811:
809:
806:
804:
801:
798:
794:
791:
790:
787:
784:
781:
778:
776:
773:
771:
768:
765:
761:
758:
757:
753:
750:
747:
744:
741:
738:
735:
732:
729:
728:
722:
720:
713:
711:
709:
705:
701:
697:
694:
690:
687:
682:
680:
676:
672:
668:
664:
660:
656:
652:
648:
643:
641:
636:
634:
630:
626:
621:
619:
615:
611:
607:
603:
599:
595:
591:
586:
584:
579:
577:
576:
571:
570:
565:
564:
559:
558:
553:
549:
545:
544:
539:
535:
531:
523:
521:
519:
510:
508:
505:
501:
493:
491:
488:
484:
479:
475:
471:
463:
458:
451:
449:
446:
442:
437:
435:
430:
426:
422:
418:
414:
410:
409:
399:
394:
386:
384:
382:
379:
375:
371:
367:
363:
355:
353:
350:
345:
342:
338:
334:
330:
326:
322:
318:
314:
310:
304:Dual incision
303:
298:
294:
291:
288:
285:
284:
283:
276:
274:
271:
268:
262:
254:
247:
242:
241:
240:
234:
232:
230:
226:
216:
209:
204:
202:
198:
196:
192:
188:
184:
180:
176:
172:
168:
164:
160:
156:
152:
148:
144:
140:
136:
132:
125:In eukaryotes
124:
122:
120:
116:
111:
109:
105:
101:
97:
93:
89:
84:
82:
78:
74:
70:
66:
62:
58:
54:
50:
46:
42:
34:
30:
19:
3447:
3428:DNA helicase
3411:8-Oxoguanine
3380:SOS response
3260:RecF pathway
3213:Excinuclease
3148:
3032:
3028:
3001:
2997:
2957:
2953:
2930:
2893:
2889:
2879:
2844:
2840:
2830:
2793:
2790:Biomolecules
2789:
2779:
2744:
2740:
2730:
2698:(1): 62–78.
2695:
2692:J. Mol. Biol
2691:
2681:
2644:
2640:
2589:
2585:
2575:
2540:
2536:
2526:
2489:
2485:
2474:
2439:
2435:
2425:
2398:
2394:
2338:
2334:
2323:
2306:
2302:
2295:
2251:
2247:
2209:
2205:
2160:
2156:
2146:
2119:
2115:
2105:
2071:(1): 47–55.
2068:
2064:
2010:
2006:
2000:
1973:
1969:
1959:
1932:
1928:
1918:
1875:
1871:
1861:
1836:
1832:
1826:
1791:
1787:
1777:
1752:
1748:
1742:
1705:
1701:
1691:
1654:
1650:
1618:
1612:
1568:
1565:PLOS Biology
1564:
1554:
1508:
1503:
1493:
1475:
1470:
1460:
1442:
1437:
1427:
1403:
1400:
1397:
1392:
1388:
1378:
1360:
1355:
1345:
1327:
1322:
1312:
1294:
1289:
1279:
1261:
1256:
1246:
1228:
1223:
1213:
1195:
1190:
1180:
1162:
1157:
1152:DNA Ligase I
1147:
1129:
1124:
1114:
1096:
1091:
1081:
1063:
1058:
1048:
1030:
1025:
1015:
997:
992:
982:
964:
959:
949:
931:
926:
916:
895:
885:
864:
854:
833:
825:
807:
802:
792:
774:
769:
759:
717:
707:
703:
699:
695:
683:
667:osteoporosis
658:
646:
644:
639:
637:
624:
622:
613:
589:
587:
582:
580:
573:
567:
561:
555:
547:
541:
533:
527:
514:
497:
483:phenotypical
467:
438:
433:
417:DNA helicase
406:
404:
370:DNA ligase I
364:) loads the
359:
346:
329:endonuclease
307:
280:
272:
263:
259:
238:
221:
199:
128:
112:
85:
40:
39:
29:
2486:Cancer Cell
1935:(1): 1–17.
1571:(6): e203.
829:(Centrin-2)
815:CAK subunit
518:lymphocytes
487:lung cancer
195:found below
161:, XPF, and
135:prokaryotes
92:DNA adducts
67:(BER), and
47:mechanism.
3554:DNA repair
3368:Regulation
3347:Photolyase
3107:DNA repair
3029:Mutat. Res
2741:Cell Cycle
2641:PLOS Genet
2537:Hum. Mutat
2509:10029/5565
2395:Cell Cycle
2248:BMC Cancer
1547:References
745:Subpathway
671:DNA damage
572:(CSB) and
478:DNA repair
462:DNA damage
441:TRCF (Mfd)
139:photolyase
131:eukaryotes
108:DNA ligase
81:base pairs
59:and other
45:DNA repair
3188:XPG/ERCC5
3173:XPD/ERCC2
2998:Mol. Cell
752:GeneCards
698:(CSA) or
689:mutations
57:radiation
3548:Category
3533:Category
3183:XPF/DDB1
3178:XPE/DDB1
3049:15913669
3020:12504014
2912:10877825
2871:17913742
2822:26287260
2771:24781187
2722:26616585
2673:25299392
2616:21612988
2567:22852219
2559:16947863
2518:16904611
2466:21571596
2417:17172862
2373:27556946
2315:11245433
2280:18267032
2226:12427537
2179:18196582
2138:15731165
2097:22754221
2043:22991719
2035:19492231
1992:11960912
1951:22493747
1910:23023028
1853:10562720
1818:19949546
1769:16464004
1734:20981145
1708:: 1–32.
1683:20725631
1657:: 1–10.
1597:16752948
1528:See also
896:Ddb2/Xpe
764:Cyclin H
742:Ortholog
736:Ortholog
686:germline
663:cachexia
635:(XPCS).
610:cachexia
297:progeria
61:mutagens
3375:SOS box
3127:AP site
2984:8836185
2890:FASEB J
2862:2190694
2813:4598778
2762:4111694
2713:4738086
2664:4191938
2607:3139823
2457:3234290
2364:5161687
2343:Bibcode
2271:2262891
2187:9456435
2088:3385179
2015:Bibcode
1942:3316453
1901:4586256
1880:Bibcode
1809:2709936
1725:2957137
1674:2915888
1588:1475692
900:Unknown
869:Unknown
838:Unknown
702:(CSB).
578:(CSA).
566:(XPG),
560:(XPF),
546:(XPD),
411:by the
376:or the
157:, XPD,
3500:FANCD2
3495:FANCD1
3255:RecBCD
3208:RAD23B
3203:RAD23A
3047:
3018:
2982:
2975:146147
2972:
2937:
2910:
2869:
2859:
2820:
2810:
2769:
2759:
2720:
2710:
2671:
2661:
2614:
2604:
2565:
2557:
2516:
2464:
2454:
2415:
2371:
2361:
2335:Nature
2313:
2278:
2268:
2254:: 50.
2224:
2185:
2177:
2136:
2095:
2085:
2041:
2033:
1990:
1949:
1939:
1908:
1898:
1851:
1816:
1806:
1767:
1732:
1722:
1681:
1671:
1625:
1595:
1585:
1447:TC-NER
1421:GTF2H3
1418:GTF2H2
1415:GTF2H1
1389:Gtf2h1
1306:RAD23B
1290:Rad23b
1284:RAD23B
1280:RAD23B
1273:RAD23A
1257:Rad23a
1251:RAD23A
1247:RAD23A
1134:TC-NER
1101:TC-NER
754:Entry
677:.(see
596:(XP),
470:cancer
452:Cancer
445:ATPase
434:E.coli
191:RAD23B
187:RAD23A
3515:FANCN
3510:FANCJ
3505:FANCI
3488:FANCM
3483:FANCL
3478:FANCG
3473:FANCF
3468:FANCE
3463:FANCC
3458:FANCB
3453:FANCA
3313:ERCC8
3308:ERCC6
3272:RAD51
3193:ERCC1
2563:S2CID
2183:S2CID
2039:S2CID
1542:(MMR)
1536:(BER)
1476:RAD14
1379:TFIIH
1295:RAD23
1262:RAD23
1240:MMS19
1229:MET18
1224:Mms19
1218:MMS19
1214:MMS19
1207:MNAT1
1191:Mnat1
1185:MNAT1
1181:MNAT1
1141:ERCC8
1130:RAD28
1125:Ercc8
1115:ERCC8
1108:ERCC6
1097:RAD26
1092:Ercc6
1082:ERCC6
1075:ERCC5
1059:Ercc5
1049:ERCC5
1042:ERCC4
1026:Ercc4
1016:ERCC4
1009:ERCC3
998:RAD25
993:Ercc3
983:ERCC3
976:ERCC2
960:Ercc2
950:ERCC2
943:ERCC1
932:RAD10
927:Ercc1
921:ERCC1
917:ERCC1
848:CETN2
834:Cetn2
826:CETN2
808:KIN28
740:Yeast
734:Mouse
708:ERCC6
704:ERCC8
700:ERCC6
696:ERCC8
693:genes
675:aging
659:ERCC5
647:ERCC5
640:ERCC4
625:ERCC3
614:ERCC2
590:ERCC2
583:ERCC1
575:ERCC8
569:ERCC6
563:ERCC5
557:ERCC4
548:ERCC3
543:ERCC2
538:ERCC1
534:ERCC1
524:Aging
425:dimer
341:ERCC1
179:ERCC1
133:than
43:is a
3401:PcrA
3357:CRY2
3352:CRY1
3287:LexA
3282:Slx4
3277:Sgs1
3250:RecA
3230:MSH2
3225:MLH1
3153:ERCC
3045:PMID
3016:PMID
2980:PMID
2935:ISBN
2908:PMID
2867:PMID
2818:PMID
2767:PMID
2718:PMID
2669:PMID
2612:PMID
2555:PMID
2514:PMID
2462:PMID
2413:PMID
2369:PMID
2311:PMID
2276:PMID
2222:PMID
2175:PMID
2134:PMID
2093:PMID
2031:PMID
1988:PMID
1947:PMID
1906:PMID
1849:PMID
1814:PMID
1765:PMID
1730:PMID
1706:2010
1679:PMID
1655:2010
1623:ISBN
1593:PMID
1509:RAD4
1480:Both
1454:XAB2
1443:SYF1
1438:Xab2
1432:XAB2
1428:XAB2
1408:Both
1404:Tfb4
1401:Ssl1
1398:Tfb1
1372:RPA2
1365:Both
1361:RFA2
1356:Rpa2
1350:RPA2
1346:RPA2
1339:RPA1
1332:Both
1328:RFA1
1323:Rpa1
1317:RPA1
1313:RPA1
1233:Both
1200:Both
1196:TFB3
1174:LIG1
1167:Both
1163:CDC9
1158:Lig1
1148:LIG1
1068:Both
1064:RAD2
1035:Both
1031:RAD1
1002:Both
969:Both
965:RAD3
936:Both
910:DDB2
890:DDB2
886:DDB2
879:DDB1
865:Ddb1
859:DDB1
855:DDB1
819:CDK7
812:Both
803:Cdk7
793:CDK7
786:CCNH
779:Both
775:CCL1
770:Ccnh
760:CCNH
673:and
665:and
502:and
381:seal
372:and
319:and
173:and
169:and
117:and
3437:WRN
3432:BLM
3396:Ogt
3198:RPA
3168:XPC
3163:XPB
3158:XPA
3037:doi
3033:577
3006:doi
2970:PMC
2962:doi
2898:doi
2857:PMC
2849:doi
2808:PMC
2798:doi
2757:PMC
2749:doi
2708:PMC
2700:doi
2696:428
2659:PMC
2649:doi
2602:PMC
2594:doi
2545:doi
2504:hdl
2494:doi
2452:PMC
2444:doi
2403:doi
2359:PMC
2351:doi
2339:537
2266:PMC
2256:doi
2214:doi
2210:509
2165:doi
2161:122
2124:doi
2083:PMC
2073:doi
2023:doi
1978:doi
1937:PMC
1896:PMC
1888:doi
1841:doi
1804:PMC
1796:doi
1757:doi
1753:106
1720:PMC
1710:doi
1669:PMC
1659:doi
1583:PMC
1573:doi
1520:XPC
1513:GGR
1504:Xpc
1498:XPC
1494:XPC
1487:XPA
1471:Xpa
1465:XPA
1461:XPA
1299:GGR
1266:GGR
1119:CSA
1086:CSB
1053:XPG
1020:XPF
987:XPB
954:XPD
903:GGR
872:GGR
841:GGR
681:).
620:).
554:),
552:XPB
540:),
504:XPC
500:XPD
362:RFC
349:XPA
337:XPF
321:XPB
317:XPD
313:XPG
229:XPE
225:XPC
183:RPA
175:CSB
171:CSA
163:XPG
159:XPE
155:XPC
151:XPB
147:XPA
55:),
49:DNA
3550::
3446::
3430::
3330:Ku
3043:.
3031:.
3014:.
3002:10
3000:.
2996:.
2978:.
2968:.
2958:24
2956:.
2952:.
2906:.
2894:14
2892:.
2888:.
2865:.
2855:.
2845:35
2843:.
2839:.
2816:.
2806:.
2792:.
2788:.
2765:.
2755:.
2745:13
2743:.
2739:.
2716:.
2706:.
2694:.
2690:.
2667:.
2657:.
2645:10
2643:.
2639:.
2624:^
2610:.
2600:.
2590:10
2588:.
2584:.
2561:.
2553:.
2541:27
2539:.
2535:.
2512:.
2502:.
2490:10
2488:.
2484:.
2460:.
2450:.
2440:10
2438:.
2434:.
2411:.
2397:.
2393:.
2381:^
2367:.
2357:.
2349:.
2337:.
2333:.
2307:61
2305:.
2274:.
2264:.
2250:.
2246:.
2234:^
2220:.
2208:.
2195:^
2181:.
2173:.
2159:.
2155:.
2132:.
2120:26
2118:.
2114:.
2091:.
2081:.
2069:18
2067:.
2063:.
2051:^
2037:.
2029:.
2021:.
2011:72
2009:.
1986:.
1974:23
1972:.
1968:.
1945:.
1931:.
1927:.
1904:.
1894:.
1886:.
1876:31
1874:.
1870:.
1847:.
1835:.
1812:.
1802:.
1792:10
1790:.
1786:.
1763:.
1751:.
1728:.
1718:.
1704:.
1700:.
1677:.
1667:.
1653:.
1649:.
1637:^
1591:.
1581:.
1567:.
1563:.
333:3'
197:.
189:,
185:,
181:,
153:,
149:,
83:.
3151:/
3125:/
3099:e
3092:t
3085:v
3051:.
3039::
3022:.
3008::
2986:.
2964::
2943:.
2914:.
2900::
2873:.
2851::
2824:.
2800::
2794:5
2773:.
2751::
2724:.
2702::
2675:.
2651::
2618:.
2596::
2569:.
2547::
2520:.
2506::
2496::
2468:.
2446::
2419:.
2405::
2399:5
2375:.
2353::
2345::
2317:.
2282:.
2258::
2252:8
2228:.
2216::
2189:.
2167::
2140:.
2126::
2099:.
2075::
2045:.
2025::
2017::
1994:.
1980::
1953:.
1933:3
1912:.
1890::
1882::
1855:.
1843::
1837:5
1820:.
1798::
1771:.
1759::
1736:.
1712::
1685:.
1661::
1631:.
1599:.
1575::
1569:4
1500:)
1496:(
1467:)
1463:(
1434:)
1430:(
1393:3
1391:-
1385:)
1381:(
1352:)
1348:(
1319:)
1315:(
1286:)
1282:(
1253:)
1249:(
1220:)
1216:(
1187:)
1183:(
1154:)
1150:(
1121:)
1117:(
1088:)
1084:(
1055:)
1051:(
1022:)
1018:(
989:)
985:(
956:)
952:(
923:)
919:(
892:)
888:(
861:)
857:(
799:)
795:(
766:)
762:(
550:(
536:(
339:–
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.