198:
35:
316:
401:(5mC) and 5hmC, 5fC is broadly distributed across the mammalian genome, although it is much more rarely occurring. The specific concentration values vary significantly depending on the cell type. 5fC can be aberrantly expressed in distinct sets of tissue that can indicate different tumor onsets and canceration.
426:
The understanding of the impact of 5fC on DNA physical properties is to date limited. Recent studies have reported contradictory findings regarding the structural impact of 5fC on DNA. On the other hand, several researchers working independently have identified 5fC to distinctly increase DNA
413:
pathway, a process that contributes to the DNA maintenance and integrity by replacing 5mC with canonical cytosine. A central dilemma regarding 5fC (and epigenetics in general) is how reader proteins recognise their substrates with such high specificity over the overwhelming background.
418:(TDG), a protein which is involved in the removal of 5fC from DNA in mammals, is especially interesting in this context. Secondly, 5fC can exist as an independent, stable modification, but its role in this context is still blurry.
366:'s research group the modified nucleoside was more recently confirmed to be relevant both as an intermediate in the active demethylation pathway and as a standalone epigenetic marker. In mammals, 5fC is formed by oxidation of
470:
Pfaffeneder, Toni; Hackner, Benjamin; Truß, Matthias; Münzel, Martin; Müller, Markus; Deiml, Christian A.; Hagemeier, Christian; Carell, Thomas (2011). "The
Discovery of 5-Formylcytosine in Embryonic Stem Cell DNA".
409:
The exact functions of 5fC have not been yet precisely defined, although it is likely to play key roles in at least two distinct frameworks. Firstly, 5fC serves as an intermediate of the active
329:
768:
Wang, Yafen; Zhang, Xiong; Zou, Guangrong; Peng, Shuang; Liu, Chaoxing; Zhou, Xiang (2019-01-22). "Detection and
Application of 5-Formylcytosine and 5-Formyluracil in DNA".
237:
517:
Bachman, Martin; Uribe-Lewis, Santiago; Yang, Xiaoping; Burgess, Heather E.; Iurlaro, Mario; Reik, Wolf; Murrell, Adele; Balasubramanian, Shankar (2015).
362:. In the context of nucleic acid chemistry and biology, it is regarded as an epigenetic marker. Discovered in 2011 in mammalian embryonic stem cells by
719:
Song, Chun-Xiao; Szulwach, Keith; Dai, Qing; Fu, Ye; Mao, Shi-Qing; Lin, Li; Street, Craig; Li, Yujing; Poidevin, Mickael; Wu, Hao; Gao, Juan (2013).
941:
Hardwick, Jack S; Ptchelkine, Denis; El-Sagheer, Afaf H; Tear, Ian; Singleton, Daniel; Phillips, Simon E V; Lane, Andrew N; Brown, Tom (2017).
212:
574:
Ito, Shinsuke; Shen, Li; Dai, Qing; Wu, Susan C.; Collins, Leonard B.; Swenberg, James A.; He, Chuan; Zhang, Yi (2011-09-02).
336:
876:
Raiber, Eun-Ang; Murat, Pierre; Chirgadze, Dimitri Y.; Beraldi, Dario; Luisi, Ben F.; Balasubramanian, Shankar (2015).
99:
176:
998:
Ngo, Thuy T. M.; Yoo, Jejoong; Dai, Qing; Zhang, Qiucen; He, Chuan; Aksimentiev, Aleksei; Ha, Taekjip (2016-02-24).
819:
Maiti, Atanu; Michelson, Anna
Zhachkina; Armwood, Cherece J.; Lee, Jeehiun K.; Drohat, Alexander C. (2013-10-23).
441:
415:
367:
1065:"Oxidized Derivatives of 5-Methylcytosine Alter the Stability and Dehybridization Dynamics of Duplex DNA"
1181:
1176:
1011:
587:
446:
193:
47:
65:
923:
821:"Divergent Mechanisms for Enzymatic Excision of 5-Formylcytosine and 5-Carboxylcytosine from DNA"
801:
701:
1153:
1102:
1084:
1045:
1027:
980:
962:
915:
897:
858:
840:
793:
785:
750:
693:
685:
621:
603:
556:
538:
496:
488:
139:
1143:
1133:
1092:
1076:
1035:
1019:
970:
954:
905:
889:
848:
832:
777:
740:
732:
677:
611:
595:
546:
530:
480:
436:
398:
260:
1120:
Dubini, Romeo C. A.; Schön, Alexander; Müller, Markus; Carell, Thomas; Rovó, Petra (2020).
1063:
Sanstead, Paul J.; Ashwood, Brennan; Dai, Qing; He, Chuan; Tokmakoff, Andrei (2020-02-20).
427:
flexibility. 5fC also curtails DNA double helix stability and increases base pair opening.
1000:"Effects of cytosine modifications on DNA flexibility and nucleosome mechanical stability"
197:
75:
1015:
591:
119:
1148:
1122:"Impact of 5-formylcytosine on the melting kinetics of DNA by 1H NMR chemical exchange"
1121:
1097:
1064:
1040:
999:
975:
942:
910:
877:
853:
820:
745:
720:
616:
575:
551:
518:
307:
576:"Tet Proteins Can Convert 5-Methylcytosine to 5-Formylcytosine and 5-Carboxylcytosine"
1170:
640:
410:
363:
355:
164:
927:
805:
721:"Genome-wide Profiling of 5-Formylcytosine Reveals Its Roles in Epigenetic Priming"
705:
781:
451:
371:
17:
736:
34:
665:
352:
287:
130:
1088:
1080:
1031:
966:
901:
844:
789:
689:
607:
542:
492:
599:
1157:
1106:
1049:
984:
919:
862:
797:
754:
697:
625:
560:
500:
484:
1138:
534:
359:
1023:
681:
666:"TET-mediated active DNA demethylation: mechanism, function and beyond"
151:
958:
893:
836:
306:
Except where otherwise noted, data are given for materials in their
110:
98:
88:
878:"5-Formylcytosine alters the structure of the DNA double helix"
943:"5-Formylcytosine does not change the global structure of DNA"
519:"5-Formylcytosine can be a stable DNA modification in mammals"
221:
InChI=1S/C5H5N3O2/c6-4-3(2-9)1-7-5(10)8-4/h1-2H,(H3,6,7,8,10)
181:
324:
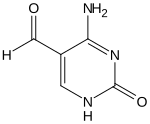
52:4-Amino-2-oxo-1,2-dihydropyrimidine-5-carbaldehyde
397:Similarly to the related cytosine modifications
163:
74:
8:
422:5fC impact on DNA structure and flexibility
196:
138:
26:
1147:
1137:
1096:
1039:
974:
947:Nature Structural & Molecular Biology
909:
882:Nature Structural & Molecular Biology
852:
744:
615:
550:
825:Journal of the American Chemical Society
473:Angewandte Chemie International Edition
462:
242:
217:
192:
224:Key: FHSISDGOVSHJRW-UHFFFAOYSA-N
118:
7:
512:
510:
1069:The Journal of Physical Chemistry B
154:
25:
314:
33:
310:(at 25 °C , 100 kPa).
292:139.11 g/mol
664:Wu, Xiaoji; Zhang, Yi (2017).
370:(5hmC) a reaction mediated by
1:
770:Accounts of Chemical Research
782:10.1021/acs.accounts.8b00543
374:. Its molecular formula is C
1198:
737:10.1016/j.cell.2013.04.001
304:
253:
233:
208:
58:
46:
41:
32:
1081:10.1021/acs.jpcb.9b11511
645:pubchem.ncbi.nlm.nih.gov
670:Nature Reviews Genetics
600:10.1126/science.1210597
523:Nature Chemical Biology
442:5-Hydroxymethylcytosine
416:Thymine-DNA glycosylase
368:5-Hydroxymethylcytosine
1126:Nucleic Acids Research
485:10.1002/anie.201103899
1004:Nature Communications
535:10.1038/nchembio.1848
447:Base excision repair
245:C1=NC(=O)NC(=C1C=O)N
48:Preferred IUPAC name
1139:10.1093/nar/gkaa589
1024:10.1038/ncomms10813
1016:2016NatCo...710813N
831:(42): 15813–15822.
682:10.1038/nrg.2017.33
592:2011Sci...333.1300I
586:(6047): 1300–1303.
29:
641:"5-Formylcytosine"
337:Infobox references
27:
1132:(15): 8796–8807.
959:10.1038/nsmb.3411
894:10.1038/nsmb.2936
837:10.1021/ja406444x
479:(31): 7008–7012.
345:Chemical compound
343:
342:
177:CompTox Dashboard
100:Interactive image
28:5-Formylcytosine
16:(Redirected from
1189:
1162:
1161:
1151:
1141:
1117:
1111:
1110:
1100:
1075:(7): 1160–1174.
1060:
1054:
1053:
1043:
995:
989:
988:
978:
938:
932:
931:
913:
873:
867:
866:
856:
816:
810:
809:
776:(4): 1016–1024.
765:
759:
758:
748:
716:
710:
709:
661:
655:
654:
652:
651:
636:
630:
629:
619:
571:
565:
564:
554:
514:
505:
504:
467:
437:5-Methylcytosine
399:5-Methylcytosine
349:5-Formylcytosine
327:
321:
318:
317:
261:Chemical formula
201:
200:
185:
183:
167:
156:
142:
122:
102:
78:
37:
30:
21:
18:5-formylcytosine
1197:
1196:
1192:
1191:
1190:
1188:
1187:
1186:
1167:
1166:
1165:
1119:
1118:
1114:
1062:
1061:
1057:
997:
996:
992:
940:
939:
935:
875:
874:
870:
818:
817:
813:
767:
766:
762:
718:
717:
713:
663:
662:
658:
649:
647:
638:
637:
633:
573:
572:
568:
516:
515:
508:
469:
468:
464:
460:
433:
424:
407:
395:
389:
385:
381:
377:
346:
339:
334:
333:
332: ?)
323:
319:
315:
311:
281:
277:
273:
269:
263:
249:
246:
241:
240:
229:
226:
225:
222:
216:
215:
204:
194:DTXSID701336487
186:
179:
170:
157:
145:
125:
105:
92:
81:
68:
54:
53:
23:
22:
15:
12:
11:
5:
1195:
1193:
1185:
1184:
1179:
1169:
1168:
1164:
1163:
1112:
1055:
990:
953:(6): 544–552.
933:
868:
811:
760:
731:(3): 678–691.
711:
676:(9): 517–534.
656:
631:
566:
529:(8): 555–557.
506:
461:
459:
456:
455:
454:
449:
444:
439:
432:
429:
423:
420:
406:
403:
394:
391:
387:
383:
379:
375:
344:
341:
340:
335:
313:
312:
308:standard state
305:
302:
301:
298:
294:
293:
290:
284:
283:
279:
275:
271:
267:
264:
259:
256:
255:
251:
250:
248:
247:
244:
236:
235:
234:
231:
230:
228:
227:
223:
220:
219:
211:
210:
209:
206:
205:
203:
202:
189:
187:
175:
172:
171:
169:
168:
160:
158:
150:
147:
146:
144:
143:
135:
133:
127:
126:
124:
123:
115:
113:
107:
106:
104:
103:
95:
93:
86:
83:
82:
80:
79:
71:
69:
64:
61:
60:
56:
55:
51:
50:
44:
43:
39:
38:
24:
14:
13:
10:
9:
6:
4:
3:
2:
1194:
1183:
1180:
1178:
1175:
1174:
1172:
1159:
1155:
1150:
1145:
1140:
1135:
1131:
1127:
1123:
1116:
1113:
1108:
1104:
1099:
1094:
1090:
1086:
1082:
1078:
1074:
1070:
1066:
1059:
1056:
1051:
1047:
1042:
1037:
1033:
1029:
1025:
1021:
1017:
1013:
1009:
1005:
1001:
994:
991:
986:
982:
977:
972:
968:
964:
960:
956:
952:
948:
944:
937:
934:
929:
925:
921:
917:
912:
907:
903:
899:
895:
891:
887:
883:
879:
872:
869:
864:
860:
855:
850:
846:
842:
838:
834:
830:
826:
822:
815:
812:
807:
803:
799:
795:
791:
787:
783:
779:
775:
771:
764:
761:
756:
752:
747:
742:
738:
734:
730:
726:
722:
715:
712:
707:
703:
699:
695:
691:
687:
683:
679:
675:
671:
667:
660:
657:
646:
642:
635:
632:
627:
623:
618:
613:
609:
605:
601:
597:
593:
589:
585:
581:
577:
570:
567:
562:
558:
553:
548:
544:
540:
536:
532:
528:
524:
520:
513:
511:
507:
502:
498:
494:
490:
486:
482:
478:
474:
466:
463:
457:
453:
450:
448:
445:
443:
440:
438:
435:
434:
430:
428:
421:
419:
417:
412:
411:demethylation
404:
402:
400:
392:
390:
373:
369:
365:
364:Thomas Carell
361:
358:derived from
357:
356:nitrogen base
354:
350:
338:
331:
326:
309:
303:
300:yellow solid
299:
296:
295:
291:
289:
286:
285:
265:
262:
258:
257:
252:
243:
239:
232:
218:
214:
207:
199:
195:
191:
190:
188:
178:
174:
173:
166:
162:
161:
159:
153:
149:
148:
141:
137:
136:
134:
132:
129:
128:
121:
117:
116:
114:
112:
109:
108:
101:
97:
96:
94:
90:
85:
84:
77:
73:
72:
70:
67:
63:
62:
57:
49:
45:
40:
36:
31:
19:
1129:
1125:
1115:
1072:
1068:
1058:
1010:(1): 10813.
1007:
1003:
993:
950:
946:
936:
888:(1): 44–49.
885:
881:
871:
828:
824:
814:
773:
769:
763:
728:
724:
714:
673:
669:
659:
648:. Retrieved
644:
634:
583:
579:
569:
526:
522:
476:
472:
465:
425:
408:
396:
393:Localization
348:
347:
59:Identifiers
1182:Nucleobases
1177:Pyrimidones
452:Epigenetics
372:TET enzymes
351:(5fC) is a
297:Appearance
254:Properties
120:CHEBI:76794
1171:Categories
650:2020-07-26
458:References
353:pyrimidine
288:Molar mass
131:ChemSpider
87:3D model (
66:CAS Number
1089:1520-6106
1032:2041-1723
967:1545-9993
902:1545-9985
845:0002-7863
790:0001-4842
690:1471-0064
639:PubChem.
608:0036-8075
543:1552-4469
493:1521-3773
405:Functions
76:4425-59-6
1158:32652019
1107:31986043
1050:26905257
985:28504696
928:10288745
920:25504322
863:24063363
806:58623597
798:30666870
755:23602153
698:28555658
626:21778364
561:26098680
501:21721093
431:See also
360:cytosine
165:10986305
1149:7470965
1098:7136776
1041:4770088
1012:Bibcode
976:5747368
911:4287393
854:3930231
746:3657391
706:3393814
617:3495246
588:Bibcode
580:Science
552:5486442
330:what is
328: (
282:
152:PubChem
140:9161502
1156:
1146:
1105:
1095:
1087:
1048:
1038:
1030:
983:
973:
965:
926:
918:
908:
900:
861:
851:
843:
804:
796:
788:
753:
743:
704:
696:
688:
624:
614:
606:
559:
549:
541:
499:
491:
325:verify
322:
238:SMILES
42:Names
924:S2CID
802:S2CID
702:S2CID
213:InChI
111:ChEBI
89:JSmol
1154:PMID
1103:PMID
1085:ISSN
1046:PMID
1028:ISSN
981:PMID
963:ISSN
916:PMID
898:ISSN
859:PMID
841:ISSN
794:PMID
786:ISSN
751:PMID
725:Cell
694:PMID
686:ISSN
622:PMID
604:ISSN
557:PMID
539:ISSN
497:PMID
489:ISSN
1144:PMC
1134:doi
1093:PMC
1077:doi
1073:124
1036:PMC
1020:doi
971:PMC
955:doi
906:PMC
890:doi
849:PMC
833:doi
829:135
778:doi
741:PMC
733:doi
729:153
678:doi
612:PMC
596:doi
584:333
547:PMC
531:doi
481:doi
182:EPA
155:CID
1173::
1152:.
1142:.
1130:48
1128:.
1124:.
1101:.
1091:.
1083:.
1071:.
1067:.
1044:.
1034:.
1026:.
1018:.
1006:.
1002:.
979:.
969:.
961:.
951:24
949:.
945:.
922:.
914:.
904:.
896:.
886:22
884:.
880:.
857:.
847:.
839:.
827:.
823:.
800:.
792:.
784:.
774:52
772:.
749:.
739:.
727:.
723:.
700:.
692:.
684:.
674:18
672:.
668:.
643:.
620:.
610:.
602:.
594:.
582:.
578:.
555:.
545:.
537:.
527:11
525:.
521:.
509:^
495:.
487:.
477:50
475:.
388:2.
1160:.
1136::
1109:.
1079::
1052:.
1022::
1014::
1008:7
987:.
957::
930:.
892::
865:.
835::
808:.
780::
757:.
735::
708:.
680::
653:.
628:.
598::
590::
563:.
533::
503:.
483::
386:O
384:3
382:N
380:5
378:H
376:5
320:N
280:2
278:O
276:3
274:N
272:5
270:H
268:5
266:C
184:)
180:(
91:)
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.