54:
63:
396:
257:
986:. It is added to running buffer and binds by intercalating between DNA base pairs. When the agarose gel is illuminated using UV light, DNA bands become visible. Intercalation of EtBr can alter properties of the DNA molecule, such as charge, weight, conformation, and flexibility. Since the mobilities of DNA molecules through the agarose gel are measured relative to a molecular weight standard, the effects of EtBr can be critical to determining the sizes of molecules.
587:
582:
678:
854:
1059:
45:
931:
2252:
739:
683:
682:
1070:
Most use of ethidium bromide in the laboratory (0.25–1 μg/mL) is below the LD50 dosage, making acute toxicity unlikely. Testing in humans and longer studies in a mammalian system would be required to fully understand the long-term risk ethidium bromide poses to lab workers, but it is clear that
906:, because the phenyl ring has been shown to project outside the intercalated bases. In fact, the phenyl group is found to be almost perpendicular to the plane of the ring system, as it rotates about its single bond to find a position where it will impinge upon the ring system minimally. Instead, the
1034:
with liver extract. However, SYBR Green I was actually found to be more mutagenic than EtBr to the bacterial cells exposed to UV (which is used to visualize either dye). This may be the case for other "safer" dyes, but while mutagenic and toxicity details are available these have not been published
978:
images recorded as photographs. Where direct viewing is needed, the viewer's eyes and exposed skin should be protected. In the laboratory the intercalating properties have long been used to minimize chromosomal condensation when a culture is exposed to mitotic arresting agents during harvest. The
1086:
The disposal of laboratory ethidium bromide remains a controversial subject. Ethidium bromide can be degraded chemically, or collected and incinerated. It is common for ethidium bromide waste below a mandated concentration to be disposed of normally (such as pouring it down a drain). A common
1050:, which has health implications of its own, including increased skin absorption of organic compounds. Despite the performance advantage of using SYBR dyes instead of EtBr for staining purposes, many researchers still prefer EtBr since it is considerably less expensive.
681:
595:
562:
1883:
Singer VL, Lawlor TE, Yue S (February 1999). "Comparison of SYBR Green I nucleic acid gel stain mutagenicity and ethidium bromide mutagenicity in the
Salmonella/mammalian microsome reverse mutation assay (Ames test)".
2200:
Mulugeta W, Wilkes J, Mulatu W, Majiwa PA, Masake R, Peregrine AS (April 1997). "Long-term occurrence of
Trypanosoma congolense resistant to diminazene, isometamidium and homidium in cattle at Ghibe, Ethiopia".
657:
1095:. Data are lacking on the mutagenic effects of degradation products. Lunn and Sansone describe more effective methods for degradation. Elsewhere, ethidium bromide removal from solutions with
1079:
Ethidium bromide is not regulated as hazardous waste at low concentrations, but is treated as hazardous waste by many organizations. Material should be handled according to the manufacturer's
1355:
Stevenson P, Sones KR, Gicheru MM, Mwangi EK (May 1995). "Comparison of isometamidium chloride and homidium bromide as prophylactic drugs for trypanosomiasis in cattle at
Nguruman, Kenya".
537:
752:
918:
environment and away from the solvent, the ethidium cation is forced to shed any water molecules that were associated with it. As water is a highly efficient fluorescence
979:
resulting slide preparations permit a higher degree of resolution, and thus more confidence in determining structural integrity of chromosomes upon microscopic analysis.
684:
1320:
1222:
1927:
Ohta T, Tokishita S, Yamagata H (May 2001). "Ethidium bromide and SYBR Green I enhance the genotoxicity of UV-irradiation and chemical mutagens in E. coli".
445:
1119:
showed universal resistance between July 1989 and
February 1993. This likely indicates a permanent loss of function in this area against the tested target,
1816:
Huang Q, Fu WL (2005). "Comparative analysis of the DNA staining efficiencies of different fluorescent dyes in preparative agarose gel electrophoresis".
1324:
1226:
1030:-based dyes are used by some researchers and there are other emerging stains such as "Novel Juice". SYBR dyes are less mutagenic than EtBr by the
162:
706:
2292:
1451:
1026:
There are alternatives to ethidium bromide which are advertised as being less dangerous and having better performance. For example, several
1147:
935:
1091:(bleach) before disposal. According to Lunn and Sansone, chemical degradation using bleach yields compounds which are mutagenic by the
1577:"Human mitochondrial DNA with large deletions repopulates organelles faster than full-length genomes under relaxed copy number control"
974:
light is harmful to eyes and skin, gels stained with ethidium bromide are usually viewed indirectly using an enclosed camera, with the
2312:
699:
2172:
2082:
911:
410:
1532:
Sigmon J, Larcom LL (October 1996). "The effect of ethidium bromide on mobility of DNA fragments in agarose gel electrophoresis".
53:
845:
is used instead. Despite its reputation as a mutagen, tests have shown it to have low mutagenicity without metabolic activation.
653:
1018:
The binding affinity of the cationic nanoparticles with DNA could be evaluated by competitive binding with ethidium bromide.
1781:
Olmsted J, Kearns DR (August 1977). "Mechanism of ethidium bromide fluorescence enhancement on binding to nucleic acids".
1328:
1230:
419:
InChI=1S/C21H19N3.BrH/c1-2-24-20-13-16(23)9-11-18(20)17-10-8-15(22)12-19(17)21(24)14-6-4-3-5-7-14;/h3-13,23H,2,22H2,1H3;1H
759:
641:
429:
InChI=1/C21H19N3.BrH/c1-2-24-20-13-16(23)9-11-18(20)17-10-8-15(22)12-19(17)21(24)14-6-4-3-5-7-14;/h3-13,23H,2,22H2,1H3;1H
2297:
2011:
1988:
1063:
1046:
for rats of over 5 g/kg, which is higher than that of EtBr (1.5 g/kg). Many alternative dyes are suspended in
783:
631:
343:
1418:
586:
1970:
983:
967:
802:
374:
2256:
993:
copy number in proliferating cells. The effect of EtBr on mitochondrial DNA is used in veterinary medicine to treat
62:
2282:
1036:
1066:
between two adenine–thymine base pairs. The intercalation is said by some to motivate a high mutagenicity of DNA.
951:
899:
Ethidium bromide's intense fluorescence after binding with DNA is probably not due to rigid stabilization of the
264:
2137:
Quillardet, P.; Hofnung, M. (April 1988). "Ethidium bromide and safety--readers suggest alternative solutions".
2307:
2272:
919:
838:
692:
252:
2302:
1121:
842:
214:
1860:
1677:"Physiological importance and identification of novel targets for the N-terminal acetyltransferase NatB"
609:
581:
574:
1936:
1893:
1737:
75:
2070:
1624:
Roy
Chowdhury A, Bakshi R, Wang J, Yildirir G, Liu B, Pappas-Brown V, et al. (December 2010).
1166:
1143:
1088:
391:
128:
1394:
2230:
1841:
1557:
1289:
1100:
1047:
1040:
955:
794:
174:
645:
2287:
2277:
2222:
2154:
2119:
2078:
2052:
1952:
1909:
1833:
1798:
1763:
1706:
1657:
1606:
1549:
1514:
1447:
1372:
1281:
1080:
990:
889:
885:
865:
818:
798:
1005:
form. This form inhibits replication of kinetoplastid DNA, which is lethal for trypanosomes.
2214:
2146:
2111:
2044:
1944:
1901:
1825:
1790:
1753:
1745:
1696:
1688:
1647:
1637:
1596:
1588:
1541:
1504:
1364:
1273:
1162:
1096:
520:
468:
352:
234:
994:
834:
787:
548:
138:
2176:
1260:
Kinabo LD (September 1993). "Pharmacology of existing drugs for animal trypanosomiasis".
1940:
1897:
1741:
395:
256:
1758:
1725:
1701:
1676:
1652:
1625:
1138:
998:
877:
730:
637:
194:
2218:
1948:
1905:
1601:
1576:
1467:
962:
can also be detected, since it usually folds back onto itself and thus provides local
2266:
2150:
2115:
2048:
1368:
1277:
943:
893:
509:
245:
31:
2234:
1845:
1561:
1293:
888:
of EtBr in aqueous solution are at 210 nm and 285 nm, which correspond to
853:
623:
1126:
873:
822:
791:
543:
1339:
1241:
1058:
316:
1989:"Executive Summary Ethidium Bromide: Evidence for Possible Carcinogenic Activity"
1642:
1310:
1212:
1692:
975:
971:
938:
and stained with ethidium bromide, which emits orange light after binding to DNA
915:
907:
862:
810:
718:
1749:
44:
1509:
1492:
1112:
1027:
491:
225:
1726:"Cationic nanoparticle as an inhibitor of cell-free DNA-induced inflammation"
1575:
Diaz F, Bayona-Bafaluy MP, Rana M, Mora M, Hao H, Moraes CT (November 2002).
1545:
1444:
Handbook of
Biological Dyes and Stains: Synthesis and Industrial Application
1092:
1031:
963:
833:, it has been commonly used since the 1950s in veterinary medicine to treat
649:
363:
1956:
1913:
1837:
1767:
1710:
1661:
1610:
1518:
2251:
2226:
2158:
2123:
2056:
1829:
1724:
Liang H, Peng B, Dong C, Liu L, Mao J, Wei S, et al. (October 2018).
1553:
1376:
1285:
1186:
930:
2210:
1802:
1592:
1269:
1156:
1116:
1103:
is recommended. Various commercial products are available for this use.
922:, the removal of these water molecules allows the ethidium to fluoresce.
881:
869:
705:
698:
691:
664:
1794:
903:
17:
1343:
1315:
1245:
1217:
1071:
ethidium bromide can cause mutations in mammalian and bacterial cells.
970:
containing nucleic acids placed on or under an ultraviolet lamp. Since
303:
265:
1152:
900:
205:
2099:
825:
with an orange colour, intensifying almost 20-fold after binding to
813:. To avoid confusion, some laboratories have used the abbreviation
729:
Except where otherwise noted, data are given for materials in their
615:
291:
1057:
1002:
929:
852:
841:
makes this treatment impractical in some areas, where the related
327:
185:
161:
151:
2100:"Ethidium bromide: destruction and decontamination of solutions"
1012:
982:
Ethidium bromide is also used during DNA fragment separation by
619:
282:
1008:
The chloride salt homidium chloride has the same applications.
959:
947:
826:
379:
966:
for the dye to intercalate. Detection typically involves a
910:
environment found between the base pairs is believed to be
514:
260 to 262 °C (500 to 504 °F; 533 to 535 K)
1493:"Ethidium DNA agarose gel electrophoresis: how it started"
989:
Ethidium bromide has also been used extensively to reduce
1626:"The killing of African trypanosomes by ethidium bromide"
676:
2035:
Hengen PN (June 1994). "Disposal of ethidium bromide".
896:, EtBr emits orange light with wavelength 605 nm.
747:
80:
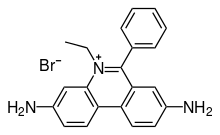
3,8-Diamino-5-ethyl-6-phenylphenanthridin-5-ium bromide
2195:
2193:
1419:"Ethidium Bromide: Swap or Not | UCSB Sustainability"
95:
2,7-Diamino-10-ethyl-9-phenylphenanthridinium bromide
92:
2,7-Diamino-10-ethyl-6-phenylphenanthridinium bromide
1675:Caesar R, Warringer J, Blomberg A (February 2006).
101:
5-Ethyl-6-phenyl-phenanthridine-3,8-diamine bromide
98:
3,8-Diamino-1-ethyl-6-phenylphenantridinium bromide
946:in molecular biology laboratories. In the case of
1305:
1303:
1015:media and used as an inhibitor for cell growth.
723:> 100 °C (212 °F; 373 K)
315:
1159:, marketed as safer and more intense DNA stains
680:
137:
2075:Hazardous Laboratory Chemicals Disposal Guide
1878:
1876:
1255:
1253:
8:
942:Ethidium bromide is commonly used to detect
880:, an isomer of which is the fluorescent dye
1087:practice is to treat ethidium bromide with
547:
1987:National Toxicology Program (2005-08-15).
1818:Clinical Chemistry and Laboratory Medicine
1001:DNA and changes their conformation to the
394:
255:
233:
36:
2017:. National Toxicology Program. 2005-08-15
1757:
1700:
1651:
1641:
1600:
1508:
950:this is usually double-stranded DNA from
351:
2077:(3rd ed.). CRC. pp. 222–223.
1178:
857:Absorption spectrum of ethidium bromide
450:
415:
390:
997:in cattle, as EtBr binds molecules of
849:Structure, chemistry, and fluorescence
246:
422:Key: ZMMJGEGLRURXTF-UHFFFAOYSA-N
213:
193:
7:
2012:"Executive Summary Ethidium Bromide"
1468:"Application Note: Ethidium Bromide"
1388:
1386:
1148:gel electrophoresis of nucleic acids
936:gel electrophoresis of nucleic acids
809:, which is also an abbreviation for
801:laboratories for techniques such as
453:CC1c2cc(N)ccc2c3ccc(N)cc3c1c4ccccc4.
432:Key: ZMMJGEGLRURXTF-UHFFFAOYAD
306:
290:
837:in cattle. The high incidence of
25:
1011:Ethidium bromide can be added to
876:moiety is generically known as a
27:DNA gel stain and veterinary drug
2250:
1155:(itself derived from ethbr) and
805:. It is commonly abbreviated as
737:
585:
580:
61:
52:
43:
2098:Lunn G, Sansone EB (May 1987).
1035:in peer-reviewed journals. The
817:for this salt. When exposed to
733:(at 25 °C , 100 kPa).
2037:Trends in Biochemical Sciences
1395:"The Myth of Ethidium Bromide"
1054:Possible carcinogenic activity
1:
2293:Quaternary ammonium compounds
2219:10.1016/s0001-706x(96)00645-6
1949:10.1016/S1383-5718(01)00155-3
1906:10.1016/s1383-5718(98)00172-7
2151:10.1016/0168-9525(88)90092-3
2116:10.1016/0003-2697(87)90419-2
2049:10.1016/0968-0004(94)90152-X
1971:"Novel Juice testing report"
1643:10.1371/journal.ppat.1001226
1369:10.1016/0001-706X(94)00080-K
1278:10.1016/0001-706x(93)90091-o
2173:"Ethidium Bromide Disposal"
1693:10.1128/EC.5.2.368-378.2006
1144:Agarose gel electrophoresis
984:agarose gel electrophoresis
934:DNA sample separated using
803:agarose gel electrophoresis
496:394.294 g/mol
2329:
1750:10.1038/s41467-018-06603-5
1393:Lowe, Derek (2016-04-18).
29:
2313:Experimental cancer drugs
1510:10.1080/15216540500380855
1491:Borst P (November 2005).
1039:for SYBR Safe reports an
786:agent commonly used as a
727:
561:
556:
530:
461:
441:
406:
121:
86:
74:
69:
60:
51:
42:
839:antimicrobial resistance
632:Precautionary statements
30:Not to be confused with
2104:Analytical Biochemistry
1976:. Newmarket Scientific.
1546:10.1002/elps.1150171003
1423:sustainability.ucsb.edu
1187:"GESTIS-Stoffdatenbank"
958:, etc. Single-stranded
1861:"Safer stains for DNA"
1581:Nucleic Acids Research
1446:. Hoboken, NJ: Wiley.
1067:
939:
914:. By moving into this
892:. As a result of this
868:, ethidium bromide is
858:
843:isometamidium chloride
687:
1830:10.1515/CCLM.2005.141
1730:Nature Communications
1075:Handling and disposal
1061:
933:
856:
686:
2259:at Wikimedia Commons
1115:Valley in southwest
1111:Trypanosomes in the
1022:Alternatives for gel
669:(fire diamond)
76:Preferred IUPAC name
2298:Phenanthridine dyes
1941:2001MRGTE.492...91O
1898:1999MRGTE.439...37S
1795:10.1021/bi00635a022
1742:2018NatCo...9.4291L
1311:"Homidium chloride"
1167:propidium monoazide
1089:sodium hypochlorite
956:restriction digests
521:Solubility in water
175:Beilstein Reference
39:
2139:Trends in Genetics
1593:10.1093/nar/gkf602
1442:Sabnis RW (2010).
1213:"Homidium bromide"
1101:ion exchange resin
1097:activated charcoal
1068:
940:
859:
760:Infobox references
688:
37:
2283:DNA intercalaters
2255:Media related to
1929:Mutation Research
1886:Mutation Research
1789:(16): 3647–3654.
1587:(21): 4626–4633.
1540:(10): 1524–1527.
1453:978-0-470-40753-0
1081:safety data sheet
1062:Ethidium bromide
991:mitochondrial DNA
890:ultraviolet light
886:Absorption maxima
829:. Under the name
819:ultraviolet light
799:molecular biology
780:homidium chloride
768:Chemical compound
766:
765:
610:Hazard statements
504:Purple-red solid
375:CompTox Dashboard
163:Interactive image
38:Ethidium bromide
16:(Redirected from
2320:
2257:Ethidium bromide
2254:
2239:
2238:
2197:
2188:
2187:
2185:
2184:
2175:. Archived from
2169:
2163:
2162:
2134:
2128:
2127:
2095:
2089:
2088:
2067:
2061:
2060:
2032:
2026:
2025:
2023:
2022:
2016:
2008:
2002:
2001:
1999:
1998:
1993:
1984:
1978:
1977:
1975:
1967:
1961:
1960:
1924:
1918:
1917:
1880:
1871:
1870:
1868:
1867:
1856:
1850:
1849:
1813:
1807:
1806:
1778:
1772:
1771:
1761:
1721:
1715:
1714:
1704:
1672:
1666:
1665:
1655:
1645:
1636:(12): e1001226.
1621:
1615:
1614:
1604:
1572:
1566:
1565:
1529:
1523:
1522:
1512:
1488:
1482:
1481:
1479:
1477:
1472:
1464:
1458:
1457:
1439:
1433:
1432:
1430:
1429:
1415:
1409:
1408:
1406:
1405:
1390:
1381:
1380:
1352:
1346:
1338:
1336:
1335:
1307:
1298:
1297:
1257:
1248:
1240:
1238:
1237:
1209:
1203:
1202:
1200:
1198:
1183:
1163:Propidium iodide
778:, chloride salt
776:homidium bromide
772:Ethidium bromide
750:
744:
741:
740:
708:
701:
694:
679:
659:
655:
651:
647:
643:
639:
625:
621:
617:
589:
584:
551:
469:Chemical formula
399:
398:
383:
381:
355:
319:
308:
294:
267:
259:
248:
237:
217:
197:
165:
141:
107:Homidium bromide
104:Ethidium bromide
65:
56:
47:
40:
21:
2328:
2327:
2323:
2322:
2321:
2319:
2318:
2317:
2308:Embryotoxicants
2273:Aromatic amines
2263:
2262:
2247:
2242:
2199:
2198:
2191:
2182:
2180:
2171:
2170:
2166:
2136:
2135:
2131:
2097:
2096:
2092:
2085:
2069:
2068:
2064:
2034:
2033:
2029:
2020:
2018:
2014:
2010:
2009:
2005:
1996:
1994:
1991:
1986:
1985:
1981:
1973:
1969:
1968:
1964:
1926:
1925:
1921:
1882:
1881:
1874:
1865:
1863:
1858:
1857:
1853:
1815:
1814:
1810:
1780:
1779:
1775:
1723:
1722:
1718:
1681:Eukaryotic Cell
1674:
1673:
1669:
1623:
1622:
1618:
1574:
1573:
1569:
1534:Electrophoresis
1531:
1530:
1526:
1503:(11): 745–747.
1490:
1489:
1485:
1475:
1473:
1470:
1466:
1465:
1461:
1454:
1441:
1440:
1436:
1427:
1425:
1417:
1416:
1412:
1403:
1401:
1399:In the Pipeline
1392:
1391:
1384:
1354:
1353:
1349:
1333:
1331:
1309:
1308:
1301:
1259:
1258:
1251:
1235:
1233:
1211:
1210:
1206:
1196:
1194:
1185:
1184:
1180:
1176:
1135:
1109:
1107:Drug resistance
1077:
1056:
1044:
1024:
995:trypanosomiasis
928:
851:
835:trypanosomiasis
788:fluorescent tag
769:
762:
757:
756:
755: ?)
746:
742:
738:
734:
713:
712:
711:
710:
703:
696:
689:
685:
677:
634:
612:
598:
577:
540:
523:
485:
481:
477:
471:
457:
454:
449:
448:
437:
434:
433:
430:
424:
423:
420:
414:
413:
402:
384:
377:
358:
338:
322:
309:
297:
277:
240:
220:
200:
177:
168:
155:
144:
131:
117:
116:
82:
81:
35:
28:
23:
22:
15:
12:
11:
5:
2326:
2324:
2316:
2315:
2310:
2305:
2300:
2295:
2290:
2285:
2280:
2275:
2265:
2264:
2261:
2260:
2246:
2245:External links
2243:
2241:
2240:
2189:
2164:
2129:
2110:(2): 453–458.
2090:
2083:
2062:
2043:(6): 257–258.
2027:
2003:
1979:
1962:
1935:(1–2): 91–97.
1919:
1872:
1851:
1824:(8): 841–842.
1808:
1773:
1716:
1687:(2): 368–378.
1667:
1630:PLOS Pathogens
1616:
1567:
1524:
1483:
1459:
1452:
1434:
1410:
1382:
1347:
1299:
1249:
1204:
1191:gestis.dguv.de
1177:
1175:
1172:
1171:
1170:
1169:, related dyes
1160:
1150:
1141:
1139:Phenanthridine
1134:
1131:
1125:isolated from
1108:
1105:
1076:
1073:
1055:
1052:
1042:
1023:
1020:
927:
924:
878:phenanthridine
850:
847:
767:
764:
763:
758:
736:
735:
731:standard state
728:
725:
724:
721:
715:
714:
704:
697:
690:
675:
674:
673:
672:
670:
661:
660:
658:P304+P340+P310
635:
630:
627:
626:
613:
608:
605:
604:
599:
594:
591:
590:
578:
573:
570:
569:
559:
558:
554:
553:
541:
536:
533:
532:
528:
527:
524:
519:
516:
515:
512:
506:
505:
502:
498:
497:
494:
488:
487:
483:
479:
475:
472:
467:
464:
463:
459:
458:
456:
455:
452:
444:
443:
442:
439:
438:
436:
435:
431:
428:
427:
425:
421:
418:
417:
409:
408:
407:
404:
403:
401:
400:
387:
385:
373:
370:
369:
366:
360:
359:
357:
356:
348:
346:
340:
339:
337:
336:
332:
330:
324:
323:
321:
320:
312:
310:
302:
299:
298:
296:
295:
287:
285:
279:
278:
276:
275:
271:
269:
261:
260:
250:
242:
241:
239:
238:
230:
228:
222:
221:
219:
218:
210:
208:
202:
201:
199:
198:
190:
188:
182:
181:
178:
173:
170:
169:
167:
166:
158:
156:
149:
146:
145:
143:
142:
134:
132:
127:
124:
123:
119:
118:
115:
114:
111:
108:
105:
102:
99:
96:
93:
89:
88:
84:
83:
79:
78:
72:
71:
67:
66:
58:
57:
49:
48:
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
2325:
2314:
2311:
2309:
2306:
2304:
2303:Staining dyes
2301:
2299:
2296:
2294:
2291:
2289:
2286:
2284:
2281:
2279:
2276:
2274:
2271:
2270:
2268:
2258:
2253:
2249:
2248:
2244:
2236:
2232:
2228:
2224:
2220:
2216:
2212:
2208:
2204:
2196:
2194:
2190:
2179:on 2015-04-15
2178:
2174:
2168:
2165:
2160:
2156:
2152:
2148:
2144:
2140:
2133:
2130:
2125:
2121:
2117:
2113:
2109:
2105:
2101:
2094:
2091:
2086:
2084:1-56670-567-3
2080:
2076:
2072:
2066:
2063:
2058:
2054:
2050:
2046:
2042:
2038:
2031:
2028:
2013:
2007:
2004:
1990:
1983:
1980:
1972:
1966:
1963:
1958:
1954:
1950:
1946:
1942:
1938:
1934:
1930:
1923:
1920:
1915:
1911:
1907:
1903:
1899:
1895:
1891:
1887:
1879:
1877:
1873:
1862:
1855:
1852:
1847:
1843:
1839:
1835:
1831:
1827:
1823:
1819:
1812:
1809:
1804:
1800:
1796:
1792:
1788:
1784:
1777:
1774:
1769:
1765:
1760:
1755:
1751:
1747:
1743:
1739:
1735:
1731:
1727:
1720:
1717:
1712:
1708:
1703:
1698:
1694:
1690:
1686:
1682:
1678:
1671:
1668:
1663:
1659:
1654:
1649:
1644:
1639:
1635:
1631:
1627:
1620:
1617:
1612:
1608:
1603:
1598:
1594:
1590:
1586:
1582:
1578:
1571:
1568:
1563:
1559:
1555:
1551:
1547:
1543:
1539:
1535:
1528:
1525:
1520:
1516:
1511:
1506:
1502:
1498:
1494:
1487:
1484:
1469:
1463:
1460:
1455:
1449:
1445:
1438:
1435:
1424:
1420:
1414:
1411:
1400:
1396:
1389:
1387:
1383:
1378:
1374:
1370:
1366:
1362:
1358:
1351:
1348:
1345:
1341:
1330:
1326:
1322:
1318:
1317:
1312:
1306:
1304:
1300:
1295:
1291:
1287:
1283:
1279:
1275:
1271:
1267:
1263:
1256:
1254:
1250:
1247:
1243:
1232:
1228:
1224:
1220:
1219:
1214:
1208:
1205:
1192:
1188:
1182:
1179:
1173:
1168:
1164:
1161:
1158:
1154:
1151:
1149:
1145:
1142:
1140:
1137:
1136:
1132:
1130:
1128:
1124:
1123:
1122:T. congolense
1118:
1114:
1106:
1104:
1102:
1098:
1094:
1090:
1084:
1082:
1074:
1072:
1065:
1060:
1053:
1051:
1049:
1045:
1038:
1033:
1029:
1021:
1019:
1016:
1014:
1009:
1006:
1004:
1000:
999:kinetoplastid
996:
992:
987:
985:
980:
977:
973:
969:
965:
961:
957:
953:
949:
945:
944:nucleic acids
937:
932:
925:
923:
921:
917:
913:
909:
905:
902:
897:
895:
891:
887:
883:
879:
875:
871:
867:
864:
861:As with most
855:
848:
846:
844:
840:
836:
832:
828:
824:
820:
816:
812:
808:
804:
800:
796:
793:
789:
785:
784:intercalating
781:
777:
773:
761:
754:
749:
732:
726:
722:
720:
717:
716:
709:
702:
695:
671:
668:
667:
663:
662:
636:
633:
629:
628:
614:
611:
607:
606:
603:
600:
597:
593:
592:
588:
583:
579:
576:
572:
571:
567:
565:
560:
555:
550:
545:
542:
539:
535:
534:
531:Pharmacology
529:
525:
522:
518:
517:
513:
511:
510:Melting point
508:
507:
503:
500:
499:
495:
493:
490:
489:
473:
470:
466:
465:
460:
451:
447:
440:
426:
416:
412:
405:
397:
393:
392:DTXSID8025258
389:
388:
386:
376:
372:
371:
367:
365:
362:
361:
354:
350:
349:
347:
345:
342:
341:
334:
333:
331:
329:
326:
325:
318:
314:
313:
311:
305:
301:
300:
293:
289:
288:
286:
284:
281:
280:
273:
272:
270:
268:
263:
262:
258:
254:
251:
249:
247:ECHA InfoCard
244:
243:
236:
232:
231:
229:
227:
224:
223:
216:
212:
211:
209:
207:
204:
203:
196:
192:
191:
189:
187:
184:
183:
179:
176:
172:
171:
164:
160:
159:
157:
153:
148:
147:
140:
136:
135:
133:
130:
126:
125:
120:
112:
109:
106:
103:
100:
97:
94:
91:
90:
85:
77:
73:
68:
64:
59:
55:
50:
46:
41:
33:
32:Ethyl bromide
19:
2206:
2203:Acta Tropica
2202:
2181:. Retrieved
2177:the original
2167:
2145:(4): 89–90.
2142:
2138:
2132:
2107:
2103:
2093:
2074:
2065:
2040:
2036:
2030:
2019:. Retrieved
2006:
1995:. Retrieved
1982:
1965:
1932:
1928:
1922:
1892:(1): 37–47.
1889:
1885:
1864:. Retrieved
1854:
1821:
1817:
1811:
1786:
1783:Biochemistry
1782:
1776:
1733:
1729:
1719:
1684:
1680:
1670:
1633:
1629:
1619:
1584:
1580:
1570:
1537:
1533:
1527:
1500:
1496:
1486:
1474:. Retrieved
1462:
1443:
1437:
1426:. Retrieved
1422:
1413:
1402:. Retrieved
1398:
1363:(2): 77–84.
1360:
1357:Acta Tropica
1356:
1350:
1332:. Retrieved
1314:
1265:
1262:Acta Tropica
1261:
1234:. Retrieved
1216:
1207:
1195:. Retrieved
1190:
1181:
1127:Boran cattle
1120:
1110:
1085:
1078:
1069:
1064:intercalated
1025:
1017:
1010:
1007:
988:
981:
964:base pairing
941:
926:Applications
898:
874:heterocyclic
860:
830:
814:
806:
792:nucleic acid
779:
775:
771:
770:
665:
601:
563:
328:RTECS number
215:ChEMBL284328
122:Identifiers
87:Other names
2213:: 205–217.
1736:(1): 4291.
1272:: 169–183.
1197:22 November
1193:(in German)
976:fluorescent
972:ultraviolet
916:hydrophobic
912:responsible
908:hydrophobic
872:. Its core
863:fluorescent
811:bromoethane
719:Flash point
596:Signal word
538:ATCvet code
501:Appearance
462:Properties
253:100.013.622
2267:Categories
2183:2006-10-03
2021:2009-09-30
1997:2009-09-30
1866:2009-12-08
1859:Madden D.
1497:IUBMB Life
1428:2023-02-08
1404:2019-02-28
1334:2021-03-14
1236:2020-09-08
1174:References
1113:Gibe River
894:excitation
821:, it will
575:Pictograms
492:Molar mass
353:059NUO2Z1L
226:ChemSpider
195:CHEBI:4883
150:3D model (
129:CAS Number
2071:Armour MA
1340:CID 11765
1242:CID 14710
1093:Ames test
1032:Ames test
866:compounds
823:fluoresce
654:P301+P312
566:labelling
364:UN number
335:SF7950000
274:214-984-6
266:EC Number
139:1239-45-8
2288:Mutagens
2278:Bromides
2235:23878484
2211:Elsevier
2073:(2003).
1957:11377248
1914:10029672
1846:27423672
1838:16201894
1768:30327464
1711:16467477
1662:21187912
1611:12409452
1562:10593378
1519:16511967
1294:27564786
1270:Elsevier
1157:GelGreen
1133:See also
1117:Ethiopia
920:quencher
882:acridine
870:aromatic
831:homidium
782:) is an
666:NFPA 704
557:Hazards
552:)
544:QP51DX03
526:~40 g/l
180:3642536
18:Ethidium
2227:9107367
2209:(3–4).
2159:3238760
2124:3605608
2057:8073504
1937:Bibcode
1894:Bibcode
1759:6191420
1738:Bibcode
1702:1405896
1653:3002999
1554:8957173
1476:6 April
1377:7676909
1344:PubChem
1316:PubChem
1286:7902656
1268:(3–4).
1246:PubChem
1218:PubChem
1083:(SDS).
753:what is
751: (
546: (
486:
304:PubChem
2233:
2225:
2157:
2122:
2081:
2055:
1955:
1912:
1844:
1836:
1803:889813
1801:
1766:
1756:
1709:
1699:
1660:
1650:
1609:
1602:135822
1599:
1560:
1552:
1517:
1450:
1375:
1292:
1284:
1153:GelRed
904:moiety
901:phenyl
748:verify
745:
602:Danger
446:SMILES
292:C11161
206:ChEMBL
70:Names
2231:S2CID
2015:(PDF)
1992:(PDF)
1974:(PDF)
1842:S2CID
1558:S2CID
1471:(PDF)
1342:from
1327:, US
1290:S2CID
1244:from
1229:, US
1003:Z-DNA
815:EthBr
797:) in
795:stain
411:InChI
368:2811
317:14710
235:14034
186:ChEBI
152:JSmol
113:EthBr
2223:PMID
2155:PMID
2120:PMID
2079:ISBN
2053:PMID
1953:PMID
1910:PMID
1834:PMID
1799:PMID
1764:PMID
1707:PMID
1658:PMID
1607:PMID
1550:PMID
1515:PMID
1478:2014
1448:ISBN
1373:PMID
1321:NCBI
1282:PMID
1223:NCBI
1199:2021
1165:and
1146:and
1048:DMSO
1037:MSDS
1028:SYBR
952:PCRs
807:EtBr
774:(or
650:P284
646:P260
642:P202
638:P201
624:H341
620:H330
616:H302
344:UNII
283:KEGG
110:EtBr
2215:doi
2147:doi
2112:doi
2108:162
2045:doi
1945:doi
1933:492
1902:doi
1890:439
1826:doi
1791:doi
1754:PMC
1746:doi
1697:PMC
1689:doi
1648:PMC
1638:doi
1597:PMC
1589:doi
1542:doi
1505:doi
1365:doi
1329:NIH
1325:NLM
1274:doi
1231:NIH
1227:NLM
1099:or
1013:YPD
968:gel
960:RNA
948:DNA
827:DNA
564:GHS
549:WHO
482:BrN
380:EPA
307:CID
2269::
2229:.
2221:.
2207:64
2205:.
2192:^
2153:.
2141:.
2118:.
2106:.
2102:.
2051:.
2041:19
2039:.
1951:.
1943:.
1931:.
1908:.
1900:.
1888:.
1875:^
1840:.
1832:.
1822:43
1820:.
1797:.
1787:16
1785:.
1762:.
1752:.
1744:.
1732:.
1728:.
1705:.
1695:.
1683:.
1679:.
1656:.
1646:.
1632:.
1628:.
1605:.
1595:.
1585:30
1583:.
1579:.
1556:.
1548:.
1538:17
1536:.
1513:.
1501:57
1499:.
1495:.
1421:.
1397:.
1385:^
1371:.
1361:59
1359:.
1323:,
1319:.
1313:.
1302:^
1288:.
1280:.
1266:54
1264:.
1252:^
1225:,
1221:.
1215:.
1189:.
1129:.
1043:50
1041:LD
954:,
884:.
656:,
652:,
648:,
644:,
640:,
622:,
618:,
568::
480:20
476:21
2237:.
2217::
2186:.
2161:.
2149::
2143:4
2126:.
2114::
2087:.
2059:.
2047::
2024:.
2000:.
1959:.
1947::
1939::
1916:.
1904::
1896::
1869:.
1848:.
1828::
1805:.
1793::
1770:.
1748::
1740::
1734:9
1713:.
1691::
1685:5
1664:.
1640::
1634:6
1613:.
1591::
1564:.
1544::
1521:.
1507::
1480:.
1456:.
1431:.
1407:.
1379:.
1367::
1337:.
1296:.
1276::
1239:.
1201:.
790:(
743:Y
707:0
700:1
693:4
484:3
478:H
474:C
382:)
378:(
154:)
34:.
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.