656:, with some but not all of the characteristics of eukaryotes, provides evidence on the transition from archaea to eukaryotes. Lokiarchaeota and the eukaryotes probably share a common ancestor, and if so, diverged roughly two billion years ago. Evidence for common ancestry, rather than an evolutionary shift from Lokiarchaeota to eukaryotes, is found in analysis of fold superfamilies (FSFs). Fold super families are evolutionarily defined domains of protein structure. It is estimated that there are around 2500 total FSFs found in nature. Utilization of Venn diagrams allowed researchers to depict distributions of FSFs of those that were shared by Archaea and Eukarya, as well as those unique to their respective kingdoms. The addition of Lokiarchaeum into the Venn groups created from an initial genomic census only added 10 FSFs to Archaea. The addition of Lokiarchaeum also only contributed to a decrease of two FSFs previously unique to Eukarya. There were still 284 FSFs found exclusively in Eukarya. Lokiarchaeota’s limited impact in changing the Venn distribution of FSFs demonstrates the lack of genes that could be traced to a common ancestor with Eukaryotes. Rather, Eukaryotic genes present in bacterial and archaeal organisms are hypothesized to be from horizontal transfer from an early ancestor of modern eukaryotes. This putative ancestor possessed crucial "starter" genes that enabled increased cellular complexity. This common ancestor, or a relative, eventually led to the evolution of eukaryotes.
579:
359:
104:
723:
utilization and the degradation of aromatic compounds. The Loki-3 subgroup was not found to utilize proteins or short chain fatty acids, even though genes for amino acid degradation were present in both subgroups. Loki-2 was found to utilize protein, as seen through activity in when proteins were provided in Loki-2 incubations. Due to the greater carbon utilization pathways of Loki-3, the subgroup is found in a more diverse range of marine sediments than Loki-2.
62:
651:
have been dated back to 3.8 billion years ago. The eukaryotes include all complex cells and almost all multicellular organisms. They are thought to have evolved between 1.6 and 2.1 billion years ago. While the evolution of eukaryotes is considered to be an event of great evolutionary significance, no
338:
The metagenomic analysis determined the presence of an organism's genome in the sample. However, the organism itself was not cultured until years later, with a
Japanese group first reporting isolation and cultivation of a Lokiarchaeota strain in 2019. Since this initial cultivation of Lokiarchaeota,
330:
site, were analysed. Specific sediment horizons, previously shown to contain high abundances of novel archaeal lineages were subjected to metagenomic analysis. Due to the low density of cells in the sediment, the resulting genetic sequence does not come from an isolated cell, as would be the case in
722:
Two major subgroups of the
Lokiarachaeota phylum are Loki-2 and Loki-3. Incubations of these two subgroups from Helgoland mud sediments were analyzed through RNA and DNA stable isotope probing to understand their respective carbon metabolisms. Loki-3 were found to be active in both organic carbon
393:
proteins. These proteins included homologs of cytoskeleton proteins, GTPases, and the oligosaccharyltransferase (OST) protein complex. Homologues for components of the endosomal sorting complex required for transport and the ubiquitin protein modifier system were also identified in
Lokiarchaeota
718:
Analysis of
Lokiarchaeon genes also showed the expression of protein-encoding open reading frames (ORFs) involving the metabolism of sugars and proteins. However, these metabolic activities vary between subgroups of Lokiarchaeota. While Lokiarchaeota subgroups have similar genetic information,
350:, the Norse shape-shifting god. The Loki of literature has been described as "a staggeringly complex, confusing, and ambivalent figure who has been the catalyst of countless unresolved scholarly controversies", an analogy to the role of Lokiarchaeota in debates about the origin of eukaryotes.
374:. Of these, roughly 32% do not correspond to any known protein, 26% closely resemble archaeal proteins, and 29% correspond to bacterial proteins. This situation is consistent with: (i) proteins from a novel phylum (with few close relatives, or none) being difficult to assign to their correct
714:
pathway. This pathway contains a series of biochemical reactions aiding in inorganic carbon utilization. In
Lokiarchaeota, the WLP is thought to be acetogenic, due to lacking the gene methyl-CoM reductase necessary for methanogenesis.
339:
members of the phylum have been reported in a diverse range of habitats. Advances in both long and short-read technologies for DNA sequencing have also aided in the recovery and identification of
Lokiarchaeota from microbial samples.
695:(from which mitochondria are thought to descend) was not observed, these features suggest that MK-D1 and its syntrophs may represent an extant example of archaea-bacteria symbiosis similar to that which gave rise to eukaryotes.
947:
Jørgensen, Steffen Leth; Hannisdal, Bjarte; Lanzen, Anders; Baumberger, Tamara; Flesland, Kristin; Fonseca, Rita; Øvreås, Lise; Steen, Ida H; Thorseth, Ingunn H; Pedersen, Rolf B; Schleper, Christa (September 5, 2012).
441:, which is a key difference between prokaryotes and eukaryotes. The presence of actin proteins and intracellular transport mechanisms provides evidence for the common ancestry between ancient Lokiarchaeota and eukarya.
882:
Spang, Anja; Saw, Jimmy H.; Jørgensen, Steffen L.; Zaremba-Niedzwiedzka, Katarzyna; Martijn, Joran; Lind, Anders E.; van Eijk, Roel; Schleper, Christa; Guy, Lionel; Ettema, Thijs J. G. (2015).
1845:
Yin, Xiuran; Cai, Mingwei; Liu, Yang; Zhou, Guowei; Richter-Heitmann, Tim; Aromokeye, David A.; Kulkarni, Ajinkya C.; Nimzyk, Rolf; Cullhed, Henrik; Zhou, Zhichao; Pan, Jie (Mar 2021).
362:
Candidatus
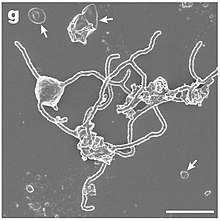
Prometheoarchaeum syntrophicum. (c) SEM of a dividing cell. Scale bar = 1 μm. (d) Cryo-EM of a single cell. Scale bar = 500 nm. White arrows indicate large membrane vesicles.
1325:
346:
analyses using a set of highly conserved protein-coding genes. Through a reference to the hydrothermal vent complex from which the first genome sample originated, the name refers to
1783:
Rodrigues-Oliveira, Thiago; Wollweber, Florian; Ponce-Toledo, Rafael I.; Xu, Jingwei; Rittmann, Simon K.-M. R.; Klingl, Andreas; Pilhofer, Martin; Schleper, Christa (January 2023).
1347:
1658:
226:. The phylum includes all members of the group previously named Deep Sea Archaeal Group, also known as Marine Benthic Group B. Lokiarchaeota is part of the superphylum
1945:
753:"Scientists glimpse oddball microbe that could help explain rise of complex life - 'Lokiarchaea', previously known only from DNA, is isolated and grown in culture"
2772:
1351:
610:
to the eukaryotes might be an intermediate step between the prokaryotic cells, devoid of subcellular structures, and the eukaryotic cells, which harbor many
1239:
624:
classifies cellular life into three domains: archaea, bacteria, and eukaryotes; the last being characterised by large, highly evolved cells, containing
1732:
331:
conventional analysis, but is rather a combination of genetic fragments. The result was a 92% complete, 1.4 fold-redundant composite genome named
1704:
691:
syntrophs. MK-D1 also seems to organize its external membrane into complex structures using genes shared with eukaryotes. While association with
398:
genes and no genes of known eukaryotic origin were detected in the metagenome from which the composite genome was extracted. Further, previous
394:
genome analysis. Sample contamination is an unlikely explanation for the unusual proteins because the recovered genes were always flanked by
1938:
1906:
809:
Caceres, Eva F.; Lewis, William H.; Homa, Felix; Martin, Tom; Schramm, Andreas; Kjeldsen, Kasper U.; Ettema, Thijs J. G. (2019-12-18).
647:
The bacteria and archaea are thought to be the most ancient of lineages, as fossil strata bearing the chemical signature of archaeal
578:
1983:
659:
In 2020, a
Japanese research group reported culturing a strain of Lokiarchaeota in the laboratory. This strain, currently named
417:. Eukaryotic protein functions found in Lokiarchaeota also include intracellular transport mechanisms. It is inferred then that
1978:
1931:
950:"Correlating microbial community profiles with geochemical data in highly stratified sediments from the Arctic Mid-Ocean Ridge"
358:
2062:
2052:
2035:
1001:"Quantitative and phylogenetic study of the Deep Sea Archaeal Group in sediments of the Arctic mid-ocean spreading ridge"
999:
Jørgensen, Steffen Leth; Thorseth, Ingunn H; Pedersen, Rolf B; Baumberger, Tamara; Schleper, Christa (October 4, 2013).
811:"Near-complete Lokiarchaeota genomes from complex environmental samples using long and short read metagenomic analyses"
103:
711:
429:
in eukaryotes. Phagocytosis is the ability to engulf and consume another particle; such ability would facilitate the
2777:
1574:
Wang, Minglei; Yafremava, Liudmila S.; Caetano-Anollés, Derek; Mittenthal, Jay E.; Caetano-Anollés, Gustavo (2007).
1468:
Lake, James A. (1988). "Origin of the eukaryotic nucleus determined by rate-invariant analysis of rRNA sequences".
389:
A small, but significant portion of the proteins (175, 3.3%) that the recovered genes code for are very similar to
668:
30:
2352:
2067:
1757:
2800:
2740:
1531:
1274:
732:
629:
2030:
1529:
Guy, Lionel; Ettema, Thijs J.G. (2011). "The archaeal 'TACK' superphylum and the origin of eukaryotes".
834:
652:
intermediate forms or "missing links" had been discovered previously. In this context, the discovery of
1373:
2313:
547:
1576:"Reductive evolution of architectural repertoires in proteomes and the birth of the tripartite world"
1479:
1418:
1109:
1101:
895:
766:
430:
2201:
522:
692:
621:
243:
2268:
2146:
2047:
2025:
2013:
1988:
1762:
1511:
1450:
822:
673:
120:
323:
taken in 2010 in the rift valley on the
Knipovich ridge in the Arctic Ocean, near the so-called
1157:
von
Schnurbein, Stefanie (November 2000). "The Function of Loki in Snorri Sturluson's "Edda"".
687:. The MK-D1 organism produces hydrogen as a metabolic byproduct, which is then consumed by the
582:
A schematic evolutionary tree of the archaea including Lokiarchaeota and the root of eukaryotes
2659:
2539:
2440:
2363:
2286:
2057:
2020:
2008:
1998:
1884:
1866:
1822:
1804:
1685:
1607:
1556:
1548:
1503:
1495:
1442:
1434:
1302:
1294:
1220:
1139:
1032:
981:
929:
911:
810:
784:
599:
591:
327:
296:
271:
239:
2667:
2597:
1972:
1874:
1858:
1812:
1796:
1675:
1667:
1634:
1597:
1589:
1540:
1487:
1470:
1426:
1409:
1286:
1210:
1200:
1166:
1129:
1119:
1092:
1022:
1012:
971:
961:
919:
903:
814:
774:
757:
698:
In 2022, the second cultured example of Lokiarchaeota was reported and the strain was named
375:
324:
300:
227:
157:
2581:
2525:
2479:
2388:
2263:
2248:
2230:
2215:
2158:
2141:
1580:
847:
607:
144:
1483:
1422:
1105:
1058:
899:
770:
402:
analysis suggested the genes in question had their origin at the base of the eukaryotic
75:
Please help update this article to reflect recent events or newly available information.
2497:
2408:
2338:
2188:
2170:
2003:
1879:
1846:
1817:
1784:
1709:
1680:
1653:
1602:
1575:
1215:
1188:
1134:
1087:
1027:
1000:
976:
949:
924:
883:
633:
598:
group composed of the Lokiarchaeota and the eukaryotes, supporting an archaeal host or
1923:
1638:
1321:
2794:
2723:
2515:
2452:
2428:
2083:
1400:
826:
683:
602:
for the emergence of the eukaryotes. The repertoire of membrane-related functions of
410:
379:
285:
analysis of a mid-oceanic sediment sample. This analysis suggests the existence of a
263:
235:
231:
2649:
2644:
2627:
2576:
2565:
2225:
2210:
2153:
2092:
1515:
1454:
644:
and all multicellular organisms such as animals, fungi, and plants are eukaryotes.
637:
625:
595:
481:
438:
434:
426:
414:
399:
378:; and (ii) existing research that suggests there has been significant inter-domain
343:
308:
282:
251:
247:
2763:
1625:
Hahn, Jürgen; Haug, Pat (1986). "Traces of Archaebacteria in ancient sediments".
2505:
2180:
2165:
1847:"Subgroup level differences of physiological activities in marine Lokiarchaeota"
632:(adenosine triphosphate, the energy currency of the cell), and a membrane-bound
395:
320:
304:
1862:
1800:
779:
752:
719:
differences in metabolic abilities explain their respective ecological niches.
1954:
1733:"We've finally gotten a look at the microbe that might have been our ancestor"
1544:
1290:
1124:
818:
678:
664:
617:
390:
1870:
1808:
1552:
1499:
1438:
1298:
1017:
915:
277:
Lokiarchaeota was introduced in 2015 after the identification of a candidate
1205:
966:
688:
611:
266:-related functions. The presence of such genes support the hypothesis of an
255:
1888:
1826:
1689:
1671:
1611:
1560:
1446:
1306:
1224:
1143:
1036:
985:
933:
788:
1507:
2757:
2042:
1378:
1275:"Lokiarchaeota: eukaryote-like missing links from microbial dark matter?"
1244:
641:
383:
1785:"Actin cytoskeleton and complex cell architecture in an Asgard archaeon"
1430:
907:
884:"Complex archaea that bridge the gap between prokaryotes and eukaryotes"
1958:
1652:
Knoll, Andrew H.; Javaux, E. J.; Hewitt, D.; Cohen, P. (29 June 2006).
1593:
267:
223:
131:
1491:
1273:
Nasir, Arshan; Kim, Kyung Mo; Caetano-Anollés, Gustavo (2015-08-01).
278:
219:
2734:
710:
Lokiarachaeota is known to have a tetrahydromethanopterin-dependent
1404:
1170:
1114:
2379:
648:
422:
403:
371:
357:
286:
1705:"Under the Sea, a Missing Link in the Evolution of Complex Cells"
2488:
1088:"Isolation of an archaeon at the prokaryote–eukaryote interface"
347:
259:
2738:
1927:
1062:
55:
1907:"Primitive Asgard Cells Show Life on the Brink of Complexity"
409:
In eukaryotes, the function of these shared proteins include
421:
may have some of these abilities. Another shared protein,
1758:"This Strange Microbe May Mark One of Life's Great Leaps"
413:
deformation, cell shape formation, and a dynamic protein
1326:
List of Prokaryotic names with Standing in Nomenclature
1059:"Newly found microbe is close relative of complex life"
34:
1187:
Ghoshdastider U, Jiang S, Popp D, Robinson RC (2015).
667:
association with two hydrogen-consuming microbes: a
2747:
2658:
2605:
2596:
2496:
2487:
2478:
2378:
2311:
2199:
2134:
2115:
2091:
2082:
1240:"Meet Loki, your closest-known prokaryote relative"
370:composite genome consists of 5,381 protein coding
1659:Philosophical Transactions of the Royal Society B
1086:Imachi, Hiroyuki; et al. (15 January 2020).
1322:"Family: Lokiarchaeota, not assigned to family"
877:
342:The Lokiarchaeota phylum was proposed based on
270:host for the emergence of the eukaryotes; the
1405:"Eukaryotic evolution, changes and challenges"
1081:
1079:
875:
873:
871:
869:
867:
865:
863:
861:
859:
857:
804:
802:
800:
798:
1939:
1352:National Center for Biotechnology Information
1268:
1266:
1264:
1262:
1182:
1180:
8:
1654:"Eukaryotic organisms in Proterozoic oceans"
1189:"In search of the primordial actin filament"
1840:
1838:
1836:
590:genome against known genomes resulted in a
18:
2735:
2602:
2493:
2484:
2088:
1946:
1932:
1924:
1633:(Archaebacteria '85 Proceedings): 178–83.
102:
93:
1878:
1816:
1679:
1601:
1214:
1204:
1133:
1123:
1113:
1026:
1016:
975:
965:
923:
778:
661:Candidatus Prometheoarchaeum syntrophicum
577:
24:This is an accepted version of this page
1372:Maximillan, Ludwig (27 December 2019).
743:
254:grouping of the Lokiarchaeota with the
20:
1374:"Evolution: A revelatory relationship"
843:
832:
513:Pallen, Rodriguez-R & Alikhan 2022
1052:
1050:
1048:
1046:
562:corrig. Imachi, Nobu & Takai 2020
552:corrig. Imachi, Nobu & Takai 2020
230:containing the phyla: Lokiarchaeota,
7:
1627:Systematic and Applied Microbiology
751:Lambert, Jonathan (9 August 2019).
49:
700:Candidatus Lokiarchaeum ossiferum
258:. The analysis revealed several
60:
628:, which help the cells produce
115:Prometheoarchaeum syntrophicum
663:strain MK-D1, was observed in
586:A comparative analysis of the
496:Rodrigues-Oliveira et al. 2023
295:. The sample was taken near a
1:
1639:10.1016/S0723-2020(86)80002-9
1905:Sokol, Joshua (2023-04-11).
1057:Rincon, Paul (May 6, 2015).
303:located at the bend between
1756:Zimmer, Carl (2020-01-15).
1731:Timmer, John (2019-08-07).
1703:Zimmer, Carl (6 May 2015).
1238:Khan, Amina (May 6, 2015).
289:of unicellular life dubbed
2817:
1863:10.1038/s41396-020-00818-5
1801:10.1038/s41586-022-05550-y
1403:; Martin, William (2006).
1348:"Candidatus Lokiarchaeota"
780:10.1038/d41586-019-02430-w
486:corrig. Spang et al. 2015
2715:
1966:
1545:10.1016/j.tim.2011.09.002
1291:10.1016/j.tim.2015.06.001
1125:10.1038/s41586-019-1916-6
1005:Frontiers in Microbiology
819:10.1101/2019.12.17.879148
669:sulfate-reducing bacteria
574:Evolutionary significance
472:Vanwonterghem et al. 2016
319:Sediments from a gravity
299:at a vent field known as
201:
196:
191:corrig. Spang et al. 2015
121:Scientific classification
119:
110:
101:
96:
69:This article needs to be
1354:(NCBI) taxonomy database
1193:Proc Natl Acad Sci U S A
1018:10.3389/fmicb.2013.00299
307:/Knipovich ridge in the
31:latest accepted revision
2353:Methanomassiliicoccales
2278:Methanonatronarchaeales
1206:10.1073/pnas.1511568112
967:10.1073/pnas.1207574109
2275:Methanonatronarchaeia
2185:"Methanofastidiosales"
1672:10.1098/rstb.2006.1843
1532:Trends in Microbiology
1279:Trends in Microbiology
842:Cite journal requires
733:List of Archaea genera
583:
363:
2557:"Methanomethylicales"
2125:"Hydrothermarchaeles"
2122:"Hydrothermarchaeia"
2116:"Hydrothermarchaeota"
813:: 2019.12.17.879148.
600:eocyte-like scenarios
581:
361:
272:eocyte-like scenarios
250:analysis disclosed a
2675:"Heimdallarchaeales"
2554:"Methanomethylicia"
2536:"Geothermarchaeales"
2357:"Natronoplasmatales"
2343:"Aciduliprofundales"
2302:"Syntropharchaeales"
1346:Sayers; et al.
1159:History of Religions
706:Physiological traits
509:Prometheoarchaeaceae
431:endosymbiotic origin
2722:Alternative views:
2325:"Thermoprofundales"
2296:"Syntropharchaeia"
2135:"Methanobacteriota"
2108:"Persephonarchaeia"
1484:1988Natur.331..184L
1431:10.1038/nature04546
1423:2006Natur.440..623E
1106:2020Natur.577..519I
908:10.1038/nature14447
900:2015Natur.521..173S
771:2019Natur.572..294L
693:alphaproteobacteria
622:three-domain system
425:, is essential for
21:Page version status
2533:Conexivisphaerales
2394:"Aenigmatarchaeia"
2360:"Sysuiplasmatales"
2269:Methanomicrobiales
2258:"Methanoliparales"
2147:Methanobacteriales
1763:The New York Times
1594:10.1101/gr.6454307
606:suggests that the
584:
364:
27:
2788:
2787:
2741:Taxon identifiers
2732:
2731:
2711:
2710:
2707:
2706:
2660:Heimdallarchaeota
2639:"Sigynarchaeales"
2592:
2591:
2560:"Nezhaarchaeales"
2540:Nitrososphaerales
2466:"Undinarchaeota"
2446:"Nanohalarchaeia"
2419:"Huberarchaeota"
2389:Aenigmatarchaeota
2374:
2373:
2364:Thermoplasmatales
2349:"Lunaplasmatales"
2346:"Gimiplasmatales"
2299:"Methanophagales"
2287:Methanosarcinales
2255:"Methanoliparia"
2220:"Mnemosynellales"
1795:(7943): 332–339.
1666:(1470): 1023–38.
1588:(11): 1572–1585.
1478:(6152): 184–186.
1417:(7084): 623–630.
1401:Embley, T. Martin
1100:(7791): 519–525.
894:(7551): 173–179.
674:Halodesulfovibrio
592:phylogenetic tree
563:
560:P. syntrophicum"
553:
538:
528:
514:
497:
487:
473:
460:
459:Spang et al. 2015
328:hydrothermal vent
297:hydrothermal vent
240:Heimdallarchaeota
213:
212:
207:
206:Spang et al. 2015
204:"Lokiarchaeales"
177:Spang et al. 2015
90:
89:
51:Phylum of archaea
2808:
2781:
2780:
2768:
2767:
2766:
2736:
2700:"Wukongarchaeia"
2692:"Borrarchaeales"
2681:"Kariarchaeales"
2672:"Gerdarchaeales"
2668:Heimdallarchaeia
2636:"Lokiarchaeales"
2620:"Hermodarchaeia"
2603:
2573:"Marsarchaeales"
2549:"Culexarchaeles"
2546:"Culexarchaeia"
2530:"Caldarchaeales"
2494:
2485:
2441:Nanohalarchaeota
2314:Thermoplasmatota
2291:Methanotrichales
2283:Methanosarcinia
2245:"Methanocellia"
2177:"Theionarchaeia"
2089:
1995:
1975:
1948:
1941:
1934:
1925:
1920:
1918:
1917:
1893:
1892:
1882:
1851:The ISME Journal
1842:
1831:
1830:
1820:
1780:
1774:
1773:
1771:
1770:
1753:
1747:
1746:
1744:
1743:
1728:
1722:
1721:
1719:
1717:
1700:
1694:
1693:
1683:
1649:
1643:
1642:
1622:
1616:
1615:
1605:
1571:
1565:
1564:
1526:
1520:
1519:
1492:10.1038/331184a0
1465:
1459:
1458:
1397:
1391:
1390:
1388:
1386:
1369:
1363:
1362:
1360:
1359:
1343:
1337:
1336:
1334:
1333:
1317:
1311:
1310:
1270:
1257:
1256:
1254:
1252:
1235:
1229:
1228:
1218:
1208:
1184:
1175:
1174:
1154:
1148:
1147:
1137:
1127:
1117:
1083:
1074:
1073:
1071:
1069:
1054:
1041:
1040:
1030:
1020:
996:
990:
989:
979:
969:
960:(42): E2846–55.
944:
938:
937:
927:
879:
852:
851:
845:
840:
838:
830:
806:
793:
792:
782:
748:
561:
551:
548:Promethearchaeum
536:
526:
512:
495:
485:
471:
458:
205:
106:
94:
85:
82:
76:
64:
63:
56:
2816:
2815:
2811:
2810:
2809:
2807:
2806:
2805:
2791:
2790:
2789:
2784:
2776:
2771:
2762:
2761:
2756:
2743:
2733:
2728:
2703:
2695:"Sifarchaeales"
2686:"Njordarchaeia"
2678:"Hodarchaeales"
2654:
2633:"Helarchaeales"
2617:"Freyrarchaeia"
2614:"Baldrarchaeia"
2588:
2582:Thermoproteales
2526:Nitrososphaeria
2510:"Korarchaeales"
2480:Proteoarchaeota
2474:
2469:"Undinarchaeia"
2422:"Huberarchaeia"
2399:"Altarchaeota"
2370:
2333:"Poseidoniales"
2307:
2264:Methanomicrobia
2249:Methanocellales
2240:"Hikarchaeales"
2231:Halobacteriales
2216:Archaeoglobales
2195:
2159:Methanococcales
2142:Methanobacteria
2130:
2111:
2103:"Hadarchaeales"
2078:
2077:
1993:
1971:
1962:
1952:
1915:
1913:
1911:Quanta Magazine
1904:
1901:
1896:
1844:
1843:
1834:
1782:
1781:
1777:
1768:
1766:
1755:
1754:
1750:
1741:
1739:
1730:
1729:
1725:
1715:
1713:
1702:
1701:
1697:
1651:
1650:
1646:
1624:
1623:
1619:
1581:Genome Research
1573:
1572:
1568:
1539:(12): 580–587.
1528:
1527:
1523:
1467:
1466:
1462:
1399:
1398:
1394:
1384:
1382:
1371:
1370:
1366:
1357:
1355:
1345:
1344:
1340:
1331:
1329:
1319:
1318:
1314:
1272:
1271:
1260:
1250:
1248:
1237:
1236:
1232:
1186:
1185:
1178:
1156:
1155:
1151:
1085:
1084:
1077:
1067:
1065:
1056:
1055:
1044:
998:
997:
993:
946:
945:
941:
881:
880:
855:
841:
831:
808:
807:
796:
750:
749:
745:
741:
729:
708:
608:common ancestor
576:
468:Lokiarchaeaceae
447:
356:
317:
192:
189:
188:
178:
175:
174:
160:
148:
145:Proteoarchaeota
134:
86:
80:
77:
74:
65:
61:
52:
47:
46:
45:
44:
43:
42:
26:
12:
11:
5:
2814:
2812:
2804:
2803:
2793:
2792:
2786:
2785:
2783:
2782:
2769:
2753:
2751:
2745:
2744:
2739:
2730:
2729:
2727:
2726:
2720:
2716:
2713:
2712:
2709:
2708:
2705:
2704:
2702:
2701:
2698:
2697:
2696:
2693:
2689:"Sifarchaeia"
2687:
2684:
2683:
2682:
2679:
2676:
2673:
2664:
2662:
2656:
2655:
2653:
2652:
2647:
2642:
2641:
2640:
2637:
2634:
2624:
2623:"Jordarchaeia"
2621:
2618:
2615:
2611:
2609:
2600:
2594:
2593:
2590:
2589:
2587:
2586:
2585:
2584:
2579:
2574:
2571:
2570:"Gearchaeales"
2563:
2562:
2561:
2558:
2552:
2551:
2550:
2544:
2543:
2542:
2537:
2534:
2531:
2523:
2522:
2521:
2520:"Hecatellales"
2513:
2512:
2511:
2502:
2500:
2498:Thermoproteota
2491:
2482:
2476:
2475:
2473:
2472:
2471:
2470:
2464:
2463:
2462:
2461:"Nanohalobiia"
2459:
2458:"Nanoarchaeia"
2449:
2448:
2447:
2437:
2436:
2435:
2434:"Micrarchaeia"
2425:
2424:
2423:
2417:
2416:
2415:
2414:"Iainarchaeia"
2405:
2404:
2403:
2397:
2396:
2395:
2384:
2382:
2376:
2375:
2372:
2371:
2369:
2368:
2367:
2366:
2361:
2358:
2355:
2350:
2347:
2344:
2339:Thermoplasmata
2336:
2335:
2334:
2330:"Poseidoniia"
2328:
2327:
2326:
2322:"Izemarchaea"
2319:
2317:
2309:
2308:
2306:
2305:
2304:
2303:
2300:
2294:
2293:
2292:
2289:
2281:
2280:
2279:
2273:
2272:
2271:
2261:
2260:
2259:
2253:
2252:
2251:
2243:
2242:
2241:
2237:"Hikarchaeia"
2235:
2234:
2233:
2223:
2222:
2221:
2218:
2207:
2205:
2202:Halobacteriota
2197:
2196:
2194:
2193:
2192:
2191:
2189:Thermococcales
2186:
2178:
2175:
2174:
2173:
2171:Methanopyrales
2163:
2162:
2161:
2151:
2150:
2149:
2138:
2136:
2132:
2131:
2129:
2128:
2127:
2126:
2119:
2117:
2113:
2112:
2110:
2109:
2106:
2105:
2104:
2100:"Hadarchaeia"
2097:
2095:
2086:
2080:
2079:
2076:
2075:
2074:
2073:
2072:
2071:
2068:Mesomycetozoea
2065:
2060:
2050:
2040:
2039:
2038:
2033:
2028:
2023:
2018:
2017:
2016:
2004:Diaphoretickes
2001:
1996:
1991:
1986:
1981:
1976:
1968:
1967:
1964:
1963:
1961:classification
1953:
1951:
1950:
1943:
1936:
1928:
1922:
1921:
1900:
1899:External links
1897:
1895:
1894:
1857:(3): 848–861.
1832:
1775:
1748:
1723:
1710:New York Times
1695:
1644:
1617:
1566:
1521:
1460:
1392:
1364:
1338:
1312:
1285:(8): 448–450.
1258:
1230:
1199:(30): 9150–1.
1176:
1171:10.1086/463618
1165:(2): 109–124.
1149:
1115:10.1101/726976
1075:
1042:
991:
939:
853:
844:|journal=
794:
742:
740:
737:
736:
735:
728:
725:
707:
704:
594:that showed a
575:
572:
571:
570:
569:
568:
567:
566:
565:
564:
541:
540:
539:
537:Wu et al. 2022
535:H. repetitus"
527:Wu et al. 2022
523:Harpocratesius
502:
501:
500:
499:
498:
494:L. ossiferum"
455:Lokiarchaeales
446:
443:
355:
352:
316:
313:
218:is a proposed
211:
210:
209:
208:
199:
198:
194:
193:
190:
187:"Lokiarchaeia"
186:
184:
180:
179:
176:
168:
166:
162:
161:
156:
154:
150:
149:
142:
140:
136:
135:
130:
128:
124:
123:
117:
116:
108:
107:
99:
98:
97:Lokiarchaeota
88:
87:
68:
66:
59:
50:
48:
39:16 August 2024
28:
22:
19:
17:
16:
15:
14:
13:
10:
9:
6:
4:
3:
2:
2813:
2802:
2801:Archaea phyla
2799:
2798:
2796:
2779:
2774:
2770:
2765:
2759:
2755:
2754:
2752:
2750:
2749:Lokiarchaeota
2746:
2742:
2737:
2725:
2721:
2718:
2717:
2714:
2699:
2694:
2691:
2690:
2688:
2685:
2680:
2677:
2674:
2671:
2670:
2669:
2666:
2665:
2663:
2661:
2657:
2651:
2648:
2646:
2643:
2638:
2635:
2632:
2631:
2629:
2625:
2622:
2619:
2616:
2613:
2612:
2610:
2608:
2607:Lokiarchaeota
2604:
2601:
2599:
2595:
2583:
2580:
2578:
2575:
2572:
2569:
2568:
2567:
2566:Thermoproteia
2564:
2559:
2556:
2555:
2553:
2548:
2547:
2545:
2541:
2538:
2535:
2532:
2529:
2528:
2527:
2524:
2519:
2518:
2517:
2516:Bathyarchaeia
2514:
2509:
2508:
2507:
2504:
2503:
2501:
2499:
2495:
2492:
2490:
2486:
2483:
2481:
2477:
2468:
2467:
2465:
2460:
2457:
2456:
2454:
2453:Nanoarchaeota
2450:
2445:
2444:
2442:
2438:
2433:
2432:
2430:
2429:Micrarchaeota
2426:
2421:
2420:
2418:
2413:
2412:
2410:
2409:Iainarchaeota
2406:
2402:"Altarchaeia"
2401:
2400:
2398:
2393:
2392:
2390:
2386:
2385:
2383:
2381:
2377:
2365:
2362:
2359:
2356:
2354:
2351:
2348:
2345:
2342:
2341:
2340:
2337:
2332:
2331:
2329:
2324:
2323:
2321:
2320:
2318:
2315:
2310:
2301:
2298:
2297:
2295:
2290:
2288:
2285:
2284:
2282:
2277:
2276:
2274:
2270:
2267:
2266:
2265:
2262:
2257:
2256:
2254:
2250:
2247:
2246:
2244:
2239:
2238:
2236:
2232:
2229:
2228:
2227:
2224:
2219:
2217:
2214:
2213:
2212:
2211:Archaeoglobia
2209:
2208:
2206:
2203:
2198:
2190:
2187:
2184:
2183:
2182:
2179:
2176:
2172:
2169:
2168:
2167:
2164:
2160:
2157:
2156:
2155:
2152:
2148:
2145:
2144:
2143:
2140:
2139:
2137:
2133:
2124:
2123:
2121:
2120:
2118:
2114:
2107:
2102:
2101:
2099:
2098:
2096:
2094:
2090:
2087:
2085:
2084:Euryarchaeota
2081:
2069:
2066:
2064:
2061:
2059:
2056:
2055:
2054:
2051:
2049:
2046:
2045:
2044:
2041:
2037:
2034:
2032:
2031:Stramenopiles
2029:
2027:
2024:
2022:
2019:
2015:
2012:
2011:
2010:
2007:
2006:
2005:
2002:
2000:
1997:
1994:(major groups
1992:
1990:
1987:
1985:
1982:
1980:
1977:
1974:
1970:
1969:
1965:
1960:
1956:
1949:
1944:
1942:
1937:
1935:
1930:
1929:
1926:
1912:
1908:
1903:
1902:
1898:
1890:
1886:
1881:
1876:
1872:
1868:
1864:
1860:
1856:
1852:
1848:
1841:
1839:
1837:
1833:
1828:
1824:
1819:
1814:
1810:
1806:
1802:
1798:
1794:
1790:
1786:
1779:
1776:
1765:
1764:
1759:
1752:
1749:
1738:
1734:
1727:
1724:
1712:
1711:
1706:
1699:
1696:
1691:
1687:
1682:
1677:
1673:
1669:
1665:
1661:
1660:
1655:
1648:
1645:
1640:
1636:
1632:
1628:
1621:
1618:
1613:
1609:
1604:
1599:
1595:
1591:
1587:
1583:
1582:
1577:
1570:
1567:
1562:
1558:
1554:
1550:
1546:
1542:
1538:
1534:
1533:
1525:
1522:
1517:
1513:
1509:
1505:
1501:
1497:
1493:
1489:
1485:
1481:
1477:
1473:
1472:
1464:
1461:
1456:
1452:
1448:
1444:
1440:
1436:
1432:
1428:
1424:
1420:
1416:
1412:
1411:
1406:
1402:
1396:
1393:
1381:
1380:
1375:
1368:
1365:
1353:
1349:
1342:
1339:
1327:
1323:
1320:J.P. Euzéby.
1316:
1313:
1308:
1304:
1300:
1296:
1292:
1288:
1284:
1280:
1276:
1269:
1267:
1265:
1263:
1259:
1247:
1246:
1241:
1234:
1231:
1226:
1222:
1217:
1212:
1207:
1202:
1198:
1194:
1190:
1183:
1181:
1177:
1172:
1168:
1164:
1160:
1153:
1150:
1145:
1141:
1136:
1131:
1126:
1121:
1116:
1111:
1107:
1103:
1099:
1095:
1094:
1089:
1082:
1080:
1076:
1064:
1060:
1053:
1051:
1049:
1047:
1043:
1038:
1034:
1029:
1024:
1019:
1014:
1010:
1006:
1002:
995:
992:
987:
983:
978:
973:
968:
963:
959:
955:
951:
943:
940:
935:
931:
926:
921:
917:
913:
909:
905:
901:
897:
893:
889:
885:
878:
876:
874:
872:
870:
868:
866:
864:
862:
860:
858:
854:
849:
836:
828:
824:
820:
816:
812:
805:
803:
801:
799:
795:
790:
786:
781:
776:
772:
768:
765:(7769): 294.
764:
760:
759:
754:
747:
744:
738:
734:
731:
730:
726:
724:
720:
716:
713:
712:Wood-Ljundahl
705:
703:
701:
696:
694:
690:
686:
685:
684:Methanogenium
681:of the genus
680:
676:
675:
671:of the genus
670:
666:
662:
657:
655:
650:
645:
643:
639:
638:nucleic acids
635:
631:
627:
623:
619:
615:
613:
609:
605:
601:
597:
593:
589:
580:
573:
559:
555:
554:
549:
546:
542:
534:
530:
529:
524:
521:
517:
516:
510:
506:
503:
493:
489:
488:
483:
480:
476:
475:
474:(MBGB, DSAG)
469:
465:
462:
461:
456:
452:
449:
448:
444:
442:
440:
436:
432:
428:
424:
420:
416:
412:
411:cell membrane
407:
405:
401:
397:
392:
387:
386:and Archaea.
385:
381:
380:gene transfer
377:
373:
369:
360:
353:
351:
349:
345:
340:
336:
334:
329:
326:
325:Loki's Castle
322:
314:
312:
310:
306:
302:
301:Loki's Castle
298:
294:
293:
288:
284:
280:
275:
273:
269:
265:
264:cell membrane
261:
257:
253:
249:
245:
241:
237:
236:Odinarchaeota
233:
232:Thorarchaeota
229:
225:
221:
217:
216:Lokiarchaeota
203:
202:
200:
195:
185:
182:
181:
172:
171:Lokiarchaeota
167:
164:
163:
159:
155:
153:Superphylum:
152:
151:
146:
141:
138:
137:
133:
129:
126:
125:
122:
118:
114:
111:SEM image of
109:
105:
100:
95:
92:
84:
72:
67:
58:
57:
54:
40:
36:
32:
25:
2748:
2650:Thorarchaeia
2645:Odinarchaeia
2628:Lokiarchaeia
2606:
2577:Sulfolobales
2226:Halobacteria
2154:Methanococci
2093:Hadarchaeota
2053:Opisthokonta
1914:. Retrieved
1910:
1854:
1850:
1792:
1788:
1778:
1767:. Retrieved
1761:
1751:
1740:. Retrieved
1737:Ars Technica
1736:
1726:
1714:. Retrieved
1708:
1698:
1663:
1657:
1647:
1630:
1626:
1620:
1585:
1579:
1569:
1536:
1530:
1524:
1475:
1469:
1463:
1414:
1408:
1395:
1383:. Retrieved
1377:
1367:
1356:. Retrieved
1341:
1330:. Retrieved
1315:
1282:
1278:
1249:. Retrieved
1243:
1233:
1196:
1192:
1162:
1158:
1152:
1097:
1091:
1066:. Retrieved
1008:
1004:
994:
957:
953:
942:
891:
887:
835:cite journal
762:
756:
746:
721:
717:
709:
699:
697:
682:
672:
660:
658:
654:Lokiarchaeum
653:
646:
626:mitochondria
616:
604:Lokiarchaeum
603:
596:monophyletic
588:Lokiarchaeum
587:
585:
557:
544:
532:
519:
508:
504:
491:
482:Lokiarchaeum
478:
467:
463:
454:
450:
439:chloroplasts
435:mitochondria
427:phagocytosis
419:Lokiarchaeum
418:
415:cytoskeleton
408:
400:phylogenetic
388:
368:Lokiarchaeum
367:
365:
344:phylogenetic
341:
337:
333:Lokiarchaeum
332:
318:
309:Arctic Ocean
292:Lokiarchaeum
291:
290:
276:
252:monophyletic
248:phylogenetic
244:Helarchaeota
215:
214:
170:
112:
91:
78:
70:
53:
38:
29:This is the
23:
2724:Wikispecies
2506:Korarchaeia
2181:Thermococci
2166:Methanopyri
1955:Prokaryotes
1385:28 December
636:containing
396:prokaryotic
354:Description
283:metagenomic
1916:2023-04-16
1769:2020-01-15
1742:2019-08-08
1358:2023-06-20
1332:2023-06-20
739:References
679:methanogen
665:syntrophic
618:Carl Woese
612:organelles
545:Candidatus
520:Candidatus
479:Candidatus
391:eukaryotic
256:eukaryotes
113:Candidatus
81:March 2018
2764:Q19868361
2048:Amoebozoa
2026:Alveolata
2014:Cryptista
1989:Eukaryota
1871:1751-7370
1809:1476-4687
1553:0966-842X
1500:0028-0836
1439:0028-0836
1299:0966-842X
916:0028-0836
827:213982145
689:symbiotic
315:Discovery
139:Kingdom:
2795:Category
2758:Wikidata
2043:Amorphea
2021:Rhizaria
2009:Hacrobia
1999:Excavata
1984:Bacteria
1889:33149207
1827:36544020
1690:16754612
1612:17908824
1561:22018741
1447:16572163
1379:Phys.org
1307:26112912
1245:LA Times
1225:26178194
1144:31942073
1037:24109477
986:23027979
934:25945739
789:31409927
727:See also
642:Protozoa
515:(MK-D1)
445:Taxonomy
384:bacteria
382:between
268:archaeal
165:Phylum:
127:Domain:
35:reviewed
2778:1655434
2719:Source:
2058:Animals
1979:Archaea
1959:Archaea
1880:8027215
1818:9834061
1681:1578724
1603:2045140
1516:4368082
1508:3340165
1480:Bibcode
1455:4396543
1419:Bibcode
1216:4522752
1135:7015854
1110:bioRxiv
1102:Bibcode
1028:3790079
1011:: 299.
977:3479504
925:4444528
896:Bibcode
767:Bibcode
634:nucleus
543:Genus "
518:Genus "
477:Genus "
224:Archaea
222:of the
183:Class:
132:Archaea
71:updated
2598:Asgard
2036:Plants
1973:Domain
1887:
1877:
1869:
1825:
1815:
1807:
1789:Nature
1688:
1678:
1610:
1600:
1559:
1551:
1514:
1506:
1498:
1471:Nature
1453:
1445:
1437:
1410:Nature
1328:(LPSN)
1305:
1297:
1251:May 9,
1223:
1213:
1142:
1132:
1112:
1093:Nature
1068:May 9,
1035:
1025:
984:
974:
932:
922:
914:
888:Nature
825:
787:
758:Nature
677:and a
649:lipids
505:Family
464:Family
404:clades
376:domain
279:genome
242:, and
228:Asgard
220:phylum
197:Order
158:Asgard
2380:DPANN
2063:Fungi
1716:8 May
1512:S2CID
1451:S2CID
823:S2CID
451:Order
423:actin
372:genes
305:Mohns
287:genus
281:in a
262:with
260:genes
2773:NCBI
2489:TACK
1885:PMID
1867:ISSN
1823:PMID
1805:ISSN
1718:2015
1686:PMID
1608:PMID
1557:PMID
1549:ISSN
1504:PMID
1496:ISSN
1443:PMID
1435:ISSN
1387:2019
1303:PMID
1295:ISSN
1253:2015
1221:PMID
1140:PMID
1070:2015
1033:PMID
982:PMID
954:PNAS
930:PMID
912:ISSN
848:help
785:PMID
437:and
366:The
348:Loki
321:core
246:. A
1875:PMC
1859:doi
1813:PMC
1797:doi
1793:613
1676:PMC
1668:doi
1664:361
1635:doi
1598:PMC
1590:doi
1541:doi
1488:doi
1476:331
1427:doi
1415:440
1287:doi
1211:PMC
1201:doi
1197:112
1167:doi
1130:PMC
1120:doi
1098:577
1063:BBC
1023:PMC
1013:doi
972:PMC
962:doi
958:109
920:PMC
904:doi
892:521
815:doi
775:doi
763:572
630:ATP
620:'s
558:Ca.
533:Ca.
492:Ca.
433:of
37:on
2797::
2775::
2760::
2630:"
2455:"
2443:"
2431:"
2411:"
2391:"
1957::
1909:.
1883:.
1873:.
1865:.
1855:15
1853:.
1849:.
1835:^
1821:.
1811:.
1803:.
1791:.
1787:.
1760:.
1735:.
1707:.
1684:.
1674:.
1662:.
1656:.
1629:.
1606:.
1596:.
1586:17
1584:.
1578:.
1555:.
1547:.
1537:19
1535:.
1510:.
1502:.
1494:.
1486:.
1474:.
1449:.
1441:.
1433:.
1425:.
1413:.
1407:.
1376:.
1350:.
1324:.
1301:.
1293:.
1283:23
1281:.
1277:.
1261:^
1242:.
1219:.
1209:.
1195:.
1191:.
1179:^
1163:40
1161:.
1138:.
1128:.
1118:.
1108:.
1096:.
1090:.
1078:^
1061:.
1045:^
1031:.
1021:.
1007:.
1003:.
980:.
970:.
956:.
952:.
928:.
918:.
910:.
902:.
890:.
886:.
856:^
839::
837:}}
833:{{
821:.
797:^
783:.
773:.
761:.
755:.
702:.
640:.
614:.
550:"
525:"
511:"
484:"
470:"
457:"
406:.
335:.
311:.
274:.
238:,
234:,
33:,
2626:"
2451:"
2439:"
2427:"
2407:"
2387:"
2316:"
2312:"
2204:"
2200:"
2070:)
1947:e
1940:t
1933:v
1919:.
1891:.
1861::
1829:.
1799::
1772:.
1745:.
1720:.
1692:.
1670::
1641:.
1637::
1631:7
1614:.
1592::
1563:.
1543::
1518:.
1490::
1482::
1457:.
1429::
1421::
1389:.
1361:.
1335:.
1309:.
1289::
1255:.
1227:.
1203::
1173:.
1169::
1146:.
1122::
1104::
1072:.
1039:.
1015::
1009:4
988:.
964::
936:.
906::
898::
850:)
846:(
829:.
817::
791:.
777::
769::
556:"
531:"
507:"
490:"
466:"
453:"
173:"
169:"
147:"
143:"
83:)
79:(
73:.
41:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.