190:. Self-replicating minicircles hold great promise for the systematic modification of stem cells and will significantly extend the potential of their plasmidal precursor forms ("parental plasmids"), the more as the principal feasibility of such an approach has amply been demonstrated for their plasmidal precursor forms.
20:
23:
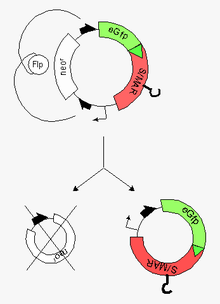
Minicircle preparation from a parental plasmid. The parental plasmid contains two recombinase target sites (black half arrows). Recombination between these sites generates the desired minicircle (bottom right) together with the miniplasmid (bottom left). The hook on the red minicircle-insert stands
180:, so they do not replicate within the target cells and the encoded genes will disappear as the cell divides (which can be either an advantage or disadvantage depending on whether the application demands persistent or transient expression). A novel addition to the field are nonviral
577:
Nehlsen, Kristina; Broll, Sandra; Kandimalla, Raju; Heinz, Niels; Heine, Markus; Binius, Stefanie; Schambach, Axel; Bode, Jürgen (5 April 2013). "Replicating
Minicircles: Overcoming the Limitations of Transient and Stable Expression Systems". In Schleef, Martin (ed.).
113:
sequences, they are less likely to be perceived as foreign and destroyed. (Typical transgene delivery methods involve plasmids, which contain foreign DNA.) The smaller size of minicircles also extends their
563:
534:
477:
428:
365:
491:
Argyros, O., Wong SP., Fedonidis C.; et al. (2011). "Development of S/MAR minicircles for enhanced and persistent transgene expression in the mouse liver".
346:
Nehlsen, K., Broll S., Bode, J. (2006). "Replicating minicircles: Generation of nonviral episomes for the efficient modification of dividing cells".
223:
138:
442:
Broll, S., Oumard A., Hahn K., Schambach A, Bode, J. . (2010). "Minicircle
Performance Depending on S/MAR-Nuclear Matrix Interactions".
595:
151:
recovery of the resulting minicircle (vehicle for the highly efficient modification of the recipient cell) and the miniplasmid by
617:
98:
295:
Dorrell, Richard G.; Nisbet, R. Ellen R.; Barbrook, Adrian C.; Rowden, Stephen J.L.; Howe, Christopher J. (2019).
622:
152:
548:
Heinz, N, Broll S, Schleef M, Baum C, Bode J (2012). "Filling a gap: S/MAR-based replicating minicircles".
557:
528:
471:
422:
359:
177:
106:
40:
516:
328:
591:
508:
459:
410:
320:
277:
583:
500:
451:
400:
392:
312:
267:
257:
182:
405:
380:
580:
Minicircle and
Miniplasmid DNA Vectors: The Future of Nonviral and Viral Gene Transfer
296:
611:
332:
520:
170:
162:
54:
141:
at the end of this process but still in bacteria. These steps are followed by the
316:
262:
245:
297:"Integrated Genomic and Transcriptomic Analysis of the Peridinin Dinoflagellate
166:
145:
95:
76:
71:
66:
58:
587:
504:
455:
126:
44:
109:
of mammalian cells, with the advantage that, since they contain no bacterial
102:
62:
47:
36:
512:
463:
414:
324:
281:
244:
Barbrook, Adrian C.; Voolstra, Christian R.; Howe, Christopher J. (2014).
199:
272:
19:
131:
115:
91:
50:
161:
The purified minicircle can be transferred into the recipient cell by
396:
187:
25:
18:
28:), which allows for autonomous replication in the recipient cell.
79:
genome is made of minicircles that encode chloroplast proteins.
246:"The Chloroplast Genome of a Symbiodinium sp. Clade C3 Isolate"
110:
125:
production of a 'parental plasmid' (bacterial plasmid with
582:. Wiley‐VCH Verlag GmbH & Co. KGaA. pp. 115–162.
381:"A robust system for production of minicircle DNA vectors"
186:
minicircles, which owe this property to the presence of a
121:
Their preparation usually follows a two-step procedure:
204:
Pages displaying short descriptions of redirect targets
224:"Kinetoplastids and Their Networks of Interlocked DNA"
118:
capacity and facilitates their delivery into cells.
169:and into a differentiated tissue by, for instance,
550:CliniBook - Nonviral Platform; Clinigene Network
144:excision of prokaryotic vector parts via two
8:
562:: CS1 maint: multiple names: authors list (
533:: CS1 maint: multiple names: authors list (
476:: CS1 maint: multiple names: authors list (
427:: CS1 maint: multiple names: authors list (
364:: CS1 maint: multiple names: authors list (
148:-target sequences at both ends of the insert
94:derivatives that have been freed from all
404:
379:Kay, M.A., He, C.-Y, Chen, Z.-H. (2010).
271:
261:
24:for a scaffold-matrix attachment region (
16:Small, circular replicating units of DNA
215:
555:
526:
469:
420:
357:
90:Minicircles are small (~4kb) circular
7:
86:experimentally-derived minicircles
14:
176:Conventional minicircles lack an
101:parts. They have been applied as
43:. They occur naturally in some
1:
153:capillary gel electrophoresis
317:10.1016/j.protis.2019.06.001
263:10.1016/j.protis.2013.09.006
639:
588:10.1002/9783527670420.ch8
505:10.1007/s00109-010-0713-3
456:10.1016/j.jmb.2009.11.066
139:site-specific recombinase
57:-derived kinetoplast of
202: – Type of plasmid
29:
178:origin of replication
61:, minicircles encode
22:
385:Nature Biotechnology
348:Gene Ther. Mol. Biol
299:Amphidinium carterae
107:genetic modification
618:Molecular genetics
30:
391:(12): 1287–1289.
105:carriers for the
630:
623:Applied genetics
602:
601:
574:
568:
567:
561:
553:
545:
539:
538:
532:
524:
488:
482:
481:
475:
467:
439:
433:
432:
426:
418:
408:
397:10.1038/nbt.1708
376:
370:
369:
363:
355:
343:
337:
336:
292:
286:
285:
275:
265:
241:
235:
234:
232:
230:
220:
205:
183:self-replicating
638:
637:
633:
632:
631:
629:
628:
627:
608:
607:
606:
605:
598:
576:
575:
571:
554:
547:
546:
542:
525:
490:
489:
485:
468:
441:
440:
436:
419:
378:
377:
373:
356:
345:
344:
340:
294:
293:
289:
243:
242:
238:
228:
226:
222:
221:
217:
212:
203:
196:
137:induction of a
88:
17:
12:
11:
5:
636:
634:
626:
625:
620:
610:
609:
604:
603:
596:
569:
540:
499:(5): 515–529.
483:
450:(5): 950–965.
434:
371:
338:
311:(4): 358–373.
287:
236:
214:
213:
211:
208:
207:
206:
195:
192:
159:
158:
157:
156:
149:
135:
87:
81:
15:
13:
10:
9:
6:
4:
3:
2:
635:
624:
621:
619:
616:
615:
613:
599:
597:9783527670420
593:
589:
585:
581:
573:
570:
565:
559:
551:
544:
541:
536:
530:
522:
518:
514:
510:
506:
502:
498:
494:
487:
484:
479:
473:
465:
461:
457:
453:
449:
445:
438:
435:
430:
424:
416:
412:
407:
402:
398:
394:
390:
386:
382:
375:
372:
367:
361:
353:
349:
342:
339:
334:
330:
326:
322:
318:
314:
310:
306:
302:
300:
291:
288:
283:
279:
274:
269:
264:
259:
255:
251:
247:
240:
237:
225:
219:
216:
209:
201:
198:
197:
193:
191:
189:
188:S/MAR-Element
185:
184:
179:
174:
172:
171:jet injection
168:
164:
154:
150:
147:
143:
142:
140:
136:
134:
133:
128:
124:
123:
122:
119:
117:
112:
108:
104:
100:
97:
93:
85:
82:
80:
78:
74:
73:
68:
64:
60:
56:
52:
49:
46:
42:
38:
35:are small (~4
34:
27:
26:S/MAR-Element
21:
579:
572:
558:cite journal
549:
543:
529:cite journal
496:
492:
486:
472:cite journal
447:
444:J. Mol. Biol
443:
437:
423:cite journal
388:
384:
374:
360:cite journal
351:
347:
341:
308:
304:
298:
290:
273:10754/563301
253:
249:
239:
229:September 2,
227:. Retrieved
218:
181:
175:
163:transfection
160:
130:
129:inserts) in
120:
89:
83:
70:
59:trypanosomes
55:mitochondria
32:
31:
493:J. Mol. Med
256:(1): 1–13.
167:lipofection
146:recombinase
96:prokaryotic
77:chloroplast
72:Amphidinium
67:RNA editing
39:) circular
33:Minicircles
612:Categories
552:: 271–277.
354:: 233–244.
210:References
127:eukaryotic
63:guide RNAs
45:eukaryotic
333:198240765
103:transgene
53:. In the
48:organelle
41:replicons
521:23986907
513:21301798
464:20004666
415:21102455
325:31415953
301:Plastid"
282:24316380
200:Episomes
194:See also
84:In vitro
406:4144359
305:Protist
250:Protist
132:E. coli
116:cloning
92:plasmid
51:genomes
594:
519:
511:
462:
413:
403:
331:
323:
280:
99:vector
75:, the
517:S2CID
329:S2CID
155:(CGE)
69:. In
592:ISBN
564:link
535:link
509:PMID
478:link
460:PMID
429:link
411:PMID
366:link
321:PMID
278:PMID
231:2019
65:for
584:doi
501:doi
452:doi
448:395
401:PMC
393:doi
313:doi
309:170
268:hdl
258:doi
254:165
165:or
111:DNA
614::
590:.
560:}}
556:{{
531:}}
527:{{
515:.
507:.
497:89
495:.
474:}}
470:{{
458:.
446:.
425:}}
421:{{
409:.
399:.
389:28
387:.
383:.
362:}}
358:{{
352:10
350:.
327:.
319:.
307:.
303:.
276:.
266:.
252:.
248:.
173:.
37:kb
600:.
586::
566:)
537:)
523:.
503::
480:)
466:.
454::
431:)
417:.
395::
368:)
335:.
315::
284:.
270::
260::
233:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.